Abstract
To date, attempts to regenerate a complete tooth, including the critical periodontal tissues associated with the tooth root, have not been successful. Controversy still exists regarding the origin of the cell source for cellular cementum (epithelial or mesenchymal). This disagreement may be partially due to a lack of understanding of the events leading to the initiation and development of the tooth roots and supportive tissues, such as the cementum. Osterix (OSX) is a transcriptional factor essential for osteogenesis, but its role in cementogenesis has not been addressed. In the present study, we first documented a close relationship between the temporal-and spatial-expression pattern of OSX and the formation of cellular cementum. We then generated 3.6 Col 1-OSX transgenic mice, which displayed accelerated cementum formation vs. WT controls. Importantly, the conditional deletion of OSX in the mesenchymal cells with two different Cre systems (the 2.3 kb Col 1 and an inducible CAG-CreER) led to a sharp reduction in cellular cementum formation (including the cementum mass and mineral deposition rate) and gene expression of dentin matrix protein 1 (DMP1) by cementocytes. However, the deletion of the OSX gene after cellular cementum formed did not alter the properties of the mature cementum as evaluated by backscattered SEM and resin-cast SEM. Transient transfection of Osx in the cementoblasts in vitro significantly inhibited cell proliferation and increased cell differentiation and mineralization. Taken together, these data support 1) the mesenchymal origin of cellular cementum (from PDL progenitor cells); 2) the vital role of OSX in controlling the formation of cellular cementum; and 3) the limited remodeling of cellular cementum in adult mice.
INTRODUCTION
Understanding the key regulators associated with the formation of tooth root tissues during development and regeneration is critical for developing therapies targeted at intentionally regenerating such tissues lost as a consequence of disease or trauma. One of the most challenging current objectives involved in tooth regeneration is defining the controllers of root formation (1). Tooth root origin is linked with two major issues. First, there are many fundamental differences between crown and root dentin formation (2–5), including the biochemical components such as collagen fibrils, dentinal tubular branches, predentin secretion rate, and morphological differences in crown and root odontoblasts. In addition, the tooth root is covered by cementum, in contrast to crown dentin, which is covered by enamel. The non-tooth structures associated with the tooth roots, i.e., the surrounding periodontal ligament (PDL) and its precursor (follicle tissues and supporting bone), may directly or indirectly contribute to normal tooth root formation.
Cementum, a thin layer of mineralized tissue, covers the entire tooth-root surface and provides a “dock” for the PDL to the cementum, thus contributing to supporting the functional PDL between the tooth and surrounding alveolar bone. A key marker for the successful regeneration of periodontal tissues is the formation of new cementum, particularly the cellular type (6). Thus, a full understanding of this tissue is paramount to developing more reproducible regenerative procedures than are currently available.
There are two major types of cementum: acellular (acellular extrinsic fiber cementum, also called “primary cementum”) and cellular (cellular mixed stratified cementum, secondary cementum or adaptive cementum). The term “adaptive cementum” is used because of its proposed function, which is to guide the tooth into its proper occlusal position during tooth eruption. Acellular cementum is located on the coronal and mid portions of the root, whereas the cellular cementum is found on the apical and interradicular portions. Developmentally, acellular cementum is formed first, followed by cellular cementum. Current theories on the origin of these tissues are conflicting and include the possibility that they are derived from different cell lineages (6).
Cementoblasts, the cells forming cementum, share many properties with osteoblasts, a cell type controlling bone formation (7,8), including the expression of genes such as bone sialoprotein (BSP) and osteocalcin (OCN), and the ability to promote mineral nodules in vitro (9). In addition, Runx2 and OSX, two key transcriptional factors in osteogenesis (10–13), are present in the cementoblasts (14), although the role of these transcriptional factors in cementogenesis is unknown since mice deficient in Runx2 (11) or OSX (12) die in utero, and cementogenesis occurs postnatally.
In contrast to our understanding of bone biology, many fundamental issues related to cementogenesis remain unsolved (8,15). For example, it is clear that osteoblasts are derived from mesenchymal progenitor cells but the origin of cementoblasts and whether or not acellular and cellular cementum are derived from a similar precursor remain controversial. One speculation is that dental follicle cells (mesenchymal-derived progenitors) differentiate directly into cementoblasts during the formation of both acellular and cellular cementum, subsequent to invading the gaps between the ruptured Hertwig’s epithelial root sheath (HERS) and attaching to root dentin. An alternative hypothesis states that the epithelial cells from HERS form cementoblast cells via an epithelial-mesenchymal transformation process (4,8,16). A recent study combining lineage tracing using Rosa 26 reporter mice and different Cre mice and expression assays concluded that HERS cells participated in the cementum development and may differentiate into cementocytes (17). Other researchers have also proposed an epithelial-mesenchymal transformation during the formation of acellular cementum and a mesenchymal derivation for cellular cementum. However, these hypotheses are not supported by solid functional studies using gain or loss research methodology.
In this study, we attempted to address the role of OSX in cementogenesis using the following four approaches: i) combinations of three different lacZ knock-in animal models plus immunohistochemistry assays to reveal the correlation of the temporal- and spatial-expression pattern of OSX with cellular cementum formation; ii) loss of function [with different Cre-loxP systems: the 2.3 kb-Cre mouse line whose Cre activity is initiated from the embryonic stage (18) or an inducible Cre mouse line (CAG-CreER) (19) with the Cre induced from postnatal day 21 by tamoxifen (TM)], and gain of function (generation of the 3.6 kb Collagen type 1-OSX Tg mice) to study the OSX-null or Tg cementum phenotype; iii) analyses of the OSX-null phenotype after the cementum is already formed by the same inducible Cre system in order to address whether the cementum is remodeled; and iv) studies focused on determining the mechanism by which OSX controls cementogenesis in vitro. Our findings support the novel concept that mesenchymal-derived PDL progenitor cells serve as the key resource for cellular cementum formation (mesenchymal origin), in which OSX, a critical transcriptional factor, plays an essential role during cementogenesis.
EXPERIMENTAL PROCEDURES
Mice and tamoxifen administration
To avoid neonatal death, mice harboring a floxed allele of OSX (Osxflox) (20) were crossed with CAG-CreER transgenic mice obtained from the Jackson laboratory, permitting a temporally regulated Cre-mediated recombination by TM (Sigma-Aldrich) (19). The offspring were further intercrossed to obtain mice homozygous for floxed OSX and heterozygous for the CMV-CreER transgene. The mice were then injected intraperitoneally with TM 1.5–3.0 mg/10 g body weight or vehicle (corn oil containing 10% ethanol) at the desired postnatal days: 1) starting at postnatal day 21 (after initiation of cellular cementum formation) for 3 weeks and sacrificed at 6 weeks; and 2) starting at 6 months for 3 weeks and sacrificed at 7 months. OSX floxed mice were also bred with the 2.3 Col 1-Cre mice (18) for the tissue-specific deletion of OSX at the embryonic stage to cover the early stage phenotype during cementogenesis. Dmp1-lacZ (21), OSX-lacZ (12) and periostin-lacZ (12,22) knock-in mice, in which Dmp1 or OSX was replaced by a lacZ reporter (12), were used for tracing when and where periostin or Dmp1 or OSX is expressed in the PDL region and cementum during postnatal development. Genotyping was determined by polymerase chain reaction (PCR) analysis of genomic DNA with primers p01: 5′-CGGGGAGTCCCGAGGACTCTG-3′ and p02: 5′-CTGTCTTCACCTCAATTCTATT-3′ to detect the wild-type allele (~400 base pairs [bp]) and OSX (~550 bp). Primers p03: 5′-CCCGCAGAACCTGAAGATG-3′ and primers p04: 5′-GACCCGGCAAAACAGGTAG-3′ were used to detect the Cre allele (~500 bp).
