Abstract
Resting human CD4 T cells are highly resistant to transfection or infection with lentiviral vectors derived from the human immunodeficiency virus (HIV LV). We now describe a flexible and efficient approach that permits effective genetic manipulation of these cells while preserving their naturally quiescent state. This technology can also be extended to primary lymphoid cultures where authentic cellular composition and functional relationships are preserved.
Keywords: Lentivirus, CD4 T cells, shRNA, Vpx, HIV, AIDS
INTRODUCTION
CD4 T cells play a central role in stimulating adaptive immune responses by interacting with various types of immune cells including B cells, cytotoxic T cells, macrophages and dendritic cells. Because of their key functions, enhanced CD4 T-cell activity produced by genetic mutations, infection, or injury generates pathogenic responses characterized by chronic inflammation and hypersensitivity disorders1–3. Conversely, compromised CD4 T-cell function produces a profound state of immunodeficiency exemplified by the infection and depletion of human CD4 T lymphocytes by the human immunodeficiency virus-1 (HIV-1).
CD4 T cells have been studied primarily in cultures of human peripheral blood because of the relative ease of access to this biological fluid. More recently, human lymphoid aggregate cultures (HLACs) derived from fresh tonsillar or splenic tissue have also been used for ex vivo analysis of CD4 T cell function4. However, a critical limitation in the study of quiescent human CD4 T cells is the lack of an experimental system that allows for either the efficient knockdown or expression of exogenous genes. A number of viral and non-viral (synthetic) methods have been tested including antibody-targeted liposomes5, CD4 aptamer-siRNA chimeras6, electroporation7, and peptide transduction8. Nevertheless, these synthetic approaches have been hampered by low efficiency, direct cell toxicity, or a lack of adequate experimental throughput. Retroviral vectors, particularly those derived from lentiviruses such as HIV-1 and SIVmac251, are an alternative and more efficient strategy for gene delivery to lymphoid cells9. However, lentiviruses are critically limited by their inability to infect resting CD4 T cells in peripheral blood and lymphoid tissues. Lentiviral infection is often aborted in these cells after viral entry as reverse transcription is initiated but progresses with a much slower kinetics that often fail to reach completion10–18. Lentiviral vectors are also commonly pseudotyped with the vesicular stomatitis virus glycoprotein G (VSV-G)2, 19 due to its broad viral tropism. However, in resting CD4 T cells, only CXCR4-tropic HIV envelope-mediated entry, but not VSV-G-mediated endocytosis, supports gene delivery by lentiviral vectors20, 21. This limitation is attributed to a lack of viral entry via the endocytosis pathway, which is active in transformed cells, antigen-presenting cells, and activated CD4 T cells, but not in quiescent primary CD4 T cells20.
RESULTS AND DISCUSSION
To relieve the resistance of primary CD4 T cells to HIV LV infection, we used the accessory lentiviral gene product protein X (Vpx), which induces proteasomal degradation of SAMHD1 in non-permissive human myeloid22 and resting CD4 T cells23, and alleviates restriction to HIV-1 in these cells. Vpx is encoded by HIV-2 and its related simian immunodeficiency virus (SIV) strains, but not by HIV-1. Vpx molecules were incorporated into viral-like particles encoding the core packaging functions of SIVmac251 (Vpx-VLPs), as previously described24. These Vpx-VLP particles are non-infectious as they do not contain any viral genetic material, but they are used to transiently deliver Vpx into target cells where they promote degradation of SAMHD1, thereby rendering the cells permissive to HIV LV infection22. In contrast to the commonly used VSV-G glycoprotein, we pseudotyped the Vpx-VLPs with the CXCR4-tropic Env of HIV-1, which supports efficient fusion of viral particles to quiescent CD4 T lymphocytes20. For the purpose of genetic modification, we used a third generation shRNA-encoding HIV LV vector pSico (plasmid for Stable RNA interference, conditional)25, bearing a EF1α:mCherry transgene expression cassette. The HIV LV particles were also pseudotyped with a CXCR4-tropic Env of HIV-1. To achieve productive infection of HIV LV particles, target cells were initially challenged with Vpx-VLPs, followed by a second infection with the HIV LV of interest after 24 hours. This sequential infection strategy allowed Vpx to establish an optimal permissive state within the target cells at the time when the HIV LV infection was performed. To facilitate a synchronized delivery of Vpx and fusion of HIV LV particles, cells and particles were subjected to high-speed spinoculation at each step (Fig. 1a).
