Abstract
Geobacter sulfurreducens, a representative of the family Geobacteraceae that predominates in Fe(III)-reducing subsurface environments, can grow by coupling the oxidation of hydrogen to the reduction of a variety of electron acceptors, including Fe(III), fumarate, and quinones. An examination of the G. sulfurreducens genome revealed two operons, hya and hyb, which appeared to encode periplasmically oriented respiratory uptake hydrogenases. In order to assess the roles of these two enzymes in hydrogen-dependent growth, Hya- and Hyb-deficient mutants were generated by gene replacement. Hyb was found to be required for hydrogen-dependent reduction of Fe(III), anthraquinone-2,6-disulfonate, and fumarate by resting cell suspensions and to be essential for growth with hydrogen and these three electron acceptors. Hya, in contrast, was not. These findings suggest that Hyb is an essential respiratory hydrogenase in G. sulfurreducens.
The metabolic activities of dissimilatory Fe(III)-reducing bacteria can influence the cycling of organic matter and minerals in the subsurface and can play a crucial role in the bioremediation of both organic and metal contamination (reviewed in references 26 and 28). The Geobacteraceae, a family of dissimilatory Fe(III)-reducing bacteria in the delta subdivision of the class Proteobacteria, has been found to dominate the microbial communities present in a diversity of subsurface environments in which Fe(III) reduction is the terminal electron-accepting process (2, 16, 25, 39, 40, 46, 47). In the environment, Fe(III) is found mainly in the form of insoluble iron oxide particles and coatings, and many species of the family Geobacteraceae have evolved the ability to reduce a variety of electron acceptors that are either insoluble or too large to be transported into the cytoplasm. These include Fe(III) and Mn(IV) oxides, humic acids, and the surfaces of the graphite electrodes of microbial fuel cells (6, 7, 27). The members of the family Geobacteraceae are also capable of reducing many soluble metals, including the radionuclide U(VI) (16, 27). In addition, fumarate and nitrate can also serve as electron acceptors for several species of the family Geobacteraceae (27). Electron donors exploited by members of the family Geobacteraceae include acetate and other short-chain fatty acids; monoaromatic compounds such as benzoate, phenol, and toluene; and in many cases hydrogen (27).
Roughly 40% of the members of the family Geobacteraceae currently available in culture are capable of using hydrogen as an electron donor (27). However, little is known about the mechanism of hydrogen-dependent growth in the family Geobacteraceae. To gain insight into this question, the genome of Geobacter sulfurreducens (34), the first Geobacter species found to use hydrogen as an electron donor (8), was searched for hydrogenase-encoding genes. Four potential hydrogenase-encoding operons were identified, two of which, hya and hyb, were found to be similar to operons encoding periplasmically oriented respiratory hydrogenases in other members of the class Proteobacteria. This report presents the results of genetic studies performed to elucidate the physiological functions of the Hya and Hyb hydrogenases and provides evidence that Hyb is essential for hydrogen-dependent growth with a variety of electron acceptors, including Fe(III).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culturing conditions.
Escherichia coli strain DH5α (supE44 ΔlacU169 φ80 lacZ ΔM150 hsdR17 recA1 endA1 gyrA96 thi-1 relA1) (51) was used for DNA manipulations. Targeted gene disruption was performed on G. sulfurreducens strain DL1 (ATCC 51573) (8, 11) to produce strains DL7 (ΔhyaSLB::kan) and DL8 (ΔhybL::cam). G. sulfurreducens strains were routinely cultured anaerobically in either acetate-fumarate or acetate-Fe(III)-citrate medium as previously described (11). Plasmids pBBR1MCS-2 (19) and pCM66 (31) were obtained from Michael Kovach and Mary Lidstrom, respectively.
DNA manipulations and reagents.
G. sulfurreducens genomic DNA was extracted with the MasterPure complete DNA and RNA purification kit (Epicentre Technologies, Madison, Wis.) or the genome DNA kit (Bio 101, Inc., Carlsbad, Calif.). Plasmid purification, PCR product purification, and gel extractions were performed with the following kits: the QIAprep Spin Miniprep kit, the Qiagen Plasmid Midi kit, the QIAquick PCR purification kit, and the QIAquick gel extraction kit (Qiagen Inc., Valencia, Calif.). Ligations, transformations into E. coli, and other routine DNA manipulations were carried out as outlined by Sambrook et al. (42). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Beverly, Mass.). Southern blot analysis was performed as previously described (11). Taq DNA polymerase (Qiagen Inc.) was used for all PCR amplifications. Unless otherwise indicated, all of the primers used to amplify G. sulfurreducens sequences were designed from the preliminary sequence of the G. sulfurreducens genome (available at www.tigr.org). Unless otherwise stated, chemicals were reagent grade or better and were purchased from the Sigma Chemical Co. (St. Louis, Mo.). Sodium deoxycholate (ultrol grade) was purchased from Calbiochem (La Jolla, Calif.).
Construction of Hya- and Hyb-deficient strains via single-step gene replacement.
