Abstract
NtcB of the cyanobacterium Synechococcus elongatus strain PCC 7942 is a LysR family protein that enhances expression of the nitrate assimilation operon (nirA operon) in response to the presence of nitrite, an intermediate of assimilatory nitrate reduction. Inactivation of ntcB in this cyanobacterium specifically abolishes the nitrite responsiveness of nirA operon expression, but under nitrate-replete conditions (wherein negative feedback by intracellularly generated ammonium prevails over the positive effect of nitrite) activity levels of the nitrate assimilation enzymes are marginally higher in the wild-type cells than in the mutant cells, raising the issue of whether the nitrite-promoted regulation has physiological importance. On the other hand, the strains carrying ntcB expressed much higher nitrate assimilation enzyme activities under nitrate-limited growth conditions than under nitrate-replete conditions whereas the ntcB-deficient strains showed levels of the enzyme activities lower than those seen under the nitrate-replete conditions. Although the ntcB mutant maintained a constant cell population in a nitrate-limited chemostat when grown as a single culture, it was diluted at a rate expected for nondividing cells when mixed with the wild-type cells and subjected to nitrate limitation in the chemostat culture system. These results demonstrated that the nitrite-promoted activation of the nitrate assimilation operon is essential for up-regulation of the nitrate assimilation activities under the conditions of nitrate limitation and for competitive utilization of nitrate.
In cyanobacteria, expression of the genes involved in nitrate assimilation, i.e., the nrt genes for the nitrate/nitrite transporter (NRT), narB for nitrate reductase (NR), and nirA for nitrite reductase (NiR), is repressed by the presence of ammonium (3, 4, 15, 17, 20, 21). Ammonium must be fixed via the glutamine synthetase-glutamate synthase cycle to exert its negative effects on transcription (3, 8, 21). Derepression of transcription by removal of ammonium from medium (or by inhibition of ammonium assimilation) results in induction of the nitrate assimilation genes, showing no requirements for nitrate or nitrite (3, 4, 8, 21). This is a part of the global nitrogen control in cyanobacteria, which involves NtcA, a Crp family protein, as the transcriptional activator (7). Tanigawa et al. recently showed by in vitro experiments that 2-oxoglutarate (2-OG), which serves as the acceptor of the newly fixed nitrogen in the glutamine synthetase-glutamate synthase cycle, activates transcription from NtcA-dependent promoters in a concentration-dependent manner (22). It has also been shown that the intracellular 2-OG concentration is low in the presence of ammonium and is increased by nitrogen deprivation (16). Taken together, these findings suggest that 2-OG acts as the coinducer of transcription of the NtcA-dependent genes, conferring ammonium sensitivity to their expression in vivo.
Besides the NtcA-dependent induction involving 2-OG as the coinducer, expression of the nitrate assimilation genes is subjected to up-regulation by nitrite, the intermediate of the nitrate assimilation pathway (2, 8, 12). Studies using Synechococcus elongatus strain PCC 7942 showed that the nitrite-promoted regulation is specific to the nitrate assimilation operon (nirA-nrtABCD-narB [designated nirA operon]) and is mediated by NtcB, a LysR family protein (1). NtcB requires the activity of NtcA to exert its positive effect on transcription (12), indicating that it is unable to promote transcription by itself. Since the activity of NtcA in transcriptional activation is down-regulated by assimilation of the ammonium generated intracellularly by nitrate reduction (21), the positive effect of nitrite is marginal in cells supplied with sufficient amounts of nitrate (8); the effect of nitrite is hence prominent in cells treated with either l-methionine-dl-sulfoximine (MSX [an inhibitor of glutamine synthetase]) or 6-diazo-5-oxo-l-norleucine (DON [an inhibitor of glutamate synthase]) to prevent ammonium assimilation (2, 8, 12). In accordance with these observations, an ntcB-deficient S. elongatus mutant (NIC1) which is defective specifically in the nitrite responsiveness of nirA operon transcription (1), showed only a small decrease in the activity levels of the nitrate assimilation enzymes during steady-state growth in nitrate-sufficient medium (19). These results raise the issue of whether or not the nitrite stimulation of the nitrate assimilation genes is of physiological importance. In the present study, we used NRT-less mutants and chemostat cultures of the cyanobacterium to investigate the physiological role of the nitrite responsiveness under nitrate-limited growth conditions. By examining the effects of inactivation of the trans-acting factor (NtcB) and modification of the cis-acting element, it was shown that the nitrite-responsiveness of nirA operon transcription is essential for high-level expression of the nitrate assimilation enzymes during growth with limiting supply of nitrate and for competitive utilization of nitrate under the nitrate-limited conditions.