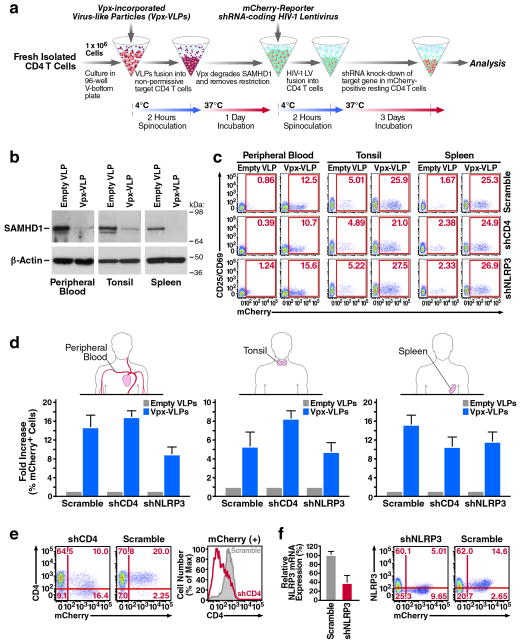
Figure 1.
Genetic manipulation of resting CD4 T cells by lentiviral vectors using sequential spinoculation of Vpx-VLP and shRNA-encoding HIV LV particles. (a) The spinoculation procedure. Fresh target cells were spinoculated with Vpx-VLPs, followed by shRNA-encoding HIV LV particles. Particles were pseudotyped with the CXCR4-tropic Env of HIV-1, which supports efficient fusion of virions to resting CD4 T cells. A detailed description is available in the Methods section (b) Spinoculation of Vpx-VLPs degrades SAMHD1 in resting CD4 T cells isolated from peripheral blood, tonsil, or spleen after 24 hours. (c) Efficient infection of resting CD4 T cells by three independent HIV LV particles indicated by mCherry transgene expression. Cells expressing mCherry remained viable (Supplementary Fig. 2) and negative for CD25/CD69 activation markers as indicated. (d) Fold increase of HIV LV infection after spinoculation with Vpx-VLPs as observed in CD4 T cells from peripheral blood, tonsil and spleen. (e) A marked decrease of surface CD4 expression in blood CD4 T cells productively infected with shCD4 HIV LV particles. (f) Reduced intracellular NLRP3 mRNA and protein expression in mCherry-positive CD4 T cells after infection with shNLRP3 HIV LV particles. Note that CD4 and NLRP3 expression were not reduced in mCherry-negative CD4 T cell populations. These data are representative of four independent experiments performed using CD4 T cells from peripheral blood, tonsil and spleen from four different donors.
Using these experimental conditions, CD4 T cells were positively isolated from fresh human peripheral blood, tonsillar, or splenic tissues, and spinoculated with Vpx-VLPs or empty VLPs, followed by infection with HIV LV particles encoding shRNA designed to silence the expression of CD4 (shCD4) or NLRP3 (shNLRP3), versus a non-silencing control scrambled sequence (Scramble). Interestingly, spinoculation of Vpx-VLPs efficiently abrogated SAMHD1 expression after 48 hours in target CD4 T cells for all of the tissues analyzed. It was at this time that HIV LV infection was in progress (Fig. 1b). Of note, at longer times (96 and 120 hours), endogenous SAMHD1 expression returned to normal levels, likely reflecting protein re-synthesis when cellular Vpx levels decline (Supplementary Fig. 1). After 4 days, the abundance of successful HIV LV infections was measured by flow cytometry according to the number of CD4 T cells expressing the mCherry transgene. Remarkably, spinoculation with Vpx-VLPs dramatically increased the number of mCherry-expressing CD4 T cells in all tested tissues (Fig. 1c). Such an increase was not observed in cultures spinoculated with empty VLPs, which remained highly resistant to HIV LV infection. The background levels of mCherry expression in these cultures represent a small fraction of naturally “primed” CD4 T cells that are permissive for HIV-1 infection10, 14, 26. Importantly, spinoculation with Vpx-VLPs did not lead to activation of resting CD4 T cells or initiation of programmed cell death, as assessed by surface expression of the CD69 and CD25 activation markers and Annexin V respectively (Fig. 1c, Supplementary Fig. 2). As previously reported27, 28, exogenous supplement of deoxynucleosides enhanced shRNA lentiviral delivery to the target cells, but not as effectively as spinoculation with Vpx-VLPs (Supplementary Fig. 3). Interestingly, the highest fold increase of HIV LV infection was observed in peripheral blood and spleen, corresponding to tissues where CD4 T cells exhibit particularly high resistance to lentiviral infection (Fig. 1d). Together, these results indicate that the loss of restriction to HIV LV infection produced by Vpx-VLPs is not associated with non-specific cellular activation, cellular stress promoting apoptosis, or matched by the addition of deoxynucleosides.