Generation of the 3.6 Col 1 OSX transgenic mice
The full-length mouse OSX cDNA and an SV40 polyadenylation signal were cloned into a mammalian expression vector containing the 3.6 kb rat type I collagen promoter, plus a 1.6 kb intron 1 (23) (graciously provided by Dr. Barbara Kream at the University of Connecticut Medical Center, USA), giving rise to the Col1a1-OSX transgene (Tg). The transgene was released by Sac II and Sal I from the vector backbone and purified for pronuclear injection. Five transgenic founders with B-6 background were generated. Genotyping was determined by PCR analysis of the genomic DNA with primers p05: 5′-GAAGCGACCACTTGAGCAAACAT-3′ and p06: 5′-TGTCCAAACTCATCAATGTATCT-3′ to detect the OSX Tg (~400 bp).
All animal protocols were approved by the Animal Welfare Committee at Texas A&M Health Science Center Baylor College of Dentistry.
Double fluorochrome labeling
To analyze the changes in the cementum deposition rate, double fluorescence labeling was performed as described previously (24,25). Briefly, subsequent to injection of TM, calcein (5 mg/kg i.p.; Sigma-Aldrich) was administered separately two times (P35 and P40). The mice were sacrificed 48 h after the second injection. The mandibles were removed and fixed in 70% ethanol for 48 h. The specimens were dehydrated through a graded series of ethanol (70–100%) and embedded in methyl-methacrylate (MMA, Buehler, Lake Bluff, IL) without decalcification. Fifty-μm sections were cut using a Leitz 1600 saw microtome. The unstained sections were viewed under epifluorescent illumination using a Nikon E800 microscope interfaced with the Bioquant software. The mean distance between the two fluorescent labels was determined and divided by the number of days between labels to calculate the deposition rate of cellular cementum formation between postnatal days 35 and 40 (μm/day).
Histology
The teeth were fixed in freshly prepared 4% paraformaldehyde in phosphate-buffered saline (pH 7.4), decalcified, and embedded in paraffin using standard histological procedures as previously described (26). The tissue blocks were cut into 4-μm thick mesio-distal serial sections to compare the tissue samples from the experimental and control groups and were then mounted on glass slides. The sections were used for H&E staining, toluidine staining, immunohistochemistry and in situ hybridization.
In situ hybridization
Mouse antisense RNAs for BSP were used for in situ hybridization as described previously (27). Briefly, the digoxigenin (DIG)-labeled mouse cRNA probes were prepared using an RNA Labeling Kit (Roche, Indianapolis, IN). The hybridization temperature was set at 55 °C and the washing temperature at 70°C, so that the endogenous alkaline phosphatase was inactivated. DIG-labeled nucleic acids were detected in an enzyme-linked immunoassay with a specific anti-DIG-AP antibody conjugate and an improved substrate that gives rise to a red signal (Vector, Burlingame, CA), according to the manufacturer’s instructions.
Isolation and photography of maxillary molars
For imaging the cellular cementum, the maxillary molar samples from the control, OSX CKO, and OSX Tg mice were extracted as described previously (28). Briefly, freshly isolated maxilla were dissected free of muscle and incubated in lysis buffer (2X SSC, 0.2% SDS, 10 mM EDTA, 1 mg/ml Proteinase K) for 1–2 days at 55°C. After the PDLs surrounding the teeth were digested, the molars were extracted under a dissection microscope and photographed.
Backscattered scanning electron microscopy (SEM) and resin-casted SEM
The mandibles were dissected and fixed in 70% ethanol at room temperature for 24 h. The tissue specimens were dehydrated in ascending concentrations of ethanol (from 70% to 100%), embedded in MMA without decalcification and sectioned through the center of the first mandibular molar using a water-cooled diamond-impregnated circular saw (Isomet, Buehler). The cut surface was polished using 1, 0.3 and 0.05 μm alumina alpha micropolish II solutions (Buehler) on a soft cloth rotating wheel (24). Each sample was placed in an ultrasonic bath between steps and immediately following the polishing steps. The dehydrated specimens were then sputter-coated with carbon and scanned with a backscattered electron detector in a JEOL JSM-6300 scanning electron microscope (JEOL, Japan). The parameters were kept constant while the backscattered SEM images were taken. Next, the carbon-coated samples were repolished, as described above. The surfaces were acid-etched with 37% phosphoric acid for 2–10 seconds, followed by 5% sodium hypochlorite for 20 minutes. The samples were then sputter-coated with gold and palladium, as described previously (29,30) and examined in the scanning electron microscope.
FITC staining (31)- Fluorescein isothiocyanate, a small molecular dye, fills in the PDL cells/fibers as well as the cementoblast and cementocyte cells but does not enter the mineral matrix. Thus, the dye provides a visual representation of the organization of the PDL and cementum under the confocal microscope. Immediately after harvesting and dissection, the jaw bones were placed in EM fixative (0.5% gluteraldehyde, 2% paraformaldehyde in 0.05 M cacodylate-sodium buffer, pH 7.4) at room temperature and then processed to obtain cortical sections and cancellous bone blocks. A cross section (300–400 μm thick) was cut with a diamond-bladed saw (Buehler, Lake Bluff, IL), and the cortical sections were then sanded and ground to a final thickness of 30–50 μm for confocal imaging.
Cell culture, cell proliferation and mineralization assay
OCCM-30 cells, an immortalized cementoblast cell line (9), were plated into 6-well plates at a density of 1.2×105 cells per well and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mmol/L L-glutamine,100 units/ml penicillin and 100 μg/ml streptomycin. A cell proliferation assay was performed by direct cell counting and the MTT (3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) method. Briefly, they were seeded into 96-well plates at 5×104 cells per well. When the cells reached 70% confluence, PEX3-Osterix plasmids were transiently transfected into OCCM-30 cells for 48 h and then trypsinized; the cells were counted daily using a hemocytometer for up to 5 days. For the MTT assay, the cells were seeded into 96-well plates with 1.5×104 cells per well; cell proliferation was determined at days 1, 3, and 5, respectively, using a MTT cell proliferation assay kit (ATCC, No. 30-1010K, Manassas, VA). For the mineralization assay, the OCCM-30 cells were cultured in 24-well plates; following transfection, 5 mM β-glycerophosphate was added to the medium on day 2. After 2 weeks in culture, the cells were fixed with ice-cold 70% ethanol/30% 1mM Hepes and stained with Alizarin red to detect calcification.
RNA isolation and quantitative PCR analyses
PEX3-Osterix plasmids or vectors only were transfected into OCCM-30 cells for 48 h. The total RNAs were extracted using Trizol reagent (Invitrogen, San Diego, CA) according to the manufacturer’s recommended protocol. The cDNA was synthesized with a reverse transcription kit II (Invitrogen). The SYBR green real-time RT-PCR technique was used to measure and analyze the BSP, OCN, OPN and ALP expression. Real-time PCR was performed using 1 μl of cDNA and 12.5 μl 2x SYBR Green Master mix in a 25 μl reaction volume with ABI PRISM 7700 (Applied Biosystems Inc.). GAPDH was used as a housekeeping gene. The primer sequences, designed using the Primer Express (version 2.0.0, Applied Biosystems), are listed in Table 1 (supplementary data).
Statistical analysis
Statistical significance was determined by an independent-sample t-test using SPSS 12.0. A P value of < 0.05 was considered statistically significant.