MATERIALS AND METHODS
Strains and general growth conditions.
A derivative of S. elongatus strain PCC 7942, which is cured of the resident small plasmid pUH24 (R2-SPc [9]; hereafter designated simply as strain PCC 7942), is the parental strain of all of the cyanobacterial strains used in this study. NIC1 is an ntcB-deficient mutant strain (ΔntcB::kan) previously described (1). NA3, an NRT-less mutant (ΔnrtABCD) constructed by deleting the nrt genes from the nirA operon (14), was the genetic background of the PnirA::luxAB reporter strains YKA1, YKA2, and YKA5 (12) (Fig. 1). NA4 (ΔnrtABCD ΔntcB::kan) was obtained from NA3 by inactivating ntcB as described for NIC1. YKA2b was a derivative of NA4 obtained by transferring the same promoter-reporter fusion as that in YKA2 to the chromosome of NA4 (Fig. 1). YKA1a was obtained by transferring the same promoter-reporter fusion as for YKA1 to the chromosome of the wild-type strain (Fig. 1). Unless otherwise stated, the cyanobacterial strains were grown photoautotrophically at 30°C under CO2-sufficient conditions in batch cultures as described previously (20). The basal medium used was a nitrogen-free medium obtained by modification of BG11 medium (18) as previously described (20). Ammonium-containing medium was prepared by addition of 3.75 mM (NH4)2SO4 to the basal medium. Nitrate-containing media were prepared by addition of 1, 15, or 60 mM of KNO3 to the basal medium. All media were buffered with 20 mM HEPES-KOH (pH 8.2). Solid media were prepared by supplementing 1.5% Bactoagar (Difco) to liquid media. When appropriate, kanamycin and spectinomycin were added to the media at 10 and 1 μg ml−1, respectively.
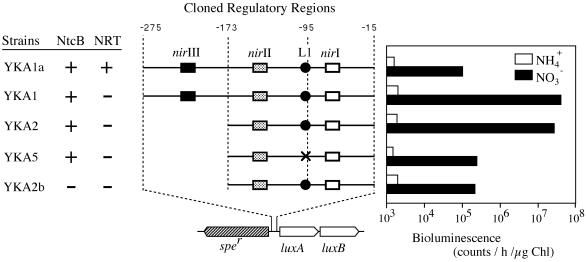
FIG. 1.
Effects of the presence of NRT, modification of the cis-acting element required for the response to nitrite, and inactivation of ntcB on expression of PnirA::luxAB transcriptional fusions in ammonium- and nitrate (60 mM)-containing media. Fragments of the nirA-nirB regulatory region having the indicated endpoints relative to the translational start site of nirA were fused individually to the luxAB gene in plasmid pYK5 (12), and the resulting PnirA::luxAB fusions were transferred to the chromosome of the wild-type strain (YKA1a), the NA3 mutant lacking NRT (YKA1, YKA2, and YKA5), and the NA4 mutant lacking NRT and NtcB (YKA2b). The three NtcA-binding sites nirI, nirII, and nirIII are indicated by open, dotted, and closed boxes, respectively. The cis-acting sequence required for the nitrite responsiveness of PnirA (L1) is indicated by filled circles. X indicates site-specific nucleotide changes in the L1 site. The bioluminescence data are the means of five measurements from cells incubated in ammonium-containing medium (open bars) and nitrate (60 mM)-containing medium (filled bars).
Expression of the nirA operon and the reporter gene fusions was induced by transfer of the ammonium-grown cells to nitrate-containing medium; the ammonium-grown cells were collected by centrifugation at 5,000 × g for 5 min at 25°C, washed twice with the basal medium by resuspension and recentrifugation, and inoculated into the nitrate-containing medium.