We next examined the efficiency of gene silencing by HIV LV particles encoding shRNAs. For these experiments, peripheral blood CD4 T cells were spinoculated with Vpx-VLPs and infected with either shCD4 or Scramble HIV LV particles. After 4 days, flow cytometry analysis revealed a marked decrease of surface CD4 expression among mCherry-positive CD4 T cells infected with shCD4 HIV LV particles (Fig. 1e). No decrease in CD4 expression occurred in either the mCherry-negative cells, or cells infected with Scramble HIV LV particles. These results support the conclusion that the decrease in CD4 expression reflected shCD4-mediated silencing in the mCherry-positive cells. We next assessed gene silencing by shNLRP3 HIV LV particles introduced into tonsillar CD4 T cells. As with peripheral blood, pronounced decreases in intracellular NLRP3 mRNA levels and protein expression were observed in the mCherry-positive subset of CD4 T cells following infection with shNLRP3 HIV LV particles (Fig. 1f). These data demonstrate the efficient and specific genetic silencing of resting CD4 T cells by shRNA-coding HIV LV particles. Importantly, pseudotyping with CXCR4-tropic HIV-1 Env allowed resting CD4 T cells to be selectively targeted for genetic silencing in fresh HLACs containing heterogeneous populations of lymphocytes, avoiding the need for prior CD4 T-cell purification (Supplementary Fig. 4).
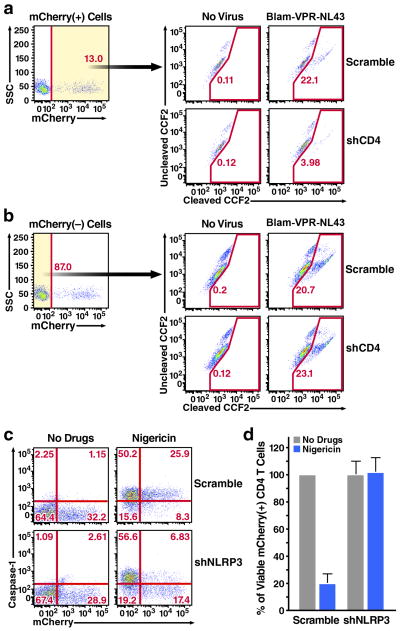
We next assessed whether the genetic manipulations produced by HIV LV particles were sufficient to affect the biological responses occurring within these target cells. We first assessed whether infection of shCD4 HIV LV particles inhibits HIV-1 fusion into resting blood CD4 T cells. Remarkably, infections with shCD4, but not Scramble HIV LV particles, significantly reduced the level of HIV-1 fusion into mCherry-positive CD4 T cells (Fig. 2a). In contrast, HIV-1 fusion was not inhibited in the mCherry-negative CD4 T cells within the same cultures (Fig. 2b), indicating that the observed inhibition was produced by the shCD4-encoding HIV LV. We next infected tonsillar CD4 T cells with shNLRP3 HIV LV particles and assessed their response to nigericin, which specifically engages NLRP3 inflammasome promoting caspase-1 activation and cell death29. Treatment with nigericin induced pronounced activation of caspase-1 activation in cultures infected with scramble HIV LV, including mCherry-positive and negative CD4 T-cells. Strikingly, nigericin treatment failed to induce caspase-1 activation in mCherry-positive CD4 T cells receiving the shNLRP3 HIV LV particles (Fig. 2c). Further, nigericin no longer induced cell death in these mCherry-positive CD4 T cells (Fig. 2d, Supplementary Fig. 5).
Figure 2.