RESULTS
Temporal- and spatial- expression pattern of OSX is closely linked to expression of DMP1, and cellular cementum formation
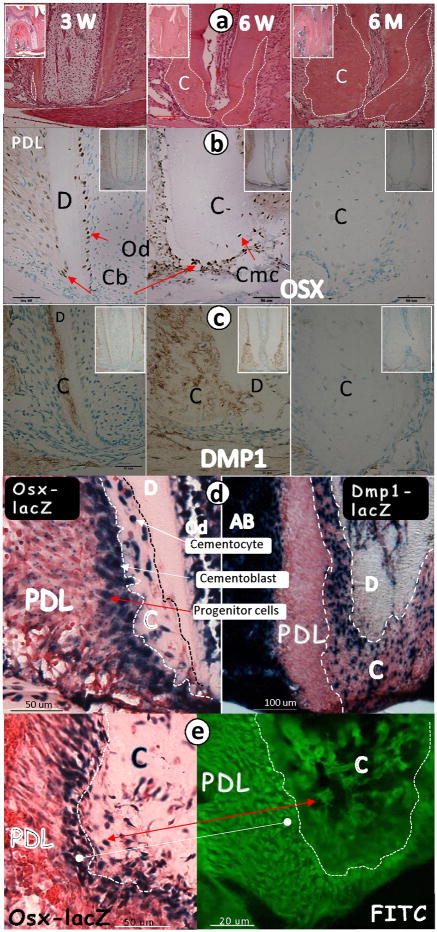
In a previous study, Hirata et al. (14) reported that OSX is expressed in 2-week-old rat cellular cementum. In the current study, we first determined the expression pattern for OSX in relation to the formation of cellular cementum during tooth eruption using tissues obtained from mice. As shown by H&E staining, cellular cementum was noted in the middle, apical region by 3 weeks (Fig 1a, left panel), with a significant increase in cementum formation by 6 weeks (Fig 1a, middle panel). By 6 months, the apical root region was fully covered by cementum (Fig 1a, right panel). Immunohistochemical staining showed that at 3 weeks, OSX was expressed in the cementoblasts lining the cellular cementum, as well as in the odontoblasts and PDL cells (Fig 1b, left panel). At 6 weeks, OSX was highly expressed in the cementoblasts and cementocytes (Fig 1b, middle panel). By 6 months, there were no OSX positive cells (Fig 1b, right panel). Similarly, DMP1, an extracellular matrix protein produced by cementocytes, displayed an expression pattern similar to that of OSX with peak intensity at weeks 3 and 6 and with no detectable signal at 6 months (Fig 1c). However, there was an important difference between the Dmp 1 and OSX expression patterns. Using OSX-lacZ and Dmp1-lacZ knock-in mice in which the lacZ expression reflects the endogenous gene signal (12,21), we noted that OSX was expressed in the PDL progenitor cells that surround the cementum with a gradient (Fig 1d, left panel), while Dmp 1 expression was absent in these cells (Fig 1d, right panel). Furthermore, the PDL cells in close proximity to the cementum were OSX-lacZ-positive (Fig 1e, left panel), which correlated with the FITC-stained PDL cells (Fig 1e, right panel). Next, we examined the temporal and spatial expression patterns of the OSX-lacZ in knock-in mice, starting from newborn to 6 months. In the newborns up to 7 days, there was no OSX signal in the HERS or follicle cells, although a strong signal was detected in the pulp and odontoblast cells (Supplementary Fig 1a–b). By 3 weeks, the OSX signal was detected in the PDL progenitor cells, cementoblasts, and cementocytes (Supplementary Fig 1c). A sharp increase in OSX expression was observed at weeks 4 and 6 in these same cell populations (Supplementary Fig 1d–e). In contrast, there were few positive cells detected in these tissues obtained from the 6-month-old mice (Supplementary Fig 1f). Importantly, the spatial expression pattern of the OSX-lacZ in the PDL progenitor cell layer was identical to that of two mesenchymal cell types of neural crest origin: the periosteal progenitor cells (forming bone, Supplementary Fig 2a) and pulp progenitor cells (forming dentin, Supplementary Fig 2b).
Fig 1. OSX expression profile correlates with cellular cementum formation during postnatal development.
a. Representative H&E staining images show a very thin layer of cellular cementum at 3 weeks (left panel); an expanded cementum layer at 6 weeks (middle panel), and a dramatically larger cementum layer at 6 months (right panel, inset); b. OSX immunohistochemistry stained images show a weak signal in PDL and cementoblast cells at 3 weeks (left panel, inset); a strong signal in PDL and cementoblast cells at 6 weeks (middle panel, inset) and no signal at 6 months (right panel, inset); c. DMP1 signal is apparent in the cemental extracellular matrix at 3 weeks (left panel, inset) and 6 weeks (middle panel, inset) but not at 6 months (right panel, inset); d. 4-wk-old lower jaws obtained from OSX-lacZ knock-in and Dmp1-lacZ knock-in mice, where lacZ is used to trace the endogenous gene (OSX, left panel; Dmp1, right panel) expression pattern, were processed for x-gal stain. OSX-lacZ is expressed in progenitor cells in the PDL region, cementoblasts and cementocytes (left panel). Dmp1 is expressed only in cementocytes and alveolar bone osteocytes (AB, right panel); e. X-gal stain reveals a OSX-lacZ signal not only in cementoblasts/cementocytes but also in progenitor cells in the PDL region with an expression gradient pattern (6-wk-old, left panel; red arrow); the FITC confocal image (6-wk-old, right panel) shows numerous cells in PDLs (likely progenitors for cementoblasts) that surround the cementum and appear to correlate with lacZ positive-stained cells on the left panel (white balls). C, cementum; D, dentin; Cb, cementoblast; Cmc, cementocyte; Od, odontoblasts; PDL, periodontal ligament.
As a whole, the above data suggest that 1) the temporal and spatial expression patterns of OSX in PDL cells, cementoblasts, and cementocytes are closely correlated with cellular cementum formation, and these OSX-positive PDL cells are most likely the progenitor cells for cellular cementum; and 2) Dmp1 expression is directly linked to the formation of cementocytes and not to the precursor cells.
Overexpression of OSX greatly accelerates the formation of cellular cementum
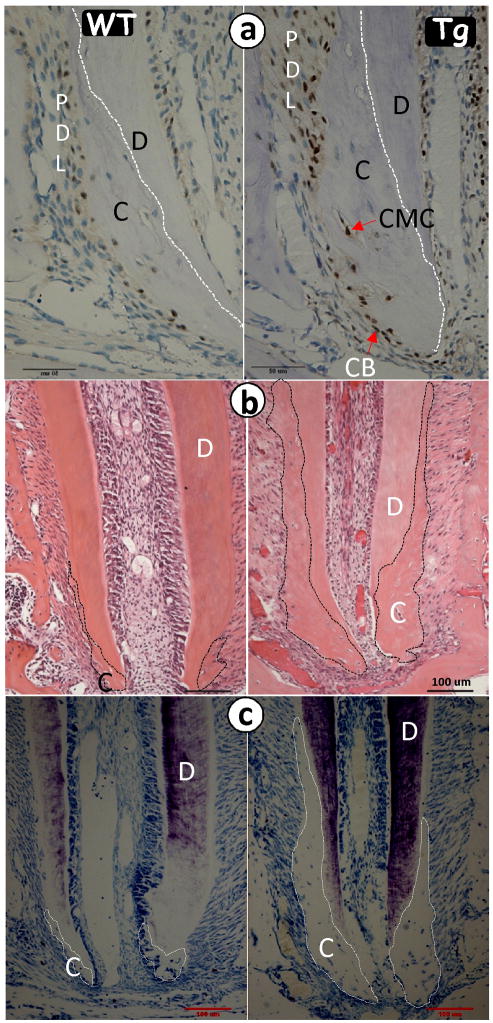
To characterize the role of OSX in cementogenesis, we first generated a transgenic mouse comprising full length OSX cDNA under the control of a 3.6 kb Col1a1 promoter, which is widely used to target genes in pulp cells, odontoblasts or osteoblasts. Thirteen independent Col1a1-OSX transgenic lines were obtained. The expression level of OSX mRNA in all transgenic mice was higher than that of the control based on real-time PCR assay (data not shown), and one of the lines was used in this study. The immunohistochemistry assay showed a higher level of OSX in the transgenic cementoblasts, cementocytes, as well as PDL cells, at 4 weeks of age (Fig 2a, right panel) compared to the age-matched wild-type (WT) mice. Images at 3 weeks obtained from H&E staining (Fig 2b, right panel) and toluidine blue staining (Fig 2c, right panel) showed more cellular cementum mass in the tissues of transgenic mice compared to age-matched WT (left panels) mice, supporting the presumption of a positive role of OSX in the acceleration of cellular cementum formation.