Continuous cultivation of cyanobacterial cells.
Continuous cultivation of cyanobacterial cells was performed (using flat glass bottles having an internal volume of 2 liters as growth vessels) essentially as described by Lehmann and Wöber (10). The cyanobacterial cells used as the inoculum were grown in batch cultures with ammonium as the nitrogen source, harvested and washed as described above, and inoculated into 1 liter of fresh medium in a growth vessel. Fresh sterile medium was continuously pumped into the vessel at a flow rate of 30 ml h−1, and the effluent was collected in a sterile bottle. The culture was aerated with air supplemented with 2% (vol/vol) CO2 and illuminated with fluorescent lamps at 200 μmol of photons m−2 s−1 under general growth conditions that were otherwise the same as those described above. Cell density in the effluent was monitored by measuring the optical density at 730 nm (OD730). When cells of the wild-type strain and the ntcB-deficient mutant (ΔntcB::kan) were mixed and subjected to continuous cultivation, serial dilutions of the effluent were plated on solid ammonium-containing medium with and without kanamycin to determine the wild-type and the mutant cell population numbers; the total cell population was obtained by counting colonies on the medium containing no kanamycin, and the mutant cell population was obtained by counting colonies on the kanamycin-containing medium. The population of the wild-type cells was obtained by subtracting the mutant cell population from the total cell population.
Measurement of in vivo bioluminescence.
For measurement of in vivo luminescence from the Synechococcus cells carrying luxAB transcriptional fusions, 1 ml of cell culture containing 0.001 to 0.5 μg of chlorophyll (Chl) was transferred to a test tube and mixed with 20 μl of 0.1% n-decanal emulsion. Bioluminescence of the cell suspension was measured with a luminometer (ARGUS-50; Hamamatsu Photonics) immediately after the addition of n-decanal. Intensity of bioluminescence was expressed in counts of photons per hour per microgram of Chl.
Other methods.
NR and NiR activities were determined at 30°C using toluene-permeabilized cells with dithionite-reduced methylviologen as the electron donor (5, 6). The rate of nitrate uptake by the cells was determined at low external nitrate concentrations (<100 μM) by monitoring the decrease in nitrate concentration in medium as previously described (14) and was regarded as the activity of the nitrate transporter (NRT). Nitrate and nitrite levels were determined with a flow-injection analyzer (NOX-1000; Tokyo Chemical Industry Co., Ltd.). Chl levels were determined as described by Mackinney (11).
RESULTS
Effects of endogenously generated nitrite on expression of PnirA::luxAB fusions in NRT-less strains grown with nitrate.
As previously observed, the reporter strain YKA1 (carrying a transcriptional fusion of the nirA operon promoter and luxAB in an NRT-deficient background) showed high-level expression of luxAB when grown with nitrate (60 mM) (12). The level of luxAB expression in YKA1 was much higher than that in YKA1a, a reporter strain constructed by introduction of the same promoter-reporter fusion into the wild-type cells (Fig. 1), suggesting that nitrate limitation due to the lack of NRT led to the high-level expression from the nirA operon promoter in YKA1. While the YKA2 strain, which differs from YKA1 in lacking the nirIII site for binding of NtcA, expressed luxAB to a level comparable to that in YKA1 as previously shown (12), the YKA5 strain, differing from YKA2 in carrying base substitutions in the cis-acting element (L1) required for the nitrite enhancement of PnirA::luxAB expression (12), expressed luciferase activity to a level slightly higher than that in YKA1a (Fig. 1). By comparing the PnirA::luxAB expression levels in cells treated with DON (an inhibitor of ammonium assimilation) alone to the results seen with DON plus nitrite, Maeda et al. previously showed that modification of the L1 site specifically abolishes the nitrite responsiveness and does not affect the basal-level expression from the nirA operon promoter, as is induced by NtcA (12). The present results therefore suggested that the positive regulation mechanism involving nitrite plays an essential role in high-level expression of PnirA::luxAB under the nitrate-limited growth conditions.