In order to create a mutant lacking Hya hydrogenase activity, recombinant PCR (35) was used to construct a 2.4-kb linear DNA fragment consisting of the first 0.1 kb of the hyaS gene preceded by 0.5 kb of upstream sequence and followed by a kanamycin resistance cassette, the last 0.1 kb of the hyaB gene, and 0.6 kb of downstream sequence. Three primary PCRs were carried out: (i) amplification of the 5′ end of the linear fragment (bp −528 to +130 of the hyaS gene) with primers mc1 (5′-GGAGCGGCTTTCTTCCTTCG-3′) and mc2 (5′-TTGCGGTGCAGAACTTCAGG-3′), (ii) amplification of the kanamycin resistance cassette from pBBR1MCS-2 with primers mc3 (5′-CCTGAAGTTCTGCACCGCAAACTGGGCTATCTGGACAAGG-3′) and mc4 (5′-GGTGGATAGCAAAGGCGATGCGAAATCTCGTGATGGCAGG-3′), and (iii) amplification of the 3′ end of the linear fragment (bp +570 to +1174 of the hyaB gene) with primers mc5 (5′-CATCGCCTTTGCTATCCACC-3′) and mc6 (5′-CGTCATGAAGGAGTGTGTCG-3′). Following recombinant PCR with the three primary PCR products serving as both templates and primers, the final 2.4-kb fragment was amplified with distal primers mc1 and mc6. PCR conditions were similar to those described by Lloyd et al. (24), except for the following modifications. Annealing was only allowed to proceed for 30 s, denaturation was performed at 94°C for 30 s, and the primer concentrations were 50 and 25 nM for the primary and recombinant PCRs, respectively.
In order to create a mutant deficient in Hyb hydrogenase activity, recombinant PCR was used to construct a 2.0-kb linear DNA fragment consisting of the first 7 bp of the hybL gene preceded by 0.6 kb of upstream sequence and followed by a chloramphenicol resistance cassette, the last 0.4 kb of the hybL gene, and 0.1 kb of downstream sequence. Three primary PCRs were carried out: (i) amplification of the 5′ end of the linear fragment (bp −585 to +7 of the hybL gene) with primers MChyd21for (5′-GCACGAACTGTCCGGCATCG-3′) and MChyd22rev (5′-TAGACATGTATTCCTCCAGAG-3′), (ii) amplification of the chloramphenicol resistance cassette from pACYC184 with primers MChyd23camfor (5′-CTCTGGAGGAATACATGTCTAAGTTGGCAGCATCACCCGACG-3′) and MChyd24camrev (5′-GTTGGCCACGAGCTTGTCCACTTATTCAGGCGTAGCACCAG-3′), and (iii) amplification of the 3′ end of the linear fragment (bp +1310 to +1813 of the hybL gene) with primers MChyd25for (5′-ACCTGGACAAGCTCGTGGCCAAC-3′) and MChyd26rev (5′-AGTTCTCTTCCAGGTGGTTG-3′). Following recombinant PCR with the three PCR products serving as both templates and primers, the final 2.0-kb fragment was amplified with distal primers MChyd21for and MChyd6rev. PCR conditions were similar to those described above except that an annealing temperature of 55°C was used.
Electroporation, mutant isolation, and genotype confirmation were performed as described by Coppi et al. (11) and Lloyd et al. (24). One each of the resulting mutants, DL7 (ΔhyaSLB::kan) and DL8 (ΔhybL::cam), was chosen as the representative strain.
Expression of the hybL gene in trans.
The complete hybL coding sequence was amplified with primers hyd2compf (5′-CCCGGATCCCTGGAGGAATACATGTC-3′ [BamHI site underlined]) and hyd2compr (5′-GGGGAATTCGGGTTCTCATTCGCTACTC-3′ [EcoRI site underlined]) under the following conditions: 94°C for 3 min followed by 25 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min and a final extension at 72°C for 10 min. The hybL amplicon was digested with BamHI and EcoRI and inserted into the corresponding sites of the broad-host-range expression vector pCM66 (31) to generate pMChybL. The hybL gene was then sequenced to screen for PCR artifacts. Following electroporation of strain DL8 (ΔhybL::cam) with pMChybL, a kanamycin-resistant transformant was isolated and designated DL8/pMChybL. The simultaneous presence of pMChybL and the ΔhybL::cam mutation in this strain was confirmed by PCR screening and plasmid isolation.
In-gel hydrogenase assay.
Membrane and soluble fractions were prepared from strains DL1 (wild type), DL7 (ΔhyaSLB::kan), and DL8 (ΔhybL::cam) under aerobic conditions essentially as described by Leang et al. (22), except that the cells were washed in 50 mM HEPES (pH 7.0)-1 mM MgSO4 and resuspended in 50 mM Tris/HCl (pH 7.5) prior to lysis. Crude membranes were resuspended in 50 mM Tris/HCl (pH 7.5), diluted to a concentration of 5 μg/μl, and partially solubilized with Triton X-114 at a final concentration of 0.5% for 15 min at room temperature. Unsolubilized membranes were pelleted by centrifugation for 45 min at 100,000 × g and 20°C. Nondenaturing polyacrylamide gel electrophoresis of the TX-114 extracts was performed aerobically as described by Magnuson et al. (30) with a final acrylamide concentration of 4% in the stacking gel and 5% in the separating gel. Sodium deoxycholate, at a final concentration of 0.7%, was present in the gel, samples, and running buffer. Nondenaturing gels were stained for hydrogenase activity (hydrogen-dependent benzyl viologen reduction) essentially in accordance with the method of Ackrell et al. (1). Staining was rendered oxygen resistant by the addition of triphenyltetrazolium chloride to a final concentration of 2 mM, and the reaction was terminated by exposure to air.
Cell suspension experiments.