Fig 2. Overexpression of OSX accelerates cementum formation.
a, Representative immunohistochemistry assay shows more OSX signals in the 2.3 Col 1-OSX transgenic (Tg) cementoblast, cementocyte and PDL cells at 4 weeks (right panel) compared to the age-matched control (WT, left panel); b. H&E stain image shows a much larger cellular cementum layer in the 2.3 Col 1-OSX Tg (right panel) compared to the age-matched control (left panel); c. The images of toluidine stain (right panel) further confirm this observation. Note that the Tg epical foramen is much narrower than that of the WT. D, dentin.
OSX is expressed in follicle cells but not in HERS, and conditional deletion of OSX dramatically slows down cementogenesis
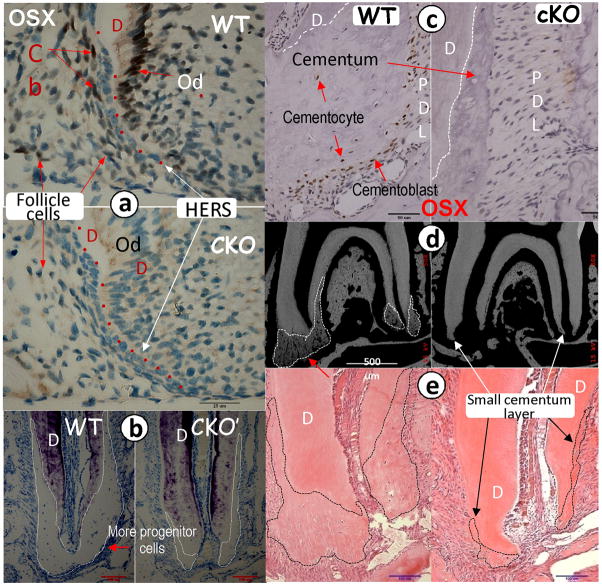
There are currently two hypotheses regarding the origins of the progenitor cells of the cementoblast: they may issue from the dental follicle cells or the HERS cells. Using immunohistochemistry, we first examined the OSX expression in these two cell populations in tissues obtained from two-week-old mice. A strong OSX signal was detected in the nuclei of the WT cementoblasts and odontoblasts, and a low reproducible OSX signal was observed in the nuclei of the dental follicle cells. Importantly, no OSX signal was detected in the HERS cells (Fig 3a). OSX is a well-documented transcriptional factor for mesenchymal-derived cells such as osteoblasts (12,32) and chondrocytes (13). Our observations strongly support the notion that cementoblasts are derived from dental follicle cells and not HERS cells, at least those involved in the formation of the cellular cementum.
Fig 3. OSX is expressed in follicle cells but not in HERS cells, and the conditional deletion (CKO) of OSX leads to a sharp reduction in cellular cementum.
The first mandibular molars were obtained from mice harboring a floxed allele of OSX and the 2.3 kb Col 1-Cre (activated from embryonic stage) (18) or the CAG-CreER transgene, which permits temporal regulation of the Cre-mediated recombination by tamoxifen (TM) (19). TM was injected (i.p.) for 3 weeks starting at 3 weeks of age, and tissues were harvested at 6 weeks. a. Representative immunohistochemistry assay shows a strong OSX signal in the wild type (WT) cementoblast (CB) and odontoblast cells (Od,) with a weak but reproducible signal in the follicle cells. Note that there is no OSX expression in the HERS cells at two weeks of age (upper panel). There was no detectable OSX signal in the CKO cementoblasts, odontoblasts, or follicle cells (lower panel); b. Toluidine stain shows a sharp reduction of the cellular cementum layer and cementoblasts, supporting cells in the CKO mice at 6 weeks of age (right panels) compared to the age-matched control (left panels); c. Immunohistochemistry assay shows a strong OSX signal in the WT cementoblasts at 6 weeks of age (left panel). There was no detectable OSX signal in the CKO cementoblasts and cementocytes (right panel); c–d. The representative images of backscattered SEM (d) and H&E stain (e) show a sharp reduction in the cellular cementum layer in the 6-week-old CKO mice crossed with CAG-CreER (right panels). D, dentin.
Because the conventional knockout of OSX leads to perinatal lethality before cementum formation (12), we studied the tissue-specific knockout effect of OSX on cellular cementum using a loxP/Cre system. Mice harboring a floxed allele of OSX (Osxflox) (20) were first crossed with the 2.3 Col 1-Cre mice, whose Cre was activated from the embryonic stage (18). Conditionally, OSX knockout (CKO) mice displayed a sharp reduction in cellular cementum mass compared to the WT controls at 6 weeks of age, as assayed by toluidine blue staining (Fig 3b, right panel). These flox mice were also crossed with CAG-CreER transgenic mice, which permitted a temporally regulated CRE-mediated recombination by TM (19). As shown in Fig 3c (right panel), injection of TM for 3 weeks starting at 3 weeks of age led to the loss of OSX signals in the cementoblasts vs. the WT control tissues, as determined by immunohistochemistry (Fig 3c, left panel). Images obtained from the backscattered SEM (Fig 3d, right panel) and H&E staining (Fig 3e, right panel) showed that conditionally removing OSX by injections of TM led to a sharp reduction of cellular cementum mass in tissues from transgenic mice vs. control tissues at 6 weeks of age. In summary, the conditional deletion of OSX by either the 2.3 Col 1-Cre or the inducible CAG-CreER has the same effect: a sharp reduction of cementum mass.
Deletion of OSX greatly reduces cementocyte number and morphology with a significant reduction in mineralization rate
We next asked whether the deletion of OSX alters the ultrastructure of the cementocytes using an acid-etched, resin-casting technique (30). In this technique, the polished surfaces of the resin-embedded cementum were etched with acid to remove the minerals, leaving a relief cast of the non-mineralized areas infiltrated by resin. This technique, therefore, revealed the morphology of the cementum system. Thus, we showed that there were fewer cementocytes in the 6-week-old CKO mice (Fig 4a, right upper panel) compared to the age-matched controls (Fig 4a, left panel). Furthermore, the dendrite numbers in the CKO cementocytes were dramatically reduced (Fig 4b, right lower panel) compared to the control (Fig 4b, left lower panel). These observations suggest that OSX plays a major role in establishing the correct architecture and organization of the cementocyte during cellular cementum formation.
Fig 4. Deletion of OSX gene leads to a reduction of the cementocyte number, dendrites, and mineralization rate.