Maeda et al. previously showed that ntcB inactivation in S. elongatus strain PCC 7942 results in specific loss of the nitrite responsiveness, as does the modification of L1 (12, 13). Since the nucleotide sequence of L1 conforms to the structure of the binding sites for LysR-type proteins, it is likely that L1 constitutes the binding site for NtcB (12). In accordance with this assumption, the level of luxAB expression in the YKA2b mutant, an ntcB-deficient derivative of YKA2, was similar to that in YKA5 under the nitrate-limited growth conditions (Fig. 1) as well as in the presence of DON (12).
Effects of ntcB mutation on activity levels of nitrate assimilation enzymes in NRT-less strains.
To examine the effects of the nitrite-responsive enhancement of nirA operon transcription on the activity levels of NR and NiR under nitrate-limited conditions, cells of the wild-type strain, the NRT-less mutant NA3, and the ntcB-deficient derivative of NA3 (NA4) were grown in nitrate (60 mM)-containing medium in batch cultures and their NR and NiR activities were compared (Table 1). While the cultures of the wild-type strain were blue-green, those of NA3 and NA4 looked yellowish green due to reduced level of phycocyanin, indicating that the mutants were under stress of nitrogen deficiency. As expected from the higher levels of PnirA::luxAB expression in the NRT-less YKA1 strain compared to the results seen with the YKA1a strain having active NRT (see above), the NA3 cells expressed higher NR and NiR activities than the wild-type cells. Presumably due to limited supply of the cofactors required for assembly of NR and NiR holoenzymes, however, the differences in the enzyme activities between NA3 and the wild-type strain were (two to three times) smaller than those observed between the luciferase activities of YKA1 and YKA1a (Fig. 1). The NR and NiR activities in the NA4 mutant were only 35 and 20% of the corresponding activities in NA3, respectively, and were even lower than those in the wild-type, nitrate-replete cells (Table 1). These results demonstrated the importance of the NtcB-dependent, nitrite-responsive enhancement of nirA operon transcription in the high-level expression of NR and NiR activities during growth with a limiting supply of nitrate.
TABLE 1.
Enzyme activities involved in nitrate assimilation in the wild-type and mutant strains grown under various nitrogen conditionsa
| Culture and strain | Genotypes | Nitrate concn | Enzyme activity
|
|||||
|---|---|---|---|---|---|---|---|---|
| μmol mg of Chl−1 h−1
|
μmol OD730−1 h−1
|
|||||||
| NRT | NR | NiR | NRT | NR | NiR | |||
| Batch cultures | ||||||||
| WTb | WT | 15 mM | 25 (3) | 185 (13) | 90 (3) | 0.15 (0.02) | 1.15 (0.09) | 0.55 (0.02) |
| WT | WT | 60 mM | —c | 196 (18) | 80 (5) | — | — | — |
| NA3 | ΔnrtABCD | 60 mM | — | 361 (63) | 243 (50) | — | — | — |
| NA4 | ΔnrtABCD ΔntcB | 60 mM | — | 129 (28) | 50 (12) | — | — | — |
| Continuous cultures | ||||||||
| WT | WT | <0.5 μM | 85 (2) | 855 (68) | 778 (31) | 0.21 (0.03) | 1.92 (0.14) | 1.75 (0.07) |
| NIC1 | ΔntcB | ∼2 μM | 51 (5) | 233 (74) | 166 (36) | 0.08 (0.01) | 0.42 (0.13) | 0.30 (0.07) |
The values are the averages from three measurements; those in parentheses are standard deviations.
WT, wild type.
—, not determined.
Growth and expression of nitrate assimilation enzymes in nitrate-limited chemostat cultures.
