Abstract
Across phyla, aging is associated with reduced sleep duration and efficiency. Both aging and sleep involve complex genetic architecture and diverse cell types and are heavily influenced by diet and environment. Therefore, understanding the molecular mechanisms of age-dependent changes in sleep will require integrative approaches that go beyond examining these two processes independently. The fruit fly, Drosophila melanogaster, provides a genetically amenable system for dissecting the molecular basis of these processes. In this review, we examine the role of metabolism and circadian rhythms in age-dependent sleep loss.
Keywords: Sleep loss, Aging, Drosophila melanogaster, Metabolism, Circadian rhythms
Introduction
Aging is characterized by changes in diverse biological processes including neural plasticity, metabolism and behavior. One consequence of aging that appears to be conserved across phyla is sleep loss. Aging results in diminished sleep efficiency and duration in humans, rodents and fruit flies [1–3]. Conversely, sleep-deprived animals or short-sleeping genetic mutants have reduced lifespans [4]. Complex genetic architecture and diverse cell types regulate both sleep and aging, making the study of age-associated changes in sleep experimentally challenging. Recent genetic studies in the fruit fly suggest the molecular basis of sleep and aging is conserved from flies to mammals [5]. Flies show age-dependent changes in sleep that are comparable to those observed in humans, but the neural basis for age-dependent changes in sleep remains unclear.
Powerful genetic tools and a simplified nervous system of approximately 200,000 neurons make Drosophila an excellent system for dissecting the molecular and neural basis of sleep and aging. A large collection of genetic mutants, genome-wide RNA interference libraries and a host of tools that allow for activating or silencing small populations of neurons provide unparalleled capabilities for genetic manipulation [6]. Flies live for only 60–80 days in standard laboratory conditions, allowing for behavioral monitoring over an entire life cycle [7]. Furthermore, gene expression can be temporally regulated allowing for adult-specific modulation of cellular function and neural circuitry [6]. Because of their small size and the ease of manipulating environmental factors such as light, temperature and food, the interactions between sleep and longevity can be investigated. Here, we briefly review studies examining the relationship between sleep and aging in Drosophila and propose future avenues for studying the molecular basis of age-dependent changes in sleep.
Sleep Studies in Drosophila
Flies display all the behavioral characteristics of sleep, including extended periods of quiescence, elevated arousal hreshold, compensatory rebound following deprivation and species-specific posture [2, 8]. Sleep in the fruit fly is typically defined as 5 min of immobility, which correlates with an elevated threshold to arousal [2, 8]. Recordings of local field potentials in the central brain reveal reduced electrical activity during behaviorally quiescent periods, suggesting that flies, like mammals, have altered physiology during sleep [9]. Many wake-promoting drugs including caffeine, modafinil, amphetamine and cocaine have comparable effects on fly and human sleep, supporting the notion that the molecular basis of sleep is highly conserved [10–12]. Also supporting this notion, pharmacological treatments of human sleep disorders target dopamine signaling, and dopamine potently suppresses sleep in Drosophila. Modafinil and amphetamine purportedly inhibit dopamine transporter function to suppress sleep in flies, while a reduction in dopamine signaling increases sleep duration and improves sleep consolidation [12, 13]. Intriguingly, at least some functions of sleep appear to be conserved from flies to mammals because sleep-deprived flies display many similar cognitive deficits to those seen in humans, including reduced memory consolidation [14].
A number of large-scale genetic screens for short-sleeping flies have identified genes that promote sleep. These genes have diverse functions and include the Shaker K+ channel, the Shaker-interacting membrane protein sleepless, the dopamine transporter fumin and the cell-cycle regulator cyclin A [15–18]. Genome-wide analysis has also been applied to identify molecular processes underlying sleep. Transcriptome analysis of wild-derived inbred Drosophila lines found genes affecting nervous system development and metabolism that correlate with sleep duration [19]. Microarray studies examining differential gene regulation in sleep-deprived flies and in flies spontaneously awake or asleep during 12-hour light/dark cycles revealed wake-associated genes involved in glutamatergic transmission, stress and immune responses, while sleep-associated genes included the glial-specific gene anachronism and enrichment for genes involved in lipid metabolism [20]. A better understanding of the genes and neurons underlying sleep regulation will be critical in determining how sleep changes with age.