Cell suspension assays were carried out essentially as previously described (22), with the following modifications. Cell suspensions prepared from mid-log-phase acetate-fumarate cultures were incubated under an 80:20 N2-CO2 atmosphere with the following combinations of electron donors and acceptors: (i) no donor, 1 mM acetate, or hydrogen and 5 mM Fe(III) chelated with nitrilotriacetic acid [Fe(III)-NTA]; (ii) no donor, 5 mM acetate, or hydrogen and 1 mM anthraquinone-2,6-disulfonate (AQDS); or (iii) no donor, 2 mM acetate, or hydrogen and 5 mM fumarate. When hydrogen was provided as the electron donor, the headspace was replaced with an 80:20 hydrogen-carbon dioxide mixture. In each case, the NaCl concentration in the basal wash medium was adjusted to keep the osmolarity constant. Incubations of cell suspensions with Fe(III)-NTA and AQDS were performed in a 10-ml volume in a 27-ml pressure tube, whereas incubations with fumarate were performed in a 25-ml volume in a 72-ml serum bottle. Fe(III)-NTA reduction and fumarate metabolism were monitored by taking 0.1-ml [Fe(III)-NTA] or 1.0-ml (fumarate) samples at 20-min intervals over 100 min. Samples taken for the analysis of fumarate metabolism were immediately filtered to remove cells, frozen, and stored at −20°C until high-pressure liquid chromatography analysis (see below). AQDS reduction was monitored by placing the pressure tube into the spectrophotometer and measuring absorbance at 436 nm at 10- to 20-min intervals for 90 min. To calculate hydrogen- and acetate-dependent reduction, the amount of reduced acceptor present in incubations in the absence of donor at each time point was averaged and subtracted from the amount of reduced acceptor present in each incubation in the presence of donor.
Hydrogen- and acetate-dependent cell growth.
Growth studies were carried out in 27-ml pressure tubes containing 10 ml of donor-free freshwater medium. The three media used were NBF medium (acetate-free NBAF [11]), FWFC medium (acetate-free FWAFC [11]), and NBQ medium (NBF medium in which 5 mM AQDS was substituted for fumarate). To measure hydrogen-dependent growth, 10 ml of hydrogen gas was injected into the headspace and a limiting concentration of acetate (0.1 mM for NBQ medium or 1 mM for NBF and FWFC media) was added as a carbon source. The concentrations of acetate in the medium during acetate-dependent growth were 10 mM for NBQ medium and 20 mM for NBF and FWFC media. Acetate was added to the media from concentrated stock solutions as needed.
Analytical techniques.
Growth of fumarate cultures was assessed by measuring turbidity at 600 nm. Cell densities of Fe(III)-citrate and AQDS cultures were determined by acridine orange staining and epifluorescence microscopy (29). Fe(II) concentrations were determined by the ferrozine assay (29). AHQDS (reduced AQDS) concentrations were determined at 436 nm for cell suspension studies (extinction coefficient = 3,500 M−1 cm−1 [5]) and 525 nm for growth studies (extinction coefficient = 471.3 M−1 cm−1, determined by mixing oxidized and reduced growth medium). AQDS reduction and turbidity were monitored by placing pressure tubes (path length = 1.5 cm) directly into a Genesys 2 spectrophotometer (Spectronics Instruments, Rochester, N.Y.). Quantitation and identification of metabolites produced by cell suspensions incubated in the presence of fumarate were performed by high-pressure liquid chromatography with an LC-10AT high-pressure liquid chromatograph (Shimadzu, Kyoto, Japan) equipped with an Aminex HPX-87H column (300 by 7.8 mm; Bio-Rad, Hercules, Calif.) equilibrated with 8 mM H2SO4 and an SPD-10VP UV detector (Shimadzu, Kyoto, Japan) set at 215 nm. The protein contents of cell fractions and cell suspensions were determined by the bicinchoninic acid method with bovine serum albumin as the standard (45).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the various proteins described and discussed in this report are as follows. G. sulfurreducens: HyaS, AAR33458; HyaL, AAR33457; HyaB, AAR33456; HyaP, AAR33455. Geobacter metallireducens: HyaS, ZP_00082073; HyaL, ZP_00082074; HyaB, ZP_00082075; HyaP, ZP_00082076. G. sulfurreducens: HybS, AAR34112; HybA, AAR34113; HybB, AAR34114; HybL, AAR34115; HybP, AAR34116; HybT, AAR34117. Wolinella succinogenes: HydA, CAA46302; HydB, CAA46303; HydC, S22406. Ralstonia eutropha membrane-bound NiFe hydrogenase: HoxK, NP_942643; HoxG, NP_942644; HoxZ, NP_942645. Rhodobacter capsulatus uptake hydrogenase: HupS, P15283; HupL, P15284; HupC, P16145. E. coli hydrogenase 2: Hyb0, Q46847; HybA, P37179; HybB, P37180; HybC, P37181. Salmonella enterica serovar Typhimurium LT2 hydrogenase 2: Hyb0, AAL22024; HybA, AAL22023; HybB, AAL22022; HybC, AAL22021. Actinobacillus pleuropneumoniae heterotetrameric uptake NiFe hydrogenase: small subunit, ZP_00134401; Fe-S cluster-containing subunit, ZP_00134402; integral membrane subunit, ZP_00134403; large subunit, ZP_00134404. Magnetospirillum magnetotacticum heterotetrameric uptake NiFe hydrogenase: small subunit, ZP_00054823; Fe-S cluster-containing subunit, ZP_00052632; 5′ end of integral membrane subunit, ZP_00054825; 3′ end of integral membrane subunit, ZP_00052631; large subunit, ZP_00052632. Magnetococcus sp. strain MC-1 heterotetrameric uptake NiFe hydrogenase: small subunit, ZP_00042948; Fe-S cluster-containing subunit, ZP_00042949; integral membrane subunit, ZP_00042950; large subunit, ZP_00042951. Desulfovibrio fructosovorans periplasmic NiFe hydrogenase: HynA, P18187; HynB, P18188.
RESULTS AND DISCUSSION
Putative hya and hyb operons of G. sulfurreducens.
The G. sulfurreducens genome (34) was searched for the presence of respiratory uptake hydrogenases with the following sequences as probes: the catalytic subunit of the periplasmic Fe hydrogenase of D. fructosovorans (hydA) (9), an Fe hydrogenase H cluster consensus sequence (pfam02906) (4), the large and small subunits (hydA and hydB) of the respiratory NiFe hydrogenase of W. succinogenes (12), and the large and small subunits (hynA and hynB) of the NiFe hydrogenase of D. fructosovorans (41). Two potential operons with significant similarity to proteobacterial respiratory uptake NiFe hydrogenases, hya and hyb, were identified.