Modulating specific CD4 T cells responses by HIV LV (a, b) Infection of shCD4 HIV LV impedes HIV-1 fusion to resting CD4 T cells isolated from blood. To test the ability of HIV-1 to fuse with infected cells, we used an HIV virion-based fusion assay that measures β-lactamase (BlaM) activity delivered to target cells upon the fusion of X4-tropic NL4-3 strain of HIV-1 containing BlaM fused to the Vpr protein (BlaM-Vpr-NL4-3)37. Inhibition of fusion was specifically observed in the mCherry-positive (a), but not in the mCherry-negative cells (b) residing in the same cultures. (c) Infection of shNLRP3 HIV LV blocks caspase-1 activation in tonsillar CD4 T cells treated with nigericin. (d) Infections with shNLRP3 HIV LV prevent caspase-1-mediated death of mCherry-positive CD4 T cells treated with nigericin. Inhibition of cell death was not inhibited by mCherry-positive Scramble HIV LV.
These findings demonstrate a new and efficient lentivirus-based approach for manipulating gene expression in resting CD4 T cells. The use of CXCR4-tropic HIV-1 Env instead of VSV-G glycoprotein allows both selective and efficient fusion of lentiviral particles into quiescent CD4 T cells without prior mitogen activation or cytokine stimulation. Activation-rest strategies have been previously used to introduce HIV LV into CD4 T cells, although this approach may alter the natural physiology of these cells, skewing them toward a memory phenotype and increasing their intrinsic state of activation. The sequential spinoculation technique that we employed allows Vpx to establish an optimal permissive state in the target cells during the early steps of HIV LV infection. Importantly, this permissive state is reversible, as SAMHD1 expression returns to normal levels in the target, which likely re-acquire their natural non-permissive state. Such a precise and synchronous response may be difficult to achieve without spinoculation (Supplementary Fig. 6)23, 30. Importantly, genetic manipulations using HIV LV has been shown with cells of myeloid origin30, but not in CD4 T cells from blood and lymphoid tissues, the principal targets of HIV-1. This technology could therefore provide a long-sought experimental tool to explore key questions in the HIV field involving non-permissive CD4 T cells, including investigating the mechanisms underlying the establishment and maintenance of latent HIV reservoirs, and identifying the innate sensors that detect abortive HIV DNA reverse transcripts in lymphoid CD4 T cells and elicit their cell death10.
MATERIALS AND METHODS
Expression plasmids
SIVmac 251 virus-like particles for Vpx delivery (Vpx-VLPs) were produced using the pSIV3+ plasmid, kindly provided by Dr. A. Cimarelli31. The pSIV3+ expression plasmid was constructed by replacing the 5′ LTR of SIVmac251 with the CMV promoter and enhancer region. The 5′ half of the env gene was also removed, leaving the REV-responsive element sequence and the 5′ and 3′ exons of the tat and rev regulatory genes intact. The 3′ LTR was replaced by a SV40 polyadenylation sequence, resulting in deletion of the 3′ end of the nef gene. Finally, the nef initiation codon was inactivated to prevent translation32. The detailed map of this construct is available at: http://www.retrovirology.com/content/5/1/50/figure/F1?highres=y. To produce empty VLPs, the vpx gene in the pSIV3+ plasmid was inactivated by converting its initiation codon to a stop codon to give pSIV3+ΔVpx, kindly provided by Dr. Jacek Skowronski33. Different shRNAs were cloned into modified versions of the pSicoR lentiviral vector, which encodes a mCherry reporter driven by an EF-1α promoter (pSicoR-mCherry).
Production of Vpx-VLPs and shRNA-coding HIV LV particles
Virus-like particles (VLPs) and HIV-1 lentiviral particles were produced from 293T cells using the standard phosphate calcium transfection protocol34. The medium was replaced after 16 hours. After 48 hours, the supernatants were collected and clarified by sedimentation, and virions were concentrated by ultracentrifugation, and stored at −80°C in 100% fetal bovine serum. For VLPs, 293T cells were transfected with 8 μg pSIV3+ and 2 μg CXCR4-tropic Env (gp160) encoding plasmid (Vpx-VLPs) or with 8 μg PSIV3+ΔVpx and 2 μg gp160 (Empty VLPs). The amount of VLPs was determined by measuring SIV p27gag levels by ELISA assay. shRNA-coding HIV-1 vectors were cloned using a modified version of the pSicoR (plasmid for Stable RNA interference, conditional) lentiviral vector25, which encodes an mCherry reporter driven by an EF-1α promoter (pSicoR-MS1)35. For cloning of shCD4-coding HIV LV the following oligos were used:
Sense: TGATCAAGAGACTCCTCAGTTTCAAGAGAACTGAGGAGTCTCTTGATCTTTTTTC;
Antisense: TCGAGAAAAAAGATCAAGAGACTCCTCAGTTCTCTTGAAACTGAGGAGTCTCTTGATCA.