Total sleep duration can be decoupled into the number of sleep bouts exhibited by an individual and the average length of each sleep bout. Efficient sleep is consolidated into longer bouts, and as flies age, sleep becomes fragmented [7]. The presence of dietary sugars potently increases consolidation of sleep into longer sleep bouts, and this is in part due to activation of sugar-sensitive sensory neurons [21]. Considerable evidence has emerged suggesting the genetic mechanisms underlying bout number and bout duration may be different. Flies mutant for the neuropeptide amnesiac have normal sleep duration but sleep is fragmented [22]. These findings suggest distinct genes regulate sleep duration and the length of individual sleep bouts. amnesiac has also been implicated in age-dependent memory decline [23], indicating that shared neuropeptide signaling may underlie age-dependent changes in sleep and memory.
Molecular Cues Underlying Age-Dependent Changes in Sleep
Screening for long-lived animals is labor-intensive, and therefore, a number of alternative assays have been developed to characterize aging, including sensitivity to the free radical-inducing drug paraquat, sensitivity to high temperature stress and screening for neurodegeneration [24, 25]. The short lifespan of the fly and ease of measuring longevity has made these approaches particularly effective for identifying genes involved in the aging process. A number of single-gene mutations have been identified that increase lifespan by as much as 50%, including numerous genes associated with metabolic function and insulin signaling [26].
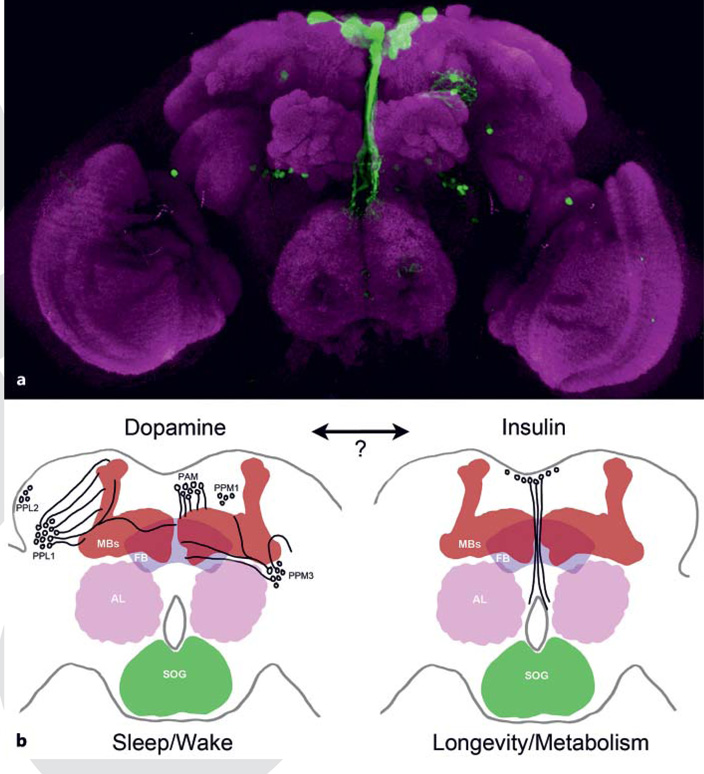
Insulin signaling is central to metabolic regulation in the fly, and insulin-producing cells within the fly brain are proposed to be integrators of nutrition and sleep (fig. 1a) [27]. Mutations in the insulin-signaling pathway including numerous insulin-like peptides, the insulin receptor and insulin receptor substrate chico are all long-lived [26]. Studies in the nematode Caenorhabditis elegans and mammals also implicate insulin in aging, suggesting a conserved role for insulin signaling in the regulation of longevity [28]. The long-lived phenotype of animals deficient for insulin signaling suggests diet modulates longevity. Indeed, dietary restriction increases lifespan in animals ranging from worms to mammals. In Drosophila, lifespan is dramatically extended by dietary restriction that consists of feeding flies food with reduced yeast content, while increased yeast content shortens lifespan [30, 31]. Adding back the amino acid methionine alone prevents the increase in longevity brought about by dietary restriction, suggesting that dietary methionine promotes aging [32]. Disruption of the insulin receptor substrate protein chico prevents lifespan extension through caloric restriction, revealing a critical role for insulin in the aging process [33]. Therefore, yeast appears to stimulate insulin signaling and promote aging.