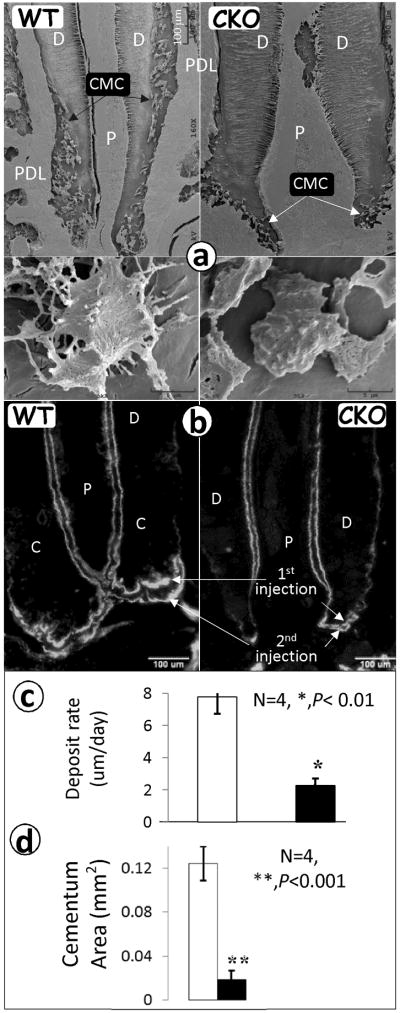
a. Representative resin-casted SEM images show a reduction in cementocyte number (right upper panel) and dendrite number (right lower panel) in the CKO mouse at 4 weeks of age compared to age-matched controls (left panels); b. fluorochrome-labeled sections of the first lower molar from WT control (left panel) and OSX CKO mice (right panel) reveal a reduced mineralization rate in the CKO cementum. The distance between the two injections reflects the cementum formation rate. The area reflects cementum mass; c–d. Quantitative data show a 4-fold reduction in the mineralization rate in the CKO cementum (c), and a significant reduction in the cellular cementum area calculated based on the fluorescence labeled area (d). Data are mean ± SEM, n=4–6, **P<0.01, ***P<0.001. C, cementum; P, pulp; and D, dentin.
Double labeling is often used for defining the mineralization status in bones (30) and teeth (24,25). To address whether or not OSX plays a role in controlling the mineralization rate in cementogenesis, a fluorochrome labeling assay was used. Subsequent to injecting TM (two times a week for 2 weeks), calcein was injected twice (5 days apart), and the animals were sacrificed two days after the second injection. As shown in Fig 4b, the fluorochrome labeling lines appeared thicker in the WT cementum area (arrows, left panel) compared to the CKO group (arrows, right panel). The distance between the two lines, reflecting the rate of cellular cementum formation, was much shorter in the CKO group (at ~2μm/day) than in the WT group (~8 um/day); this difference was statistically significant (p < 0.01; Fig 4c). We also measured the total labeling area in both groups. The labeled cellular cementum area in the CKO was reduced to a much greater extent (4-fold) compared to the control and was highly significant (p < 0.001; Fig 4d). These data suggest that the decreased mineralization rate associated with the CKO cementocytes accounts for the observed decrease in cementum mass.
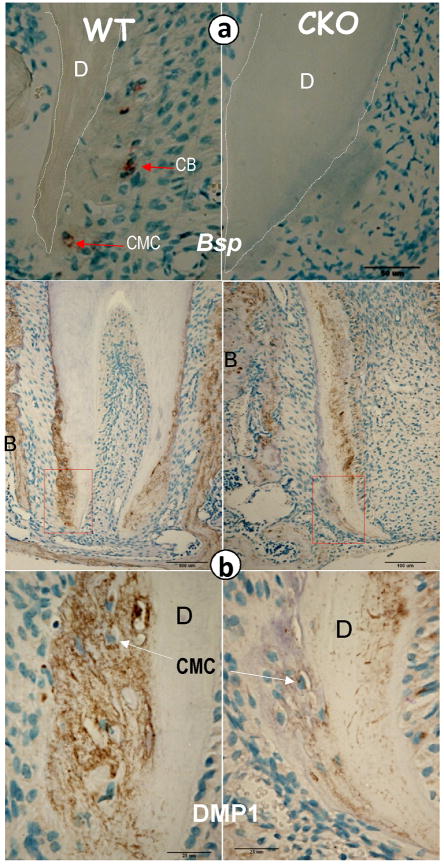
Deletion of OSX changes the levels of extracellular matrix proteins important for regulation of cellular cementum
To address the potential mechanisms by which deletion of OSX leads to the above morphological changes in cellular cementum, we evaluated the expression of two key markers of cementum (BSP and DMP1). In he OSX CKO cementum, the mRNA levels of BSP (Fig 5a, right panel) were reduced in comparison with that of the age-matched controls. Importantly, DMP1, a selective marker for cementocyte was highly expressed in both cementocytes (Fig 5c, left panel) and osteocytes (Supplementary Fig 3, left panel); deletion of OSX led to a dramatic reduction of DMP1 in the cementocytes (Fig 5c, right panel) and osteocytes (Supplementary Fig 3, right panel). Note that DMP1 was largely undetectable in the control dentin, since the dentin is fully formed and the odontoblasts are not active at the time points studied. In contrast, there was a strong DMP1 signal in the OSX CKO dentin, i.e., the dentin was not fully formed. All of these observations demonstrate the vital role of OSX in controlling the molecular event during cementogenesis.
Fig 5. Deletion of Osx gene changes molecular markers which are important for cementogenesis.
a, Representative in situ hybridization assay shows a sharp reduction in of BSP (right panel) in Osx CKO cementoblasts and cementocytes compared to age matched control (left panel); b, Representative immunohistochemistry assay shows a sharp reduction of DMP1 (right panels) in Osx CKO′ cementoblast (CB) and cementocyte (CMC) cells compared to the age matched control (left panels). BSP, bone sialoprotein; DMP1, dentin matrix protein 1; D, dentin; and B, bone.
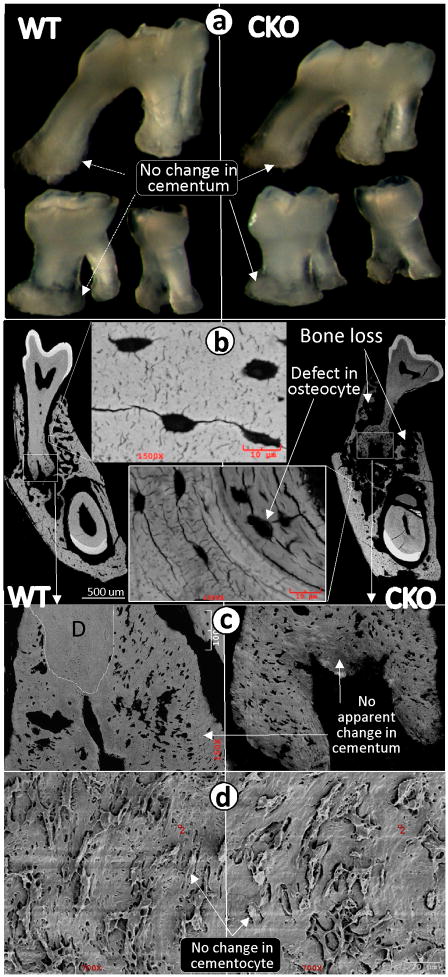
There appears to be no apparent cementum phenotype when OSX is deleted in 6-month-old mice
Cementum and bone share many characteristics in both gene expression profiles and cell morphologies. For example, cementocytes and osteocytes are the only cells buried in mineralized tissues. It is also known that bone remodels. However, existing studies on cementum indicate that there is limited or no remodeling, unless the teeth are stressed (e.g., exaggerated occlusal forces). In the present study, we attempted to address the cementum-remodeling issue by deleting OSX after the cementum is fully formed. Specifically, TM was injected into the OSXfx/CAG-CreER inducible mice twice a week starting at 6 months of age, and the animals were sacrificed at 7 months of age. The photographs of cellular cementum showed no apparent difference between the CKO and the control molars (Fig 6a), which was further confirmed by backscattered SEM (Fig 6b–c, right panels) and resin-casted SEM images (Fig 6d, right panel). In contrast, there was striking bone loss plus a defect in osteocyte maturation and mineralization in the OSX CKO bone vs. control tissues (Fig 6b, right panel). These data support the notion that cementum, unlike bone, does not undergo remodeling under normal conditions.