The putative hya operon (Fig. 1A) consists of four genes, hyaS, hyaL, hyaB, and hyaP, encoding a large subunit, a small subunit with an N-terminal twin-arginine motif (50, 52) (SRRDFLK), a hydrophobic type b cytochrome, and a maturation protease, respectively. Thus, Hya appears to be a member of a family of heterotrimeric membrane-bound uptake hydrogenases with periplasmic active sites (48) that includes the respiratory hydrogenases of W. succinogenes, (12), R. eutropha (18, 44), and a variety of nitrogen-fixing members of the class Proteobacteria. The HyaS, HyaL, and HyaB subunits of G. sulfurreducens Hya are 53 to 57, 63 to 65, and 46 to 51% similar to their respective homologs in R. eutropha and R. capsulatus (23, 38), a nitrogen-fixing member of the alpha subdivision of the class Proteobacteria. They are slightly less similar, 58.2, 59.8, and 39.3%, to the subunits of HydABC of W. succinogenes. (Global pairwise alignments were performed with the algorithm of Needleman and Wunsch [36] and scored with the blosum62 matrix [15].)
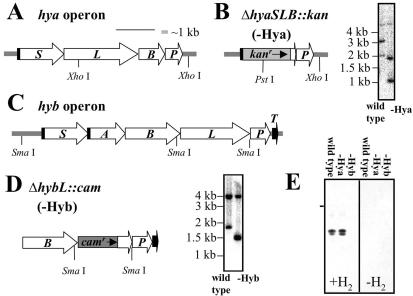
FIG. 1.
Structures and mutagenesis of putative hya and hyb operons. (A) Structure of putative hya operon. Twin-arginine motifs are indicated as vertical black bars. (B) Structure of disrupted hya operon and confirmation of Hya-deficient mutant genotype by Southern blotting. Wild-type and Hya-deficient G. sulfurreducens genomic DNAs were cleaved with PstI and XhoI, blotted, and probed with the 2.4-kb DNA fragment used to introduce the ΔhyaSLB::kan mutation into the genome (see Materials and Methods). Expected radiolabeled bands are 5.28, 3.03, and 2.04 kb for the wild-type strain and 5.28, 1.88, and 1.02 kb for the mutant. (C) Structure of putative hyb operon. Twin-arginine motif sequences are indicated as vertical black bars. (D) Structure of disrupted hyb operon and confirmation of Hyb-deficient mutant genotype by Southern blotting. Wild-type and Hyb-deficient G. sulfurreducens genomic DNAs were cleaved with SmaI, blotted, and probed with the 2.0-kb DNA fragment used to introduce the ΔhybL::cam mutation into the genome (see Materials and Methods). Expected radiolabeled bands are 4.02 and 1.76 kb for the wild-type strain and 4.02 and 1.41 kb for the mutant. (E) In-gel hydrogenase activity of Triton X-114 extracts of membranes prepared from acetate-fumarate cultures of the wild-type and Hya- and Hyb-deficient strains. Nondenaturing polyacrylamide gels were stained for hydrogen-dependent benzyl viologen reductase activity as described in Materials and Methods. Twenty micrograms of solubilized membranes was loaded per lane. Lanes 1 to 3 were incubated under hydrogen, while lanes 4 to 6 were incubated under nitrogen. The horizontal bar indicates the interface of the stacking and resolving gels.
A search of the partial genome sequence of the closely related species G. metallireducens (available at www.jgi.doe.gov) revealed the presence of a homologous hya operon. The three subunits of G. metallireducens Hya are 90 to 95% similar to their G. sulfurreducens counterparts. Because G. metallireducens does not appear to be capable of growth with hydrogen as an electron donor (27), this result suggested that the physiological function of Hya might not be related to hydrogen-dependent energy generation.
The subunits and maturation proteases of the three Geobacter Hya hydrogenases have sizes and features typical of the family of heterotrimeric uptake hydrogenases (48), with one notable exception. The small subunits of the periplasmically oriented heterotrimeric uptake hydrogenases, as well as those of the periplasmic heterodimeric NiFe hydrogenases, of Desulfovibrio species contain 10 conserved cysteine residues and 1 conserved histidine residue that serve as ligands for three iron-sulfur clusters (37, 49). Surprisingly, both of the Geobacteraceae family HyaS subunits contain an aspartate residue in place of one of the conserved cysteine residues that in other hydrogenases ligates the proximal [4Fe-4S] cluster (CXXD versus CXXC). This cysteine-to-aspartate substitution may have implications for the catalytic activity of Hya. When the analogous cysteine residue of the small subunit of the heterotrimeric respiratory hydrogenase of Azotobacter vinelandii (HoxGKZ) was mutated to serine, the ability of the hydrogenase to catalyze hydrogen oxidation was nearly eliminated (2% of that of the wild type), whereas its ability to catalyze hydrogen evolution was relatively unaffected (22% of that of the wild type) (32).