Fig. 1.
Insulin-producing cells in the Drosophila brain. a Confocal image depicts approximately 20 insulin-producing cells, genetically labeled with green fluorescent protein (green), which produce and secrete insulin-like peptides. These cells are involved in the regulation of sleep, metabolism and aging. The brain is colabeled with a general synaptic neuropil marker (magenta). b The schematic depicts a subset of the dopamine neurons (left) and insulin-producing cells (right) in the fly brain. Dopamine is a central regulator of sleep-wake regulation, while insulin regulates metabolism and longevity. It is possible that interactions between these two systems regulate age-dependent changes in Drosophila sleep. The major neuropils in the fly brain are labeled as follows: FB = fan-shaped body; AL = antennal lobe; SOG = subesophageal ganglion; MBs = mushroom bodies. PAM = ■■■; PPL1 = ■■■; PPL2 = ■■■; PPM1 = ■■■; PPM3 = ■■■.
There is abundant evidence that sensory cues derived from the presence of dietary macronutrients modulate both sleep and aging. Removal of all olfactory cues blocks the effect of yeast on longevity in dietary-restricted animals. In fact, removal of a single gustatory receptor, Gr63a, extends lifespan, indicating that the smell of food alone can promote aging [34]. The effect of caloric restriction, dietary methionine and sensory perception of yeast on age-associated changes in sleep remains unknown, and it will be interesting to determine whether these manipulations prevent age-dependent sleep changes in addition to increasing longevity.
While there is a clear link between diet and longevity, the relationship between diet and age-related changes in sleep is less clear. Aged flies have dramatically altered sleeping patterns including decreased sleep duration and reduced sleep bout length, suggesting that in flies, like humans, aging prevents sleep consolidation [2, 7]. Moreover, flies that are selected for short and fragmented sleep display phenotypes associated with increased aging, as well as shortened lifespan [35]. Treating young flies with the free radical-inducing drug paraquat results in reduced sleep consolidation similar to that observed in aged flies, leading to speculation that free radicals are responsible for age-dependent changes in sleep [7]. There is also evidence suggesting the dietary changes that promote aging also modulate sleep. Raising flies on a high-calorie diet results in cardiac dysfunction, diminishes longevity and accelerates age-related sleep fragmentation [36, 37].
Aging may modulate sleep indirectly by disrupting functionally linked neural systems. In addition to sleep, circadian rhythms and associated molecular clocks are disrupted in aged animals. A key feature of circadian clocks is transcriptional synchrony of clock gene expression and neural activity between the primary pacemaker neurons, and disruption of core circadian induces sleep loss. Aged flies have reduced strength of circadian rhythms and a lengthening of the free-running circadian clock [38, 39]. The molecular basis for altered circadian rhythms and whether this is responsible for age-related sleep loss remains unclear. One study reported that aged flies displayed head-specific reductions in the oscillation of some, but not all, circadian genes [38]. These findings raise the possibility that the reduced robustness of transcriptional rhythms result in prolonged locomotor rhythms [38]. Another study found weak transcriptional rhythms in the periphery and strong rhythms in central brain neurons [39]. Surprisingly, inhibition of protein kinase A in central clock neurons rescued the age-dependent loss of circadian rhythms and fragmentation of sleep, suggesting elevated protein kinase A signaling may disrupt sleep and circadian rhythms in aged animals [39]. Furthermore, genetic manipulation of nutrient-sensing components of the insulin pathway, such as target of rapamycin and AKT, alters circadian rhythms [40]. Given that shared genes and neurons regulate aging, sleep and circadian rhythms, it will be informative to further study the relationship between transcriptional regulation of circadian function and sleep in aged animals.