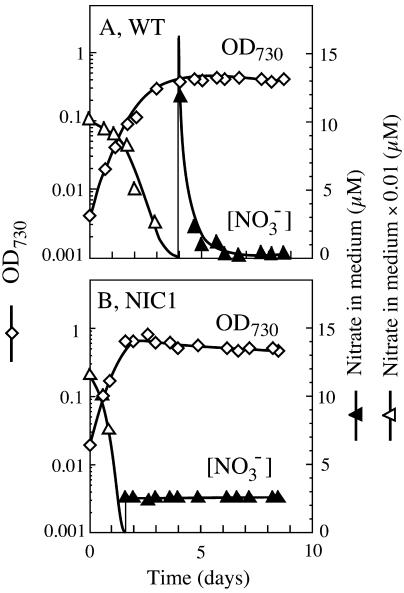
Growth of Synechococcus cells on nitrate with a constant stress of nitrogen deficiency was achieved also by means of a chemostat. With constant dilution with fresh nitrate (1 mM)-containing medium at a rate of 0.03 h−1, both the wild-type and the mutant cultures reached a steady state within several days after inoculation in which a constant culture density and a low nitrate concentration were maintained (Fig. 2). The wild-type cells in the chemostat cultures were yellow-green due to a low level of phycocyanin content (Fig. 3). The mutant cultures were green-tinged yellow (Fig. 3), showing that the cellular phycocyanin content is lower than the wild-type level. These results indicated that the mutant cells were more severely stressed with nitrogen deficiency than the wild-type cells. The nitrate concentration in medium was 2 to 3 μM in the cultures of the NIC1 mutant and <0.5 μM in the cultures of the wild-type cells. These results indicated that the ntcB mutant has a smaller capacity to utilize low levels of nitrate in medium.
FIG. 2.
Growth of the wild-type strain (A) and the ntcB mutant NIC1 (B) of S. elongatus strain PCC 7942 in continuous cultures with a limited supply of nitrate. Ammonium-grown cells were collected by centrifugation, suspended in nitrate-containing medium, and inoculated into 1 liter of nitrogen-free medium. Sterile fresh medium containing 1 mM nitrate was continuously pumped into the vessel at a dilution rate of 0.03 h−1, and the effluent was collected axenically for measurements of cell density (diamonds) and nitrate concentration (triangles).
FIG. 3.
Appearance of the nitrate-limited chemostat cultures of the wild-type strain (WT) and the ntcB mutant NIC1 of S. elongatus strain PCC 7942.
When expressed on the Chl basis, the NRT, NR, and NiR activities of the wild-type cells in the nitrate-limited chemostat cultures were 3 to 4, 4 to 5, and 8 to 10 times higher than those of the cells grown under nitrate-replete conditions in batch cultures, respectively (Table 1). The NRT, NR, and NiR activities of the ntcB mutant in the chemostat cultures were much lower than the corresponding wild-type levels (60, 27, and 21% of the latter values, respectively) (Table 1). Measurements of the turbidity at 730 nm, however, revealed that Chl-to-turbidity ratios in the chemostat cultures (2.2 and 1.8 μg ml−1 [OD unit]−1 for the wild-type strain and the mutant, respectively) were much lower than those seen with the wild-type level in batch cultures (6.2 μg ml−1 [OD unit]−1), indicating that cellular Chl content in the former cultures had been decreased due to nitrogen stress. Thus, the high activity levels of the nitrate assimilation enzymes in the chemostat-grown wild-type cultures were to be ascribed partly to the low level of Chl content of the cells. Even when expressed on the basis of turbidity, nevertheless, the enzyme activities in the chemostat-grown wild-type cells were higher than the corresponding activities in the nitrate-replete batch cultures (Table 1). The activity levels in the ntcB mutant were, on the other hand, lower than those in the wild-type batch cultures (Table 1). These results confirmed the results obtained with the NRT-less strains, i.e., that the NtcB-dependent, nitrite-responsive enhancement of nirA operon expression is required for up-regulation of the nitrate assimilation activities in nitrate-limited cells.
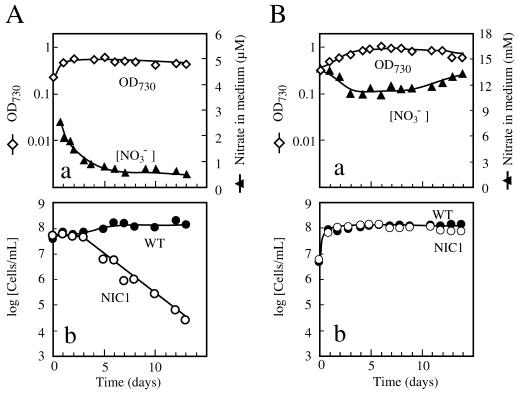
Competition between the wild-type and ntcB mutant strains.