For cloning of shNLRP3-coding HIV LV the following oligos were used:
Sense: TGAAATGGATTGAAGTGAAATTCAAGAGATTTCACTTCAATCCATTTCTTTTTTC;
Antisense: TCGAGAAAAAAGAAATGGATTGAAGTGAAATCTCTTGAATTTCACTTCAATCCATTTCA.
To generate the shRNA HIV LV particles, 293T cells were co-transfected with 10μg pSicoR-mCherry shRNA constructs, 9μg HIV-based packaging construct NL4-3 8.9136, and 2μg gp160 encoding plasmids. The lentiviral particle stocks were quantitated by measuring p24gag levels by ELISA assay (1 ng p24gag equals approximately 2 × 106 viral particles).
Culture of primary cells
Peripheral blood mononuclear cells (PBMCs) from healthy blood donors (Blood Centers of the Pacific, San Francisco, CA) were purified by Ficoll-Hypaque density gradients, followed by negative selection with CD14+ microbeads and positive selection with CD4+ microbeads (Miltenyi, Bergisch Gladbach, Germany) or negative selection with EasySep Human CD4+ T Cell Enrichment Kit (Stemcell) (Supplementary Fig. 7). The purified resting CD4+ T cells were cultured in RPMI 1640 medium supplemented with 10% FBS, L-glutamine (2 mM), penicillin (50 U/ml), and streptomycin (50 μg/ml). Human tonsil or spleen tissue was obtained from the National Disease Research Interchange and the Cooperative Human Tissue Network. Human lymphoid aggregate cultures (HLACs) were prepared using tonsillar or splenic tissue as previously described10. CD4 T cells were isolated from HLACs by positive selection using CD4 microbeads (Miltenyi) and cultured overnight in 96-well U-bottomed polystyrene plates (1 X 106 cells/well) in medium (200 μl/well) consisting of RPMI 1640 supplemented with 15% heat-inactivated fetal bovine serum, 100 mg/ml gentamicin, 200 mg/ml ampicillin, 1 mM sodium pyruvate, 1% nonessential amino acids (Mediatech), 2 mM L-glutamine, and 1% fungizone (Invitrogen).
Two-step spinoculation procedure
Fresh CD4 T cells, human aggregate lymphoid cultures (HLAC), or peripheral blood lymphocytes (PBLs), were cultured in a V-bottom 96-well plate (1×106 cells/well) and chilled on ice for 15 min. Vpx-VLPs pseudotyped with the CXCR4-tropic Env of HIV-1 were then added (100 ng p24gag/100μl) to each well and mixed with cold cells for an additional 15 minutes. The cold temperature allows particle-cell attachment but prevents fusion. Particle-coated cells are then tightly packed into a pellet by high-speed centrifugation (1200g) for 2 hours at 4°C. This step brings the viral particles in close apposition to the cell membranes, promoting a uniform and high level of attachment. Immediately after centrifugation cells are cultured at 37°C as a pellet. This step facilitates a coordinated fusion of the attached viruses, generating a pulse of virion entry into the target cells. After 24 hours at 37°C, Vpx-containing target cells are spinoculated again with shRNA-coding HIV LV particles coted with CXCR4-tropic Env of HIV-1 as described above. After 3 days of incubation, the cultures are analyzed. Of note, the enhancement by Vpx-VLPs using regular infection was not as effective as spinoculation (Supplementary Fig. 6). The background levels of mCherry expression in these cultures without Vpx-VLPs enhancement represent a small fraction of naturally “primed” CD4 T cells that are permissive for HIV LV infection, which can be blocked by reverse transcriptase inhibitor (NNRTI) efavirenz (Supplementary Fig. 8).