Future Areas of Speculation
While progress has been made in understanding the biological basis of sleep and aging, less is known about how these processes interact. There are a number of central questions regarding the molecular basis of age-related changes in sleep that remain unanswered. While aged flies display reduced sleep and increased sleep fragmentation, the functional and physiological significance of these changes is unclear. While mammalian aging is accompanied by a reduction in slow-wave sleep, which is particularly important for consolidation of memories and synaptic homeostasis [41, 42], distinct phases of sleep have not been identified in Drosophila. Currently, fly sleep is predominantly defined by periods of behavioral quiescence, though many studies examine arousal threshold and sleep rebound. The development of improved assays for physiological and behavioral analysis of sleep in the fruit fly may allow for a better understanding of how sleep changes with age.
Age-related sleep loss and fragmentation may involve altered expression of genes central to sleep regulation. Alternatively, global transcriptional changes may underlie age-associated changes in sleep. A more comprehensive understanding of the genetic architecture underlying sleep-wake regulation and the associated changes in neural activity may help to address these questions. For example, it has been proposed that insulin release from the neurosecretory insulin-producing cells regulates nutrient-dependent changes in sleep [27]. Supporting the findings that insulin is a critical regulator of sleep, the neurotransmitter octopamine promotes wakefulness by signaling insulin-secreting neurosecretory cells in the fly brain [43]. Given the role of insulin signaling in aging, this raises the possibility that altered function of the insulin-producing cells may underlie age-associated changes in sleep [43]. While there is clearly a role for insulin signaling in regulating longevity and age-dependent changes in sleep, the location of sleep neurons targeted by insulin remains unclear. Dopamine is central to sleep-wake regulation in flies and mammals, and dopamine neurons densely innervate the Drosophila mushroom bodies, which are central to sleep. Thus, identifying whether insulin confers age-related changes in sleep through targeting the same brain regions that regulate sleep in young animals, such as dopamine neurons, the mushroom bodies or other novel sleep circuitry, will be informative (fig. 1b). Interactions between insulin and dopamine signaling appear to be conserved in mammals. Insulin signaling is impaired in diabetes and is associated with decreased memory and cognition. Diabetic induced rats display decreased levels of dopamine with a concomitant increase in hippocampal dopamine receptor expression [44]. If this interaction is conserved in Drosophila, it is possible that insulin release from the insulin-producing cells may regulate dopamine signaling and provide a link between aging, metabolic state and sleep-wake regulation.
Finally, much of the neural circuitry and synaptic changes underlying age-associated sleep loss remains to be determined. One proposed function of sleep in flies and humans is the maintenance of synaptic homeostasis through a global depotentiation of synapses [45]. Evidence suggests synaptic homeostasis is essential for the synaptic plasticity that underlies memory formation but may also underlie a more general mechanism of neuronal maintenance. The effect of aging on the modulation of synaptic proteins remains unclear. Examining the effects of sleep on synaptic homeostasis in aging flies may unveil novel mechanisms for age-dependent changes in sleep. For example, it is possible that reduced synaptic homeostasis in aged animals underlies interactions between sleep, memory and aging. Age-related changes in neural morphology and, likely, connectivity, have been noted between dorsal paired medial (DPM) neurons and their targets in the mushroom bodies [23]. These neural loci have been implicated in both memory formation and sleep. In fact, blocking transmitter output from DPM neurons results in fragmented sleep, raising the possibility that age-dependent changes in DPM function underlie sleep fragmentation [22]. As additional neural circuitry underlying sleep becomes known, the application of techniques examining the physiological function of these neurons in old and young flies may reveal neural mechanisms underlying age-dependent sleep loss.
In summary, sleep is modulated by both aging and metabolism. Aging has also been implicated in age-dependent disruption of circadian rhythms and sleep consolidation. Insulin signaling intermediates that promote aging also modulate circadian rhythms, raising the possibility that insulin signaling underlies age-dependent changes in sleep-wake regulation. Likewise, dietary changes that promote aging also modulate sleep. However, the molecular and neural links between diet and age-related changes in sleep remain unclear. The ease of measuring sleep and aging in flies, along with powerful genetic tools, will aid future investigations into the molecular and neural basis of age-related sleep loss.