The second putative G. sulfurreducens hydrogenase-encoding operon, hyb, appears to encode a periplasmically oriented membrane-bound NiFe hydrogenase with four subunits: (i) HybS, a small subunit with an N-terminal twin-arginine motif (SRRDFMK); (ii) HybA, a second iron-sulfur cluster-containing subunit with an N-terminal twin-arginine motif (TRRDFLK); (iii) HybB, an integral membrane subunit; and (iv) HybL, a large subunit. Similar gene clusters encoding heterotetrameric NiFe hydrogenases can be found in the genomes of two members of the alpha subdivision of the class Proteobacteria, Magnetococcus sp. MC-1 and M. magnetotacticum, as well as at least three members of the gamma subdivision of the class Proteobacteria, E. coli, S. enterica serovar Typhimurium, and A. pleuropneumoniae. However, to date, only one heterotetrameric membrane-bound NiFe hydrogenase, Hyd2 of E. coli, has been genetically and biochemically characterized (3, 33, 43). E. coli Hyd2 is a membrane-bound respiratory hydrogenase with a periplasmically oriented active site that is required for the hydrogen-dependent reduction of a variety of electron acceptors and is essential for growth in the presence of hydrogen and fumarate. The four G. sulfurreducens Hyb subunits are ca. 50 to 70% similar to the subunits of the heterotetrameric hydrogenases encoded in the five gene clusters listed above, with the highest degree of similarity occurring between the various large (70 to 74%) and small (60 to 68%) subunits. Overall, the subunits of G. sulfurreducens Hyb are slightly more similar to those of their counterparts in the alpha subdivision of the class Proteobacteria.
The organization of the 5′ ends of the various gene clusters encoding heterotetrameric hydrogenases is the same; genes encoding the four hydrogenase subunits are followed by a maturation protease gene. In the E. coli hyb operon, three additional processing genes, hybE, hybF, and hybG, are present (10, 17). In contrast, the maturation protease gene, hybP, of G. sulfurreducens is followed by a short open reading frame, hybT, and a putative Rho-independent transcriptional terminator (identified with Transterm software [13]). On the basis of the presence of a conserved MttA domain (pfam02416 [4]) at its N terminus, we propose that hybT may encode a homolog of TatA, a critical component of the Tat secretory pathway (50, 52). A homologous hyb operon was not identified in the partial genome sequence of G. metallireducens.
Alignment of the various subunits of G. sulfurreducens Hyb with those of E. coli Hyd2 revealed that all of the defining features of the subunits of Hyd2 (33, 43) were present. However, HybB, the integral membrane subunit of G. sulfurreducens Hyb, contains a hydrophilic 35-amino-acid insertion that is not found in the integral membrane subunit of E. coli Hyd2 or any of other heterotetrameric hydrogenases examined.
Construction and preliminary analysis of Hya- and Hyb-deficient mutants.
In order to elucidate the roles of Hya and Hyb during hydrogen-dependent growth, Hya and Hyb knockout mutants were constructed via homologous recombination (Fig. 1C and D). A Hya-deficient mutant (DLMC1 [ΔhyaSLB::kan]) was constructed by replacing a 3.25-kb stretch of sequence encompassing 90% of hyaS, all of hyaL, and 83% of hyaB with a kanamycin resistance cassette. A Hyb-deficient mutant (DLMC2 [ΔhybL::cam]) was constructed by replacing the N-terminal 77% of the HybL coding sequence with a chloramphenicol resistance cassette. The genotypes of these two mutants were confirmed by Southern blotting (Fig. 1B and D) and PCR screening (data not shown).
In-gel hydrogenase assays were performed on membrane fractions prepared from acetate-fumarate-grown wild-type and hydrogenase-deficient cultures (Fig. 1E). Electrophoresis and solubilization conditions were optimized to maximize the rate of hydrogen-dependent staining. All of the hydrogen-dependent staining in the membrane fraction was found to be dependent on the presence of Hyb (lane 1 versus lane 3). Hyb was found to migrate as three species, a very minor diffuse band with low electrophoretic mobility, and two intense, rapidly migrating species. The relative intensity of these three species was found to be strongly dependent on the solubilization conditions and the presence or absence of specific detergents during electrophoresis (data not shown), suggesting that Hyb may dissociate either in gel or during solubilization. In the case of Hyd2 of E. coli, multiple species were visualized in nondenaturing gels until it was purified to near homogeneity, when it was found to be a heterodimer of the large and small subunits (3, 33).
We have been unable to detect Hya-specific hydrogen uptake activity in either the soluble (data not shown) or the membrane fraction (Fig. 1E) of acetate-fumarate-grown cells either in gel or in vitro. This may be due either to insufficient expression of Hya during growth on acetate-fumarate medium or because the enzyme is inactive under the conditions tested to date.
Phenotype of the Hya- and Hyb-deficient mutants.
Resting cell suspensions were prepared from acetate-fumarate cultures of wild-type and Hya- and Hyb-deficient G. sulfurreducens and tested for the ability to reduce three electron acceptors: Fe(III)-NTA, AQDS, and fumarate (Table 1). Surprisingly, the rate of fumarate reduction by the wild-type strain, measured as succinate production, was significantly lower than that of either Fe(III)-NTA or AQDS reduction. In addition, in contrast to Fe(III) and AQDS reduction, succinate production was higher in the presence of acetate than in the presence of hydrogen. One factor that may have contributed to the latter finding is that in the presence of acetate, a portion of the succinate (25%) can be produced by oxidation of acetate via the tricarboxylic acid cycle instead of direct reduction of fumarate (14).
TABLE 1.
Reduction of Fe(III)NTA, AQDS, and fumarate by cell suspensions
| Suspensiona and electron donor | Mean productionc (μmol/mg/min) ± SD
|
||
|---|---|---|---|
| Fe(II)-NTA | AHQDSb | Succinate | |
| Wild type | |||
| Acetate | 0.84 ± 0.17 | 0.64 ± 0.03 | 0.182 ± 0.002 |
| Hydrogen | 1.48 ± 0.22 | 1.29 ± 0.19 | 0.091 ± 0.010 |
| Hya deficient | |||
| Acetate | 1.24 ± 0.05 | 0.81 ± 0.01 | 0.189 ± 0.001 |
| Hydrogen | 1.83 ± 0.10 | 1.46 ± 0.05 | 0.072 ± 0.013 |
| Hyb deficient | |||
| Acetate | 0.95 ± 0.04 | 0.62 ± 0.05 | 0.187 ± 0.011 |
| Hydrogen | 0.00 ± 0.03 | 0.02 ± 0.01 | 0.004 ± 0.004 |
Resting cell suspensions were prepared from late-log-phase acetate-fumarate cultures as described in Materials and Methods.