SAMHD1 protein analysis
CD4 T cells were lysed after 48 hrs VLPs treatment in RIPA buffer (150mM NaCl, 1% Nonidet P-40 (vol/vol), 0.5% AB-deoxycholate (vol/vol), 0.1% SDS (vol/vol), 50mM Tris-HCl (pH 8), 1 mM DTT), and EDTA-free Protease Inhibitor (Roche Applied Science) and preceded for SDS-PAGE immunoblotting analysis. The primary antibodies used were 1/1000 dilution of rabbit polyclonal anti-SAMHD1 (Sigma, Cat # SAB2102077),
FACS analysis and gating strategy
CD4 T cells were washed in FACS buffer (PBS supplemented with 2 mM EDTA and 2% fetal bovine serum), stained with APC-conjugated anti-CD4 or anti-CD25/CD69 (all from BD Pharmingen) and fixed in 1% paraformaldehyde. Data were collected on a FACS Calibur (BD Biosciences) and analyzed with Flowjo software (Treestar). The percentages of mCherry(+) CD4 T cells were defined based on uninfected control samples. In some experiments, mCherry(+) CD4 T cells were purified by flow cytometry on a FACSAria II cell sorter (BD Biosciences). After cell sorting, >99% cells were mCherry positive.
Quantitative real-time RT-PCR
Total RNA was purified from presorted mCherry(+) CD4 T cells with an RNeasy kit (Qiagen). Purified RNA was reverse-transcribed with Oligo(dT)20 and the SuperScript III first-stand synthesis system (Invitrogen). Real-time PCR was performed with a QuantiTect Probe PCR kit (Qiagen) on a 7900HT fast real-time PCR system (Applied Biosystems). Thermal cycling consisted of 15 min denaturation at 95 °C, followed by 50 cycles of 15 s at 95 °C and 60 s at 60 °C. The data were normalized relative to GAPDH as an endogenous control. PCR assays were conducted with predeveloped TaqMan assay primers and probes (NLRP3: Hs00918082_m1; GAPDH: Hs03929097_g1; Life Technology).
Intracellular NLRP3 staining
For fixation and antibody staining, tonsil CD4 T cells were seeded in a 96-well V-bottom plate at 1 ×106 cells/100 μl/well. Cells were fixed for 60 min at room temperature with 2% paraformaldehyde and permeabilized by washing 2 times in 1× BD Perm/Wash buffer (BD Biosciences, 554723). Cells were stained with DyLight 488-conjugated anti-NLRP3 antibody (1:20, Abcam, #ab139884) in 1× BD Perm/Wash buffer at 4°C for 30 minutes in the dark. The staining was then monitored by flow cytometry.
Nigericin treatment, cell viability measurement and intracellular caspase-1 activation
For stimulating the pyroptosis death pathway, tonsil CD4 T cells were treated with 5μM nigericin (Sigma, Cat. # N7143) for 12 h at 37°C. The potassium ionophore nigericin mediates an electroneutral exchange of intracellular K+ ions for extracellular protons, providing a second inflammatory stimulus, which results in the NLRP3-mediated activation of caspase-1. After 12hrs the cells were collected and assessed for cell viability and levels of caspase-1 activity. To determine the cell viability, a standard number of fluorescent beads (Flow-Count Fluorospheres, Beckman Coulter) were added to each cell-suspension sample before data acquisition. The cell-suspension samples were analyzed using flow cytometry. The relative cell viability is normalized based on the number of fluorescent beads acquired. To determine intracellular activation of caspase-1, Fluorescent Labeled Inhibitors of Caspases (FLICA) probe assays (ImmunoChemistry Technologies #9122) were performed. FLICA probes were added directly to the cell culture media, incubated for 15 min at 37°C, and washed five times with PBS supplemented with 2 mM EDTA and 2% fetal bovine serum. FLICA probes are cell-permeable and covalently bind to the active caspase-1. After washing, FLICA fluorescent signal is specifically retained within cells containing active caspase-1 while the reagent is washed away in cells lacking active caspase-1.
HIV virion-based fusion assay
The HIV virion-based fusion assay was performed as previously described37. Briefly, resting CD4+ T cells (1 × 106) were incubated with BlaM-Vpr containing virions (100 ng of p24gag) at 37°C for 2 hr, washed in CO2-independent medium (Gibco BRL), and loaded with CCF2/AM dye as described by the manufacturer (Invitrogen). 2 ml of CCF2/AM (1 mM) was mixed with 8 ml of 0.1% acetic acid containing 100 mg/ml Pluronic-F127R and 1ml of RPMI to constitute the loading solution. Cells were incubated in 100 ml of loading solution for 1 hr at room temperature. After two washes with RPMI, the BlaM reaction was conducted for 16 h at room temperature in 200 ml of CO2 independent media supplemented with 10% FBS and 2.5 mM probenecid, a nonspecific inhibitor of anion transport (Sigma Pharmaceuticals). Finally, the cells were washed once in RPMI and fixed in a 2% solution of paraformaldehyde. The change in emission fluorescence of CCF2 after cleavage by the BlaM-Vpr chimera was monitored by flow cytometry with LSR2 (Becton Dickinson, San Jose, CA). Data were collected with DiVa software and analyzed with FlowJo software (Treestar, San Carlos, CA).