References
- 1.Pandi-Perumal SR, Seils LK, Kayumov L, Ralph MR, Lowe A, Moller H, Swaab DF. Senescence, sleep, and circadian rhythms. Ageing Res Rev. 2002;1:559–604. doi: 10.1016/s1568-1637(02)00014-4. [DOI] [PubMed] [Google Scholar]
- 2.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 3.Hasan S, Dauvilliers Y, Mongrain V, Franken P, Tafti M. Age-related changes in sleep in inbred mice are genotype dependent. Neurobiol Aging. 2012;33:195.e13–195.e26. doi: 10.1016/j.neurobiolaging.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Bushey D, Hughes KA, Tononi G, Cirelli C. Sleep, aging, and lifespan in Drosophila. BMC Neurosci. 2010;11:56. doi: 10.1186/1471-2202-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sehgal A, Mignot E. Genetics of sleep and sleep disorders. Cell. 2011;146:194–207. doi: 10.1016/j.cell.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venken KJ, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. Proc Natl Acad Sci USA. 2006;103:13843–13847. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 9.Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol. 2002;12:1934–1940. doi: 10.1016/s0960-9822(02)01300-3. [DOI] [PubMed] [Google Scholar]
- 10.Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 11.Wu MN, Ho K, Crocker A, Yue Z, Koh K, Sehgal A. The effects of caffeine on sleep in Drosophila require PKA activity, but not the adenosine receptor. J Neurosci. 2009;29:11029–11037. doi: 10.1523/JNEUROSCI.1653-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendricks JC, Kirk D, Panckeri K, Miller MS, Pack AI. Modafinil maintains waking in the fruit fly Drosophila melanogaster. Sleep. 2003;26:139–146. doi: 10.1093/sleep/26.2.139. [DOI] [PubMed] [Google Scholar]
- 13.Lebestky T, Chang JS, Dankert H, Zelnik L, Kim YC, Han KA, Wolf FW, Perona P, Anderson DJ. Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron. 2009;64:522–536. doi: 10.1016/j.neuron.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Yu F, Guo A. Sleep deprivation specifically impairs short-term olfactory memory in Drosophila. Sleep. 2009;32:1417–1424. doi: 10.1093/sleep/32.11.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogulja D, Young MW. Control of sleep by cyclin A and its regulator. Science. 2012;335:1617–1621. doi: 10.1126/science.1212476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 19.Harbison ST, Carbone MA, Ayroles JF, Stone EA, Lyman RF, Mackay TF. Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat Genet. 2009;41:371–375. doi: 10.1038/ng.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirelli C, LaVaute TM, Tononi G. Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem. 2005;94:1411–1419. doi: 10.1111/j.1471-4159.2005.03291.x. [DOI] [PubMed] [Google Scholar]
- 21.Linford NJ, Chan TP, Pletcher SD. Re-patterning sleep architecture in Drosophila through gustatory perception and nutritional quality. PLoS Genet. 2012;8:e1002668. doi: 10.1371/journal.pgen.1002668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, Guo F, Lu B, Guo A. Amnesiac regulates sleep onset and maintenance in Drosophila melanogaster. Biochem Biophys Res Commun. 2008;372:798–803. doi: 10.1016/j.bbrc.2008.05.119. [DOI] [PubMed] [Google Scholar]
- 23.Tamura T, Chiang AS, Ito N, Liu HP, Horiuchi J, Tully T, Saitoe M. Aging specifically impairs amnesiac-dependent memory in Drosophila. Neuron. 2003;40:1003–1011. doi: 10.1016/s0896-6273(03)00732-3. [DOI] [PubMed] [Google Scholar]
- 24.Lin YJ, Seroude L, Benzer S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- 25.Kretzschmar D, Hasan G, Sharma S, Heisenberg M, Benzer S. The Swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J Neurosci. 1997;17:7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paaby AB, Schmidt PS. Dissecting the genetics of longevity in Drosophila melanogaster. Fly (Austin) 2009;3:29–38. doi: 10.4161/fly.3.1.7771. [DOI] [PubMed] [Google Scholar]