AHQDS is the reduced form of AQDS.
Data are means of triplicate incubations, except for measurements of wild-type reduction of AQDS and Fe(III)-NTA, which are means of six incubations.
In the presence of hydrogen, Hya-deficient suspensions reduced the three acceptors at rates comparable to those of the wild type, whereas the Hyb-deficient suspensions could not reduce any of the three acceptors (Table 1). Thus, Hyb was found to be essential for hydrogen-dependent reduction of a variety of acceptors by acetate-fumarate-grown cell suspensions.
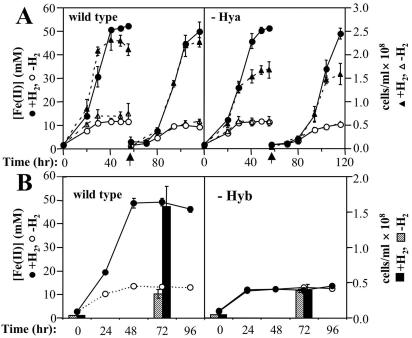
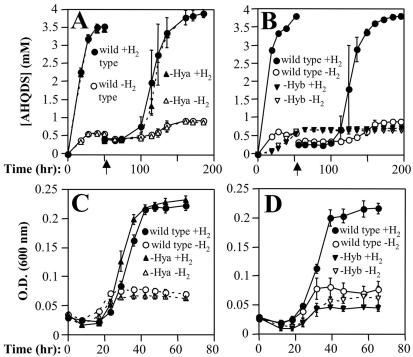
Longer-term growth studies yielded results that were analogous to those of the cell suspension studies. Both of the hydrogenase-deficient strains could be cultured in medium containing acetate as the electron donor and either Fe(III)-citrate, AQDS, or fumarate as the electron acceptor (data not shown). The wild-type and Hya-deficient strains were capable of sustained hydrogen-dependent growth in medium containing a small amount of acetate as a source of organic carbon and either Fe(III)-citrate, AQDS, or fumarate as the electron acceptor (Fig. 2 and 3). When Fe(III)-citrate was provided as the electron acceptor, the final cell yield for the Hya-deficient mutant was ∼25% less that than that of the wild type. The Hyb-deficient mutant was incapable of hydrogen-dependent growth in the presence of these three electron acceptors (Fig. 2 and 3). In addition, the Hyb-deficient mutant was also found to be unable to grow with hydrogen and poorly crystalline Fe(III) oxide as the electron acceptor (data not shown). To confirm that the phenotype of the Hyb-deficient mutant was due to the absence of the HybL subunit, a hybL expression vector was constructed. Expression of the hybL gene in trans restored the ability of the mutant to grow with hydrogen and AQDS as the electron donor (data not shown). Thus, Hyb was essential for hydrogen-dependent growth in the presence of three electron acceptors.
FIG. 2.
Growth of wild-type and hydrogenase-deficient strains with Fe(III)-citrate as the electron acceptor. (A) Hydrogen-dependent growth of the wild-type and Hya-deficient strains. (B) Hydrogen-dependent growth of the wild-type and Hyb-deficient strains. The experiment was initiated with 2.5% inocula from early-stationary-phase acetate-Fe(III)-citrate-grown cultures. A limiting concentration of acetate (1 mM) was provided as a carbon source. In each panel, wild-type and mutant cultures were inoculated and incubated in parallel. When two transfers are shown, the time of the second transfer (2.5%) is indicated by a vertical arrow. Data are the mean ± standard deviation of triplicate cultures.
FIG. 3.
Hydrogen-dependent growth of wild-type and hydrogenase-deficient strains in the presence of AQDS and fumarate. Acetate-fumarate-grown cultures were washed in isotonic basal wash medium (22) prior to inoculation to eliminate carryover of acetate and fumarate. Cultures were supplied with a small amount of acetate as a carbon source (0.1 mM for AQDS curves [A and B] and 1 mM for fumarate curves [C and D]). For each panel, wild-type and mutant cultures were inoculated and incubated in parallel. Data are the mean ± standard deviation of triplicate cultures. When two transfers are shown, the time of the second transfer (5%) is indicated by a vertical arrow. Cell densities at time zero were ∼6 × 106/ml for AQDS growth curves (A and B) and ∼2.5 × 106/ml for fumarate growth curves (C and D). For AQDS growth curves, final cell densities at the end of first and second transfers were 4.87 × 107 ± 0.5 × 107 and 3.6 × 107 ± 1.06 × 107/ml, respectively, for the wild-type strain and 3.81 × 107 ± 0.41 × 107 and 2.88 × 107 ± 0.25 × 107/ml, respectively, for the Hya-deficient strain. O.D., optical density.
Insights into the physiological function of Hya and Hyb.
The finding that Hyb was required for hydrogen-dependent reduction of Fe(III), AQDS, and fumarate indicates that Hyb plays a central role in hydrogen-dependent respiration in G. sulfurreducens. Thus, the physiological function of Hyb may be to transfer electrons from hydrogen to the menaquinone pool, where they can be redistributed to a variety of reductases. Examination of the hydrogenase content of the genome of the closely related species G. metallireducens further supports this hypothesis. G. metallireducens is unable to grow with hydrogen as an electron donor, and its genome contains homologs of each of the hydrogenases encoded in the G. sulfurreducens genome except for Hyb. This result confirms the importance of Hyb for growth with hydrogen as the electron donor and suggests that it may be possible to use the presence or absence of a Hyb ortholog as a criterion for predicting whether other members of the family Geobacteraceae have the ability to couple hydrogen oxidation to cell growth.