Supplementary Material
Acknowledgments
Funding for this project was provided by: the UCSF/Robert John Sabo Trust Award (Doitsh); NIH/NIAID R21 AI102782 (Greene), and Martin Delaney CARE Collaboratory NIH/NIAID U19 AI096113 (Greene). We thank the UCSF-GIVI CFAR for providing core services that facilitated these studies (CFAR P30 AI027763).
We thank Dr. Matthew Spindler in the laboratory of Dr. Bruce Conklin for the generous gift of pSico-mCherry HIV LV vector. We thank Dr. Marielle Cavrois, Marianne Gesner, and Jaime Tawney for assistance with flow cytometry. We also thank Gary Howard and Anna Lisa Lucido for editorial assistance, John C.W. Carroll, Giovanni Maki, and Teresa Roberts for graphics arts, and Robin Givens and Sue Cammack for administrative assistance. Special thanks to Jason Neidleman for stimulating discussions and technical advice.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on Gene Therapy website (http://www.nature.com/gt)
References
- 1.Bikker A, Hack CE, Lafeber FP, van Roon JA. Interleukin-7: a key mediator in T cell-driven autoimmunity, inflammation, and tissue destruction. Current pharmaceutical design. 2012;18(16):2347–56. doi: 10.2174/138161212800165979. [DOI] [PubMed] [Google Scholar]
- 2.Berger G, Durand S, Goujon C, Nguyen XN, Cordeil S, Darlix JL, et al. A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat Protoc. 2011;6(6):806–16. doi: 10.1038/nprot.2011.327. [DOI] [PubMed] [Google Scholar]
- 3.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–98. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glushakova S, Baibakov B, Margolis LB, Zimmerberg J. Infection of human tonsil histocultures: a model for HIV pathogenesis. Nat Med. 1995;1(12):1320–2. doi: 10.1038/nm1295-1320. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki H, Zelphati O, Hildebrand G, Leserman L. CD4 and CD7 molecules as targets for drug delivery from antibody bearing liposomes. Exp Cell Res. 1991;193(1):112–9. doi: 10.1016/0014-4827(91)90544-5. [DOI] [PubMed] [Google Scholar]
- 6.Tan JT, Ernst B, Kieper WC, LeRoy E, Sprent J, Surh CD. Interleukin (IL)-15 and IL-7 jointly regulate homeostatic proliferation of memory phenotype CD8+ cells but are not required for memory phenotype CD4+ cells. The Journal of experimental medicine. 2002;195(12):1523–32. doi: 10.1084/jem.20020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan E, Varro R, Sepulveda H, Ember JA, Apgar J, Wilson J, et al. Cytometric bead array: a multiplexed assay platform with applications in various areas of biology. Clinical immunology (Orlando, Fla) 2004;110(3):252–66. doi: 10.1016/j.clim.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 8.Geginat J, Sallusto F, Lanzavecchia A. Cytokine-driven proliferation and differentiation of human naive, central memory and effector memory CD4+ T cells. Pathologie-biologie. 2003;51(2):64–6. doi: 10.1016/s0369-8114(03)00098-1. [DOI] [PubMed] [Google Scholar]
- 9.Budagian V, Bulanova E, Orinska Z, Pohl T, Borden EC, Silverman R, et al. Reverse signaling through membrane-bound interleukin-15. The Journal of biological chemistry. 2004;279(40):42192–201. doi: 10.1074/jbc.M403182200. [DOI] [PubMed] [Google Scholar]
- 10.Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, Santiago ML, et al. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell. 2010;143(5):789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61(2):213–22. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y, Zhang H, Siliciano JD, Siliciano RF. Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J Virol. 2005;79(4):2199–210. doi: 10.1128/JVI.79.4.2199-2210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swiggard WJ, O’Doherty U, McGain D, Jeyakumar D, Malim MH. Long HIV type 1 reverse transcripts can accumulate stably within resting CD4+ T cells while short ones are degraded. AIDS Res Hum Retroviruses. 2004;20(3):285–95. doi: 10.1089/088922204322996527. [DOI] [PubMed] [Google Scholar]