- 27.Broughton SJ, Slack C, Alic N, Metaxakis A, Bass TM, Driege Y, Partridge L. DILP-producing median neurosecretory cells in the Drosophila brain mediate the response of lifespan to nutrition. Aging Cell. 2010;9:336–346. doi: 10.1111/j.1474-9726.2010.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 29.Spiegel K, Knutson K, Leproult R, Tasali E, Van Cauter E. Sleep loss: a novel risk factor for insulin resistance and type 2 diabetes. J Appl Physiol. 2005;99:2008–2019. doi: 10.1152/japplphysiol.00660.2005. [DOI] [PubMed] [Google Scholar]
- 30.Bass TM, Grandison RC, Wong R, Martinez P, Partridge L, Piper MD. Optimization of dietary restriction protocols in Drosophila. J Gerontol A Biol Sci Med Sci. 2007;62:1071–1081. doi: 10.1093/gerona/62.10.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Libert S, Zwiener J, Chu X, Vanvoorhies W, Roman G, Pletcher SD. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- 32.Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clancy DJ, Gems D, Hafen E, Leevers SJ, Partridge L. Dietary restriction in long-lived dwarf flies. Science. 2002;296:319. doi: 10.1126/science.1069366. [DOI] [PubMed] [Google Scholar]
- 34.Poon PC, Kuo TH, Linford NJ, Roman G, Pletcher SD. Carbon dioxide sensing modulates lifespan and physiology in Drosophila. PLoS Biol. 2010;8:e1000356. doi: 10.1371/journal.pbio.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seugnet L, Suzuki Y, Thimgan M, Donlea J, Gimbel SI, Gottschalk L, Duntley SP, Shaw PJ. Identifying sleep regulatory genes using a Drosophila model of insomnia. J Neurosci. 2009;29:7148–7157. doi: 10.1523/JNEUROSCI.5629-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamazaki M, Tomita J, Takahama K, Ueno T, Mitsuyoshi M, Sakamoto E, Kume S, Kume K. High calorie diet augments age-associated sleep impairment in Drosophila. Biochem Biophys Res Commun. 2012;417:812–816. doi: 10.1016/j.bbrc.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 37.Birse RT, Choi J, Reardon K, Rodriguez J, Graham S, Diop S, Ocorr K, Bodmer R, Oldham S. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 2010;12:533–544. doi: 10.1016/j.cmet.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rakshit K, Krishnan N, Guzik EM, Pyza E, Giebultowicz JM. Effects of aging on the molecular circadian oscillations in Drosophila. Chronobiol Int. 2012;29:5–14. doi: 10.3109/07420528.2011.635237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo W, Chen WF, Yue Z, Chen D, Sowcik M, Sehgal A, Zheng X. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell. 2012;11:428–438. doi: 10.1111/j.1474-9726.2012.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng X, Sehgal A. AKT and TOR signaling set the pace of the circadian pacemaker. Curr Biol. 2010;20:1203–1208. doi: 10.1016/j.cub.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tononi G. Slow wave homeostasis and synaptic plasticity. J Clin Sleep Med. 2009;5:S16–S19. [PMC free article] [PubMed] [Google Scholar]
- 42.Cirelli C. Brain plasticity, sleep and aging. Gerontology. 2012;58:441–445. doi: 10.1159/000336149. [DOI] [PubMed] [Google Scholar]
- 43.Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron. 2010;65:670–681. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson R, Krishnakumar A, Paulose CS. Enhanced dopamine D1 and D2 receptor gene expression in the hippocampus of hypoglycaemic and diabetic rats. Cell Mol Neurobiol. 2009;29:365–372. doi: 10.1007/s10571-008-9328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tononi G, Cirelli C. Sleep function and synaptic homeostasis. Sleep Med Rev. 2006;10:49–62. doi: 10.1016/j.smrv.2005.05.002. [DOI] [PubMed] [Google Scholar]