Fig 6. Conditional deletion of OSX by tamoxifen (TM) injections after cementum has fully formed has little effect on cementum formation but dramatically alters bone remodeling.
a. Representative photomicrographs of three maxillary molars (1st on the top and 2nd/3rd on the bottom) show no apparent difference between the WT (left panel) and the CKO (right panel) mice that received TM injections (two times a week for 3 weeks starting at 6 months; sacrifice was at 7 months); b. The backscattered SEM images obtained from the mandibles show dramatic alveolar bone loss and defects in osteocyte morphology in the CKO mice (right and lower images); c–d. The images of backscattered SEM (c) and resin-casted cementocytes (d) revealed no apparent difference between the WT (left panels) and the CKO cellular cementum mass and cementocyte morphology (right panels). (D=dentin).
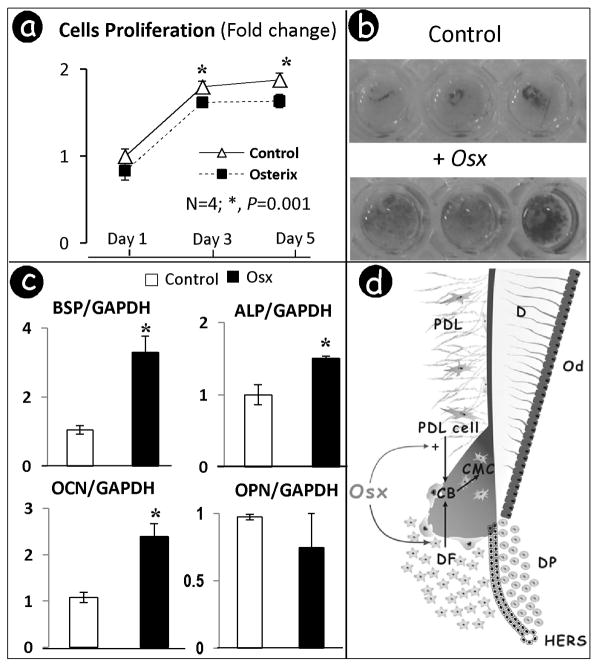
Overexpression of OSX in cementoblasts inhibits cell proliferation but accelerates cell differentiation and mineralization in vitro
Next, we addressed the potential role of OSX in controlling the proliferation, differentiation and mineralization by cementoblasts in vitro. PEX3-Osterix plasmids were transiently transfected into a cementoblast cell line (33), whose transient transfection efficiency was ~29%± 4.7 based on pcDNA3-GFPtopaz-positive cell number estimation (data not shown). The cells were harvested at days 1, 3 and 5. Quantitative assay showed that OSX significantly inhibited cell proliferation (Fig 7a). Alizarin red staining showed the acceleration of mineralization by OSX (Fig 7b). Real-time PCR was used to further address the molecular mechanisms involved in OSX-mediated mineral formation. Specific markers of cementogenesis were determined after 48 h transfection with PEX3-Osterix or the empty vector. As shown in Fig 7c, the transcripts for BSP, OCN, and ALP significantly increased with a fold range between 1.5 and 3.5 higher than that of the control group, although there was no difference in OPN mRNA. These changes are similar to the changes observed in bone cells (32).
Fig 7. Overexpression of OSX in cementoblasts inhibited cell proliferation, induced the formation of mineralized nodules and stimulated differentiation.
a. The MTT assay shows a significant inhibition of cementoblast proliferation at days 3 and 5 after transient transfection with PEX3-Osterix plasmids; b. Representative photo images of Alizarin red assay reveals acceleration of mineralization after OSX transfection; c. Overexpression of OSX significantly increases expressions of BSP, OCN, and ALP, differentiation markers for cementoblast. Levels of different differentiation markers were normalized with those of the loading control GAPDH in three independent experiments. *, p <0.05; d. working hypothesis: OSX is initially expressed at low levels during cementogenesis in dental follicle cells, which increases as cellular cementum formation is being initiated. OSX controls cellular cementum formation by inhibiting proliferation of cells in the local region (PDL and follicle cells), while accelerating differentiation of follicle and PDL cells toward a cementoblast (Cb)/cementocyte (CMC) phenotype. Similar to dentinogenesis, but in contrast to osteogenesis, there appears to be a lack of remodeling in cementum. PDL, periodontal ligament; DF, dental follicle; DP, dental papilla; HERS, Hertwig’s epithelial root sheath cells.
DISCUSSION
Do cementoblasts associated with the formation of cellular cementum originate from mesenchymal cells (dental follicle cells) or epithelium cells (HERS cells)? As mentioned in the Introduction, there are currently two hypotheses on the origin of cellular cementoblasts: they may arise from the dental mesenchymal cells or from the HERS cells. The mesenchymal (“Classic”) hypothesis proposes that the dental follicle cells migrate to the HERS cells on the root surface, disrupt the epithelial structure, and differentiate into cementoblasts that form both acellular and cellular cementum matrices (34–38). The epithelial origin hypothesis suggests an epithelial-mesenchymal transformation of the HERS cells, which then differentiate into the cementoblasts involved in forming acellular cementum and possibly cellular cementum as well. In addition, some researchers have proposed an epithelial origin for acellular cementum and a mesenchymal origin for cellular cementum (4,8,39–41). There is evidence supporting both hypotheses, which have attracted great interest for several decades since this information is important for defining the factors involved in regenerating these tissues. However, the evidence for both of these hypotheses remains inconclusive, so the debate is ongoing. A lack of progress in this research area is due to the following: (i) the research results obtained are difficult for comparisons due to the use of different reporter mice, in vitro cultures, and tissue structure analyses under light and electron microscopy; (ii) no genes have been identified as exclusive markers for cementogenesis; and (iii) cementum formation occurs after birth, and often the genes knocked down in mice are lethal at the neonatal stage and/or the genes affect crown development, and thus, roots do not form.
To better address this issue, we took a unique approach, including a) using OSX-lacZ knock-in mice and mapping the expression pattern of OSX from newborn (before cellular cementum formation) to 6 months of age when no new cellular cementum formation was noted (Fig 1d–e; Supplementary Fig 1); b) comparing unique expression patterns in 3 lacZ knock-in mice: periostin, a gene highly expressed in PDL but not in cellular cementum; OSX, a transcriptional factor expressed in PDL progenitor cell layers surrounding the cellular cementum, which is different from that of periostin-positive cells in PDL (Supplementary Fig 5); and DMP1, an extracellular matrix protein expressed selectively by cementocytes (Fig 1c–d; Supplementary Fig 3); and c) using FITC-stained imaging, which demonstrated that the PDL cells in close proximity to cellular cementum correlated with OSX-lacZ-positive PDL cells (Fig 1e). These data support the concept that PDL progenitor cells are the key cell resource for cellular cementum, which is consistent with the OSX-lacZ expression pattern in two mesenchymal-derived cells: periosteum (the progenitors for periosteal bone formation) (Supplementary Fig 5a) and pulp cells (the progenitors for dentin formation) (Supplementary Fig 5b).
Furthermore, we selected loss- and gain-of-function approaches to study cellular cementum origin and cellular cementum formation targeting OSX, based on two factors: 1) both cementoblasts and osteoblasts share many common characteristics (8,42); and 2) OSX is an essential transcriptional factor for mesenchymal-derived osteoblasts (12,13). Using the OSX floxed mice crossed with either the 2.3 Col 1A1-Cre [activated from embryonic through postnatal development stage (18) or the CAG inducible Cre model (13)], we reported a dramatic decrease in cellular cementum in both models, a severe change in cementocyte number and morphology, and a significant reduction in cementum-formation rate and mass. Moreover, overexpression of OSX controlled by the 3.6 Col 1A1 promoter resulted in an increase in cementum mass. Importantly, there was no apparent change in the HERS cells. Thus, these in vivo data not only support the mesenchymal hypothesis, but also document OSX as a key transcriptional factor that directly regulates the formation of cellular cementum.