The genetic studies presented herein indicate that Hya, unlike Hyb, is not essential for coupling hydrogen oxidation to the reduction of Fe(III), AQDS, or fumarate under the conditions tested. These findings are consistent with the presence of a Hya ortholog in the genome of G. metallireducens, which cannot grow with hydrogen as an electron donor. Elucidation of the physiological role of Hya requires extensive further investigation, including a detailed study of Hya expression, defining growth conditions and media that maximize Hya expression, and creating an in vitro or in vivo assay for Hya activity. Our failure to detect Hya activity or a clear Hya phenotype may be due to a lack of Hya expression or to a low Hya expression level under the growth conditions used in this study. It is also possible that Hya is involved in the reduction of electron acceptors that were not tested in this study. Hyd1 of E. coli, also a heterotrimeric periplasmically oriented uptake hydrogenase, was found to preferentially reduce high potential electron acceptors such as ferricyanide and oxygen and to be unable to reduce low potential acceptors such as benzyl and methyl viologen (20, 21).
In summary, the studies presented herein indicate that Hyb, which is closely related to the Hyd2 respiratory hydrogenase of E. coli, may serve as the principal respiratory hydrogenase of G. sulfurreducens. The physiological role of Hya, in contrast, remains unclear.
Acknowledgments
This work was supported by the Office of Science (BER), U.S. Department of Energy (grants DE-FC02-02ER63446 and DE-FGO2-01ER63145). All G. metallireducens genome sequence data were provided by the U.S. Department of Energy Joint Genome Institute (www.jgi.doe.gov).
We thank Mary E. Lidstrom for providing plasmid pCM66, Lorrie Adams for skilled technical assistance, and the other members of our laboratory for encouragement, expertise, and editorial assistance.
REFERENCES
- 1.Ackrell, B. A., R. N. Asato, and H. F. Mower. 1966. Multiple forms of bacterial hydrogenases. J. Bacteriol. 92:828-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballantine, S. P., and D. H. Boxer. 1986. Isolation and characterisation of a soluble active fragment of hydrogenase isoenzyme 2 from the membranes of anaerobically grown Escherichia coli. Eur. J. Biochem. 156:277-284. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. Eddy, S. Griffiths-Jones, K. Howe, M. Marshall, and E. Sonnhammer. 2002. The Pfam Protein Families Database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer, M., K. Walter, and H. Simon. 1996. Purification and partial characterisation of a reversible artificial mediator accepting NADH oxidoreductase from Clostridium thermoaceticum. Eur. J. Biochem. 239:686-691. [DOI] [PubMed] [Google Scholar]
- 6.Bond, D. R., D. E. Holmes, L. M. Tender, and D. R. Lovley. 2002. Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483-485. [DOI] [PubMed] [Google Scholar]
- 7.Bond, D. R., and D. R. Lovley. 2003. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 69:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caccavo, F., Jr., D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz, and M. J. McInerney. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60:3752-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casalot, L., C. E. Hatchikian, N. Forget, P. de Philip, Z. Dermoun, J. P. Bélaïch, and M. Rousset. 1998. Molecular study and partial characterization of iron-only hydrogenase in Desulfovibrio fructosovorans. Anaerobe 4:45-55. [DOI] [PubMed] [Google Scholar]
- 10.Casalot, L., and M. Rousset. 2001. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9:228-237. [DOI] [PubMed] [Google Scholar]
- 11.Coppi, M. V., C. Leang, S. J. Sandler, and D. R. Lovley. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dross, F., V. Geisler, R. Lenger, F. Theis, T. Krafft, F. Fahrenholz, E. Kojro, A. Duchene, D. Tripier, K. Juvenal, et al. 1992. The quinone-reactive Ni/Fe-hydrogenase of Wolinella succinogenes. Eur. J. Biochem. 206:93-102. [DOI] [PubMed] [Google Scholar]
- 13.Ermolaeva, M. D., H. G. Khalak, O. White, H. O. Smith, and S. L. Salzberg. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27-33. [DOI] [PubMed] [Google Scholar]
- 14.Galushko, A. S., and B. Schink. 2000. Oxidation of acetate through reactions of the citric acid cycle by Geobacter sulfurreducens in pure culture and in syntrophic coculture. Arch. Microbiol. 174:314-321. [DOI] [PubMed] [Google Scholar]
- 15.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hube, M., M. Blokesch, and A. Bock. 2002. Network of hydrogenase maturation in Escherichia coli: role of accessory proteins HypA and HybF. J. Bacteriol. 184:3879-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kortluke, C., K. Horstmann, E. Schwartz, M. Rohde, R. Binsack, and B. Friedrich. 1992. A gene complex coding for the membrane-bound hydrogenase of Alcaligenes eutrophus H16. J. Bacteriol. 174:6277-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 20.Laurinavichene, T. V., and A. A. Tsygankov. 2001. H2 consumption by Escherichia coli coupled via hydrogenase 1 or hydrogenase 2 to different terminal electron acceptors. FEMS. Microbiol. Lett. 202:121-124. [DOI] [PubMed] [Google Scholar]
- 21.Laurinavichene, T. V., N. A. Zorin, and A. A. Tsygankov. 2002. Effect of redox potential on activity of hydrogenase 1 and hydrogenase 2 in Escherichia coli. Arch. Microbiol. 178:437-442. [DOI] [PubMed] [Google Scholar]
- 22.Leang, C., M. V. Coppi, and D. R. Lovley. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185:2096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leclerc, M., A. Colbeau, B. Cauvin, and P. M. Vignais. 1988. Cloning and sequencing of the genes encoding the large and the small subunits of the H2 uptake hydrogenase (hup) of Rhodobacter capsulatus. Mol. Gen. Genet. 214:97-107. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd, J. R., C. Leang, A. L. Hodges Myerson, M. V. Coppi, S. Cuifo, B. Methe, S. J. Sandler, and D. R. Lovley. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369:153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lonergan, D. J., H. L. Jenter, J. D. Coates, E. J. Phillips, T. M. Schmidt, and D. R. Lovley. 1996. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 178:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovley, D. R. 2000. Fe(III) and Mn(IV) reduction, p. 3-30. In D. R. Lovley (ed.), Environmental microbe-metal interactions. ASM Press, Washington, D.C.