When equal numbers of the wild-type and the mutant cells were mixed and subjected to growth under nitrate-limited conditions in the continuous-culture system, the cell population of the wild-type strain remained constant whereas that of the mutant decreased exponentially, giving a straight line on a logarithmic scale (Fig. 4A, panel b). The slope of the line was −0.302 day−1 (Fig. 4A, panel b), −0.297 day−1, and −0.261 day−1 in three separate experiments. These figures were close to that expected for dilution of nondividing cells at a dilution rate of 0.03 h−1, i.e., −0.313 day−1. The mutant cells were thus virtually unable to grow in the presence of the wild-type cells under the nitrate-limited conditions. When mixed cultures were subjected to continuous cultivation under nitrate-replete conditions, the proportion of the mutant cells in the total cell population was almost constant (Fig. 4B, panel b). These results demonstrated that the nitrite-responsive, NtcB-dependent enhancement of nirA operon transcription is essential for nitrate assimilation and growth of S. elongatus strain PCC 7942 in competitive environments with a limiting supply of nitrate.
FIG. 4.
Competition between the wild-type strain and the ntcB mutant of S. elongatus strain PCC 7942 in mixed continuous cultures with (A) and without (B) limitation of nitrate. Ammonium-grown cells of the two strains were collected by centrifugation and washed with nitrogen-free medium by resuspension and recentrifugation. Equal amounts of the cells of the two strains were inoculated together into 1 liter of nitrogen-free medium (A) and nitrate (15 mM)-containing medium (B), and sterile fresh media containing 1 mM (A) and 15 mM (B) of nitrate was continuously pumped into the vessels at a dilution rate of 0.03 h−1. The effluents were collected axenically for measurements of total cell density (open diamonds) and nitrate concentration (closed triangles) and for counting of the wild-type (closed circles) and the mutant (open circles) cell populations.
DISCUSSION
Because of the negative effect of assimilation of internally generated ammonium, the positive effect of nitrite on expression of the nirA operon and the nitrate assimilation enzymes is not obvious during growth of S. elongatus strain PCC 7942 in medium containing sufficient amounts of nitrate (1, 8, 19). Under nitrate-limited conditions, by contrast, the nitrite-responsive enhancement of nirA operon leads to a large increase in the activities of the nitrate assimilation enzymes (Table 1). Thus, the nitrite stimulation enables allotment of more nitrogen for synthesis of the nitrate assimilation enzymes in nitrogen-deficient cells than in nitrate-replete cells. Since nitrite is produced from nitrate, the regulatory system provides an effective mechanism to express high activities of the nitrate assimilation enzymes when the substrate is present in the medium and is limiting the growth. In the nitrate-limited cells, ammonium production is presumed to be too slow to cause negative feedback but the same cells are producing required amounts of nitrite sufficient for the enhancement of transcription. It is therefore deduced that the positive regulation system has very high sensitivity for nitrite. The molecular mechanism of the nitrite sensing is being investigated.
The stimulation by nitrite and NtcB of nitrate assimilation activities is not essential for growth of the cells in a single culture even under nitrate-limited conditions (Fig. 2B). However, it is essential for competitive utilization of limiting amounts of nitrate (Fig. 4A). In a previous study, Aichi and Omata also showed that NtcB is required for induction of the nitrate assimilation operon after replenishment of nitrate to nitrogen-starved cultures of S. elongatus strain PCC 7942 (1). Nitrate is the most common form of combined inorganic nitrogen in the largely nitrogen-deficient natural environment. Cyanobacteria having no N2-fixing ability are therefore daily exposed to nitrate-limited or nitrate-deficient growth conditions. The nitrite-responsive positive regulation of nirA operon expression would be essential for growth and survival of the cells in the natural environment, although its ecophysiological importance is not obvious in the nitrate-sufficient media commonly used for laboratory cultures.
Acknowledgments
We thank Takahira Ogawa (Sojo University, Kumamoto, Japan) for helpful advice about chemostat experiments.