With existing data, and the new data presented here, are we able to definitively exclude epithelial involvement in cementogenesis? The answer is no, as Huang et al showed, using a K14-Cre (reflecting the epithelial origin) and Rosa26R cross in mice, that a limited number of cells were β-gal positive, suggesting that during root development a small subset of HERS cells may undergo transformation to cementoblasts (1). Interestingly, the same research group reported a greater number of β-gal positive cells in cellular cementum when crossing Wnt1-Cre (reflecting the mesenchymal origin) and Rosa26R mice vs K14-Cre cross with Rosa26R mice (17). These findings, coupled with our results demonstrating a mesenchymal origin for cementoblasts, suggest that while a subset of HERS cells may undergo transformation, a large proportion of cementoblasts are of mesenchymal origin. Thus, it seems apparent that lineage tracing approaches alone do not allow for a conclusion that cementoblasts are of epithelial origin. Importantly, existing data suggest a very prominent role for HERS, vs simply undergoing apoptosis, during root development. Steele-Perkins et al have shown the mice null for Nfic, a transcription factor, develop tooth crowns but with poorly developed roots as their major phenotype (43). Further studies by Huang et al demonstrated that HERS cells, through BMP mediated Smad-Shh signaling, activate Nfic in pre-odontoblasts to promote root dentin formation and subsequently continue the formation of root dentin/cementum (44). The new results presented here provide significant insights as to the key regulators of root formation, while it is clear that further studies are needed to define the cell type and to map the genes and proteins modulating root (including cementum) formation.
Bone remodeling (formation and resorption) occurs throughout our lifespan. In contrast, cementum is thought to undergo minimal remodeling (45). However, cementoblasts, like osteoblasts, contain PTH1R, a receptor for PTHrP that regulates bone remodeling. Further, cementoblasts can support osteoclast differentiation via control of OPG and RANKL in vitro (46). By deleting Osx in bone and cementum in the same adult flox/flox mice and using comparable analyses for both tissues, our approach now provides clear data that mature cementum in mice undergoes limited, if any, remodeling under normal conditions. In contrast, there was striking bone loss and a marked defect in osteocyte morphology with the deletion of OSX in adult mice (also see Supplementary Fig 4). This loss-of-function assay supports early studies of tissues obtained from human subjects: cementum does not remodel under normal conditions (45).
Previous studies indicated that OSX controls bone formation through two different mechanisms: inhibition of cell proliferation via blocking Wnt/β-catenin signaling and acceleration of cell differentiation through upregulation of BSP and OCN (32). The current studies using immunohistochemistry assays to evaluate protein and gene expression of DMP1, BSP and OCN in tissues from Osx conditional knockout and transgenic mice, coupled with in vitro assays using cementoblasts, and real-time RT-PCR/mineralization/cell proliferation assay, support a similar mechanism by which OSX controls cementogenesis: a decrease in cell proliferation and an increase in cell differentiation and mineralization.
In summary, these data support the mesenchymal hypothesis for the derivation of cellular cementum and further identify the vital role of OSX in modulating the formation of cellular cementum (see working hypothesis, Fig 7d). However, this model, which provides a powerful framework for future studies of cementoblast maturation and mineralization, has certain limitations. For example, the genes specific for early vs. more mature cementocytes have not been fully characterized, and the morphological characteristics of early vs. mature cementocytes are still largely unknown. The effect of the loss of OSX on HERS cells is not clear, although there seems to be no apparent change in the morphology of these cells. Regardless, this work will stimulate more interest in searching for the role of OSX in forming cellular cementum with the potential for applying the knowledge gained to develop therapies for regenerating the periodontal structure.
Supplementary Material
Acknowledgments
This study was supported by NIH grants to JQF (DE018486) and by the National Natural Science Foundation of China to ZC (81170933).
Authors’ roles: Study design: JQF, YX, and BD; Study conduct: ZC, HZ, XZ, and XH; Data collection: TG. Data analysis: YR. Data interpretation: ZC, MS and JQF; Drafting manuscript: ZC. Revising manuscript content: MS and JQF. Approving final version of manuscript: ZC, MS and JQF.
Footnotes
There is no conflict of interest.
References
- 1.Fong HK, Foster BL, Popowics TE, Somerman MJ. The crowning achievement: getting to the root of the problem. J Dent Educ. 2005;69(5):555–570. [PubMed] [Google Scholar]
- 2.Ten Cate AR. A fine structural study of coronal and root dentinogensis in the mouse: observations on the so-called ‘von Korff fibres’ and their contribution to mantle dentine. J Anat. 1978;125(Pt 1):183–197. [PMC free article] [PubMed] [Google Scholar]
- 3.Beertsen W, Niehof A. Root-analogue versus crown-analogue dentin: a radioautographic and ultrastructural investigation of the mouse incisor. Anat Rec. 1986;215(2):106–118. doi: 10.1002/ar.1092150204. [DOI] [PubMed] [Google Scholar]
- 4.Thomas HF. Root formation. Int J Dev Biol. 1995;39(1):231–237. [PubMed] [Google Scholar]
- 5.Andujar MB, Couble P, Couble ML, Magloire H. Differential expression of type I and type III collagen genes during tooth development. Development. 1991;111(3):691–698. doi: 10.1242/dev.111.3.691. [DOI] [PubMed] [Google Scholar]
- 6.Beertsen W, VandenBos T, Everts V. Root development in mice lacking functional tissue non-specific alkaline phosphatase gene: inhibition of acellular cementum formation. J Dent Res. 1999;78(6):1221–1229. doi: 10.1177/00220345990780060501. [DOI] [PubMed] [Google Scholar]
- 7.Somerman MJ, Shroff B, Agraves WS, Morrison G, Craig AM, Denhardt DT, Foster RA, Sauk JJ. Expression of attachment proteins during cementogenesis. J Biol Buccale. 1990;18 (3):207–214. [PubMed] [Google Scholar]
- 8.Bosshardt DD. Are cementoblasts a subpopulation of osteoblasts or a unique phenotype? J Dent Res. 2005;84(5):390–406. doi: 10.1177/154405910508400501. [DOI] [PubMed] [Google Scholar]
- 9.D’Errico JA, Berry JE, Ouyang H, Strayhorn CL, Windle JJ, Somerman MJ. Employing a transgenic animal model to obtain cementoblasts in vitro. J Periodontol. 2000;71 (1):63–72. doi: 10.1902/jop.2000.71.1.63. [DOI] [PubMed] [Google Scholar]
- 10.Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes & Development. 1999;13(8):1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 12.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, Zhang Z, Feng JQ, Dusevich VM, Sinha K, Zhang H, Darnay BG, de Crombrugghe B. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A. 2010;107(29):12919–12924. doi: 10.1073/pnas.0912855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirata A, Sugahara T, Nakamura H. Localization of runx2, osterix, and osteopontin in tooth root formation in rat molars. J Histochem Cytochem. 2009;57(4):397–403. doi: 10.1369/jhc.2008.952192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster BL, Popowics TE, Fong HK, Somerman MJ. Advances in defining regulators of cementum development and periodontal regeneration. Curr Top Dev Biol. 2007;78:47–126. doi: 10.1016/S0070-2153(06)78003-6. [DOI] [PubMed] [Google Scholar]