- 27.Lovley, D. R. 2000. Fe(III)- and Mn(IV)-reducing prokaryotes. In M. Dworkin, S. Falkow, K. Rosenberg, H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed. [Online.] Springer-Verlag, Inc., New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 28.Lovley, D. R. 2001. Reduction of iron and humics in subsurface environments, p. 193-217. In K. F. Fredrickson and M. Fletcher (ed.), Subsurface microbiology and biogeochemistry. Wiley-Liss Inc., New York, N.Y.
- 29.Lovley, D. R., and E. J. P. Phillips. 1988. Novel mode of microbial energy-metabolism: organic-carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magnuson, T. S., A. L. Hodges-Myerson, and D. R. Lovley. 2000. Characterization of a membrane-bound NADH-dependent Fe3+ reductase from the dissimilatory Fe3+-reducing bacterium Geobacter sulfurreducens. FEMS Microbiol. Lett. 185:205-211. [DOI] [PubMed] [Google Scholar]
- 31.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 32.McTavish, H., L. A. Sayavedra-Soto, and D. J. Arp. 1995. Substitution of Azotobacter vinelandii hydrogenase small-subunit cysteines by serines can create insensitivity to inhibition by O2 and preferentially damages H2 oxidation over H2 evolution. J. Bacteriol. 177:3960-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menon, N. K., C. Y. Chatelus, M. Dervartanian, J. C. Wendt, K. T. Shanmugam, H. D. Peck, Jr., and A. E. Przybyla. 1994. Cloning, sequencing, and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J. Bacteriol. 176:4416-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Methe, B. A., K. E. Nelson, J. A. Eisen, I. T. Paulsen, W. Nelson, J. F. Heidelberg, D. Wu, M. Wu, N. Ward, M. J. Beanan, R. J. Dodson, R. Madupu, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, M. Gwinn, J. F. Kolonay, S. A. Sullivan, D. H. Haft, J. Selengut, T. M. Davidsen, N. Zafar, O. White, B. Tran, C. Romero, H. A. Forberger, J. Weidman, H. Khouri, T. V. Feldblyum, T. R. Utterback, S. E. Van Aken, D. R. Lovley, and C. M. Fraser. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967-1969. [DOI] [PubMed] [Google Scholar]
- 35.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 36.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 37.Przybyla, A. E., J. Robbins, N. Menon, and H. D. Peck, Jr. 1992. Structure-function relationships among the nickel-containing hydrogenases. FEMS Microbiol. Rev. 8:109-135. [DOI] [PubMed] [Google Scholar]
- 38.Richaud, P., P. M. Vignais, A. Colbeau, R. L. Uffen, and B. Cauvin. 1990. Molecular biology studies of the uptake hydrogenase of Rhodobacter capsulatus and Rhodocyclus gelatinosus. FEMS Microbiol. Rev. 7:413-418. [DOI] [PubMed] [Google Scholar]
- 39.Roling, W. F., B. M. van Breukelen, M. Braster, B. Lin, and H. W. van Verseveld. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rousset, M., Z. Dermoun, C. E. Hatchikian, and J. P. Belaich. 1990. Cloning and sequencing of the locus encoding the large and small subunit genes of the periplasmic [NiFe]hydrogenase from Desulfovibrio fructosovorans. Gene 94:95-101. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., J. E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sargent, F., S. P. Ballantine, P. A. Rugman, T. Palmer, and D. H. Boxer. 1998. Reassignment of the gene encoding the Escherichia coli hydrogenase 2 small subunit—identification of a soluble precursor of the small subunit in a hypB mutant. Eur. J. Biochem. 255:746-754. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz, E., A. Henne, R. Cramm, T. Eitinger, B. Friedrich, and G. Gottschalk. 2003. Complete nucleotide sequence of pHG1: a Ralstonia eutropha H16 megaplasmid encoding key enzymes of H2-based ithoautotrophy and anaerobiosis. J. Mol. Biol. 332:369-383. [DOI] [PubMed] [Google Scholar]
- 45.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 46.Snoeyenbos-West, O. L., K. P. Nevin, R. T. Anderson, and D. R. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153-167. [DOI] [PubMed] [Google Scholar]
- 47.Stein, L. Y., M. T. La Duc, T. J. Grundl, and K. H. Nealson. 2001. Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ. Microbiol. 3:10-18. [DOI] [PubMed] [Google Scholar]
- 48.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 49.Volbeda, A., M. H. Charon, C. Piras, E. C. Hatchikian, M. Frey, and J. C. Fontecilla-Camps. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580-587. [DOI] [PubMed] [Google Scholar]
- 50.Weiner, J. H., P. T. Bilous, G. M. Shaw, S. P. Lubitz, L. Frost, G. H. Thomas, J. A. Cole, and R. J. Turner. 1998. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93:93-101. [DOI] [PubMed] [Google Scholar]
- 51.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, L. F., B. Ize, A. Chanal, Y. Quentin, and G. Fichant. 2000. Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J. Mol. Microbiol. Biotechnol. 2:179-189. [PubMed] [Google Scholar]