This work was supported by a grant-in-aid for Scientific Research in Priority Areas (13206027) and in part by a grant-in-aid for Specially Promoted Research (13CE2005) and by The 21st Century COE Program from Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Aichi, M., and T. Omata. 1997. Involvement of NtcB, a LysR family transcription factor, in nitrite activation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 179:4671-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aichi, M., N. Takatani, and T. Omata. 2001. Role of NtcB in activation of the nitrate assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 183:5840-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, Y., and C. P. Wolk. 1997. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J. Bacteriol. 179:258-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frías, J. E., E. Flores, and A. Herrero. 1997. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 179:477-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrero, A., E. Flores, and M. G. Guerrero. 1981. Regulation of nitrate reductase levels in the cyanobacteria Anacystis nidulans, Anabaena sp. strain 7119, and Nostoc sp. strain 6719. J. Bacteriol. 145:175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrero, A., and M. G. Guerrero. 1986. Regulation of nitrite reductase in the cyanobacterium Anacystis nidulans. J. Gen. Microbiol. 132:2463-2468. [Google Scholar]
- 7.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi, H., M. Aichi, I. Suzuki, and T. Omata. 1996. Positive regulation by nitrite of the nitrate assimilation operon in the cyanobacteria Synechococcus sp. strain PCC 7942 and Plectonema boryanum. J. Bacteriol. 178:5822-58525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhlemeier, C. J., A. A. M. Thomas, A. van der Ende, R. W. van Leen, W. E. Borrias, C. A. M. J. J. van den Hondel, and G. A. van Arkel. 1983. A host-vector system for gene cloning in the cyanobacterium Anacystis nidulans R2. Plasmid 10:156-163. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann, M., and G. Wöber. 1978. Continuous cultivation in a chemostat of the phototrophic prokaryote, Anacystis nidulans, under nitrogen-limiting conditions. Mol. Cell. Biochem. 19:155-163. [DOI] [PubMed] [Google Scholar]
- 11.Mackinney, G. 1941. Absorption of light by chlorophyll solutions. J. Biol. Chem. 140:315-322. [Google Scholar]
- 12.Maeda, S., Y. Kawaguchi, T. Ohe, and T. Omata. 1998. cis-acting sequences required for NtcB-dependent, nitrite-responsive positive regulation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 180:4080-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda, S., and T. Omata. 2004. A novel gene (narM) required for expression of nitrate reductase activity in the cyanobacterium Synechococcus elongatus strain PCC7942. J. Bacteriol. 186:2107-2114. [DOI] [PMC free article] [PubMed]
- 14.Maeda, S., and T. Omata. 1997. Substrate-binding lipoprotein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in the transport of nitrate and nitrite. J. Biol. Chem. 272:3036-3041. [DOI] [PubMed] [Google Scholar]
- 15.Merchán, F., K. L. Kindle, M. J. Llama, J. L. Serra, and E. Fernández. 1995. Cloning and sequencing of the nitrate transport system from the thermophilic, filamentous cyanobacterium Phormidium laminosum: comparative analysis with the homologous system from Synechococcus sp. PCC 7942. Plant Mol. Biol. 28:759-766. [DOI] [PubMed] [Google Scholar]
- 16.Muro-Pastor, M. I., J. C. Reyes, and F. J. Florencio. 2001. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 276:38320-38328. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto, T., K. Inoue-Sakamoto, and D. A. Bryant. 1999. A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 181:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanier, R. Y., R. Kunisawa, M. Mandel, and G. Cohen-Bazire. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35:171-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki, I., N. Horie, T. Sugiyama, and T. Omata. 1995. Identification and characterization of two nitrogen-regulated genes of the cyanobacterium Synechococcus sp. strain PCC7942 required for maximum efficiency of nitrogen assimilation. J. Bacteriol. 177:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki, I., H. Kikuchi, S. Nakanishi, Y. Fujita, T. Sugiyama, and T. Omata. 1995. A novel nitrite reductase gene from the cyanobacterium Plectonema boryanum. J. Bacteriol. 177:6137-6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki, I., T. Sugiyama, and T. Omata. 1993. Primary structure and transcriptional regulation of the gene for nitrite reductase from the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 34:1311-1320. [Google Scholar]
- 22.Tanigawa, R., M. Shirokane, S. Maeda, T. Omata, K. Tanaka, and H. Takahashi. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA 99:4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]