- 14.Swiggard WJ, Baytop C, Yu JJ, Dai J, Li C, Schretzenmair R, et al. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. Journal of virology. 2005;79(22):14179–88. doi: 10.1128/JVI.79.22.14179-14188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai J, Agosto LM, Baytop C, Yu JJ, Pace MJ, Liszewski MK, et al. Human immunodeficiency virus integrates directly into naive resting CD4+ T cells but enters naive cells less efficiently than memory cells. Journal of virology. 2009;83(9):4528–37. doi: 10.1128/JVI.01910-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pace MJ, Graf EH, Agosto LM, Mexas AM, Male F, Brady T, et al. Directly infected resting CD4+T cells can produce HIV Gag without spreading infection in a model of HIV latency. PLoS Pathog. 2012;8(7):e1002818. doi: 10.1371/journal.ppat.1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Marsh JW. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science. 2001;293(5534):1503–6. doi: 10.1126/science.1061548. [DOI] [PubMed] [Google Scholar]
- 18.Spina CA, Guatelli JC, Richman DD. Establishment of a stable, inducible form of human immunodeficiency virus type 1 DNA in quiescent CD4 lymphocytes in vitro. Journal of virology. 1995;69(5):2977–88. doi: 10.1128/jvi.69.5.2977-2988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musso T, Calosso L, Zucca M, Millesimo M, Ravarino D, Giovarelli M, et al. Human monocytes constitutively express membrane-bound, biologically active, and interferon-gamma-upregulated interleukin-15. Blood. 1999;93(10):3531–9. [PubMed] [Google Scholar]
- 20.Agosto LM, Yu JJ, Liszewski MK, Baytop C, Korokhov N, Humeau LM, et al. The CXCR4-tropic human immunodeficiency virus envelope promotes more-efficient gene delivery to resting CD4+ T cells than the vesicular stomatitis virus glycoprotein G envelope. Journal of virology. 2009;83(16):8153–62. doi: 10.1128/JVI.00220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu D, Wang W, Yoder A, Spear M, Wu Y. The HIV envelope but not VSV glycoprotein is capable of mediating HIV latent infection of resting CD4 T cells. PLoS Pathog. 2009;5(10):e1000633. doi: 10.1371/journal.ppat.1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–7. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18(11):1682–9. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Negre D, Mangeot PE, Duisit G, Blanchard S, Vidalain PO, Leissner P, et al. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 2000;7(19):1613–23. doi: 10.1038/sj.gt.3301292. [DOI] [PubMed] [Google Scholar]
- 25.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101(28):10380–5. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreisberg JF, Yonemoto W, Greene WC. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J Exp Med. 2006;203(4):865–70. doi: 10.1084/jem.20051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plesa G, Dai J, Baytop C, Riley JL, June CH, O’Doherty U. Addition of deoxynucleosides enhances human immunodeficiency virus type 1 integration and 2LTR formation in resting CD4+ T cells. Journal of virology. 2007;81(24):13938–42. doi: 10.1128/JVI.01745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korin YD, Zack JA. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. Journal of virology. 1998;72(4):3161–8. doi: 10.1128/jvi.72.4.3161-3168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440(7081):228–32. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 30.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29(6):848–62. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Goujon C, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC) Gene Ther. 2006;13(12):991–4. doi: 10.1038/sj.gt.3302753. [DOI] [PubMed] [Google Scholar]
- 32.Derdouch S, Gay W, Negre D, Prost S, Le Dantec M, Delache B, et al. Reconstitution of the myeloid and lymphoid compartments after the transplantation of autologous and genetically modified CD34+ bone marrow cells, following gamma irradiation in cynomolgus macaques. Retrovirology. 2008;5:50. doi: 10.1186/1742-4690-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–61. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 35.Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. The Journal of biological chemistry. 2011;286(42):36427–37. doi: 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grivel JC, Elliott J, Lisco A, Biancotto A, Condack C, Shattock RJ, et al. HIV-1 pathogenesis differs in rectosigmoid and tonsillar tissues infected ex vivo with CCR5- and CXCR4-tropic HIV-1. AIDS (London, England) 2007;21(10):1263–72. doi: 10.1097/QAD.0b013e3281864667. [DOI] [PubMed] [Google Scholar]
- 37.Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol. 2002;20(11):1151–4. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.