- 16.Orban B. The epithelial network in the periodontal membrane. J Am Dent Assoc. 1952;44 (6):632–635. doi: 10.14219/jada.archive.1952.0112. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, Bringas P, Jr, Slavkin HC, Chai Y. Fate of HERS during tooth root development. Dev Biol. 2009;334(1):22–30. doi: 10.1016/j.ydbio.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M, Kream BE. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004;48(7):645–653. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244(2):305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 20.Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T, de Crombrugghe B. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci U S A. 2005;102(41):14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng JQ, Huang H, Lu Y, Ye L, Xie Y, Tsutsui TW, Kunieda T, Castranio T, Scott G, Bonewald LB, Mishina Y. The Dentin matrix protein 1 (Dmp1) is specifically expressed in mineralized, but not soft, tissues during development. J Dent Res. 2003;82(10):776–780. doi: 10.1177/154405910308201003. [DOI] [PubMed] [Google Scholar]
- 22.Rios H, Koushik SV, Wang HY, Wang J, Zhou HM, Lindsley A, Rogers R, Chen Z, Maeda M, Kruzynska-Frejtag A, Feng JQ, Conway SJ. periostin null mice exhibit dwarfism, incisor enamel defects, and an early-onset periodontal disease-like phenotype. Molecular and Cellular Biology. 2005;25(24):11131–11144. doi: 10.1128/MCB.25.24.11131-11144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braut A, Kalajzic I, Kalajzic Z, Rowe DW, Kollar EJ, Mina M. Col1a1-GFP transgene expression in developing incisors. Connect Tissue Res. 2002;43(2–3):216–219. doi: 10.1080/03008200290001078. [DOI] [PubMed] [Google Scholar]
- 24.Miller SC, Omura TH, Smith LJ. Changes in dentin appositional rates during pregnancy and lactation in rats. J Dent Res. 1985;64(8):1062–1064. doi: 10.1177/00220345850640080701. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y, Ye L, Yu S, Zhang S, Xie Y, McKee MD, Li YC, Kong J, Eick JD, Dallas SL, Feng JQ. Rescue of odontogenesis in Dmp1-deficient mice by targeted re-expression of DMP1 reveals roles for DMP1 in early odontogenesis and dentin apposition in vivo. Dev Biol. 2007;303(1):191–201. doi: 10.1016/j.ydbio.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fen JQ, Zhang J, Dallas SL, Lu Y, Chen S, Tan X, Owen M, Harris SE, MacDougall M. Dentin matrix protein 1, a target molecule for Cbfa1 in bone, is a unique bone marker gene. J Bone Miner Res. 2002;17(10):1822–1831. doi: 10.1359/jbmr.2002.17.10.1822. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y, Yuan B, Qin C, Cao Z, Xie Y, Dallas SL, McKee MD, Drezner MK, Bonewald LF, Feng JQ. The biological function of DMP1 in osteocyte maturation is mediated by its 57 kDa C-terminal fragment. J Bone Miner Res. 2010 doi: 10.1002/jbmr.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye L, MacDougall M, Zhang S, Xie Y, Zhang J, Li Z, Lu Y, Mishina Y, Feng JQ. Deletion of dentin matrix protein-1 leads to a partial failure of maturation of predentin into dentin, hypomineralization, and expanded cavities of pulp and root canal during postnatal tooth development. The Journal of biological chemistry. 2004;279(18):19141–19148. doi: 10.1074/jbc.M400490200. [DOI] [PubMed] [Google Scholar]
- 29.Martin DM, Hallsworth AS, Buckley T. A method for the study of internal spaces in hard tissue matrices by SEM, with special reference to dentine. Journal of microscopy. 1978;112(3):345–352. doi: 10.1111/j.1365-2818.1978.tb00085.x. [DOI] [PubMed] [Google Scholar]
- 30.Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, Yu X, Rauch F, Davis SI, Zhang S, Rios H, Drezner MK, Quarles LD, Bonewald LF, White KE. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet. 2006;38(11):1310–1315. doi: 10.1038/ng1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciani C, Doty SB, Fritton SP. An effective histological staining process to visualize bone interstitial fluid space using confocal microscopy. Bone. 2009;44(5):1015–1017. doi: 10.1016/j.bone.2009.01.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Cho K, Huang Y, Lyons JP, Zhou X, Sinha K, McCrea PD, de Crombrugghe B. Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc Natl Acad Sci U S A. 2008;105(19):6936–6941. doi: 10.1073/pnas.0710831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mada Y, Miyauchi M, Oka H, Kitagawa M, Sakamoto K, Iizuka S, Sato S, Noguchi K, Somerman MJ, Takata T. Effects of endogenous and exogenous prostaglandin E2 on the proliferation and differentiation of a mouse cementoblast cell line (OCCM-30) J Periodontol. 2006;77(12):2051–2058. doi: 10.1902/jop.2006.060148. [DOI] [PubMed] [Google Scholar]
- 34.Paynter KJ, Pudy G. A study of the structure, chemical nature, and development of cementum in the rat. Anat Rec. 1958;131(2):233–251. doi: 10.1002/ar.1091310207. [DOI] [PubMed] [Google Scholar]
- 35.Cho MI, Garant PR. Ultrastructural evidence of directed cell migration during initial cementoblast differentiation in root formation. J Periodontal Res. 1988;23(4):268–276. doi: 10.1111/j.1600-0765.1988.tb01371.x. [DOI] [PubMed] [Google Scholar]
- 36.Diekwisch TG. The developmental biology of cementum. Int J Dev Biol. 2001;45(5–6):695–706. [PubMed] [Google Scholar]
- 37.Ten Cate AR. The development of the periodontium--a largely ectomesenchymally derived unit. Periodontol 2000. 1997;13:9–19. doi: 10.1111/j.1600-0757.1997.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 38.Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127(8):1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 39.Slavkin HC. Towards a cellular and molecular understanding of periodontics. Cementogenesis revisited. J Periodontol. 1976;47(5):249–255. doi: 10.1902/jop.1976.47.5.249. [DOI] [PubMed] [Google Scholar]
- 40.Slavkin HC, Bringas P, Jr, Bessem C, Santos V, Nakamura M, Hsu MY, Snead ML, Zeichner-David M, Fincham AG. Hertwig’s epithelial root sheath differentiation and initial cementum and bone formation during long-term organ culture of mouse mandibular first molars using serumless, chemically-defined medium. J Periodontal Res. 1989;24(1):28–40. doi: 10.1111/j.1600-0765.1989.tb00854.x. [DOI] [PubMed] [Google Scholar]
- 41.Bosshardt DD, Nanci A. Hertwig’s epithelial root sheath, enamel matrix proteins, and initiation of cementogenesis in porcine teeth. J Clin Periodontol. 2004;31(3):184–192. doi: 10.1111/j.0303-6979.2004.00473.x. [DOI] [PubMed] [Google Scholar]
- 42.Ouyang H, McCauley LK, Berry JE, Saygin NE, Tokiyasu Y, Somerman MJ. Parathyroid hormone-related protein regulates extracellular matrix gene expression in cementoblasts and inhibits cementoblast-mediated mineralization in vitro. J Bone Miner Res. 2000;15(11):2140–2153. doi: 10.1359/jbmr.2000.15.11.2140. [DOI] [PubMed] [Google Scholar]
- 43.Steele-Perkins G, Butz KG, Lyons GE, Zeichner-David M, Kim HJ, Cho MI, Gronostajski RM. Essential role for NFI-C/CTF transcription-replication factor in tooth root development. Mol Cell Biol. 2003;23(3):1075–1084. doi: 10.1128/MCB.23.3.1075-1084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang X, Xu X, Bringas P, Jr, Hung YP, Chai Y. Smad4-Shh-Nfic signaling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J Bone Miner Res. 2010;25(5):1167–1178. doi: 10.1359/jbmr.091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zander HA, Hurzeler B. Continuous cementum apposition. J Dent Res. 1958;37(6):1035–1044. doi: 10.1177/00220345580370060301. [DOI] [PubMed] [Google Scholar]
- 46.Boabaid F, Berry JE, Koh AJ, Somerman MJ, McCcauley LK. The role of parathyroid hormone-related protein in the regulation of osteoclastogenesis by cementoblasts. J Periodontol. 2004;75(9):1247–1254. doi: 10.1902/jop.2004.75.9.1247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.