Abstract
Autophagy is an evolutionarily conserved catabolic process involved in several physiological and pathological processes1,2. Although primarily cytoprotective, autophagy can also contribute to cell death; it is thus important to understand what distinguishes the life or death decision in autophagic cells3. Here, we report that induction of autophagy is coupled to reduction of histone H4 lysine 16 acetylation (H4K16ac) through downregulation of the histone acetyltransferase hMOF/KAT8/MYST1, and demonstrate that this histone modification regulates the outcome of autophagy. At a genome-wide level we find that H4K16 deacetylation is associated predominantly with the downregulation of autophagy-related genes. Antagonizing H4K16ac downregulation upon autophagy induction results in the promotion of cell death. Our findings establish that alteration in a specific histone posttranslational modification during autophagy, affects the transcriptional regulation of autophagy-related genes and initiates a regulatory feedback loop, which serves as a key determinant of survival versus death responses upon autophagy induction.
Autophagy is a catabolic process that results in the autophagosome-dependent lysosomal degradation of bulk cytoplasmic contents, abnormal protein aggregates, and excess or damaged organelles1,2. This process involves a series of dynamic membrane-rearrangements mediated by a core set of autophagy-related (ATG) proteins4. Although autophagy is primarily a protective process for the cell, it can also play a role in cell death3; however, it is not clear what distinguishes the life or death decision5. Protein acetylation, in particular acetylation of ULK1 and ATG proteins, has emerged as a regulator of autophagy6–9. Accumulating evidence has established sirtuin 1 (SIRT1), a NAD+-dependent deacetylase, as a player in this process7,10. However, SIRT1 is not always required for autophagy to occur. Indeed, while SIRT1 overexpression is sufficient to increase the basal level of autophagy and is required for starvation-induced autophagy, it is not necessary for rapamycin-induced autophagy7,11. SIRT1 has a wide range of non-histone targets, but lysine 16 on histone H4 (H4K16) is its primary histone target12,13. The histone acetyltransferase hMOF/KAT8/MYST1 is necessary and sufficient for the bulk of H4K16 acetylation and thereby antagonizes the enzymatic activity of SIRT113–15. Since SIRT1 has been linked to both autophagy and epigenetic chromatin changes, this encouraged us to investigate the role of covalent histone modifications in autophagy. As SIRT1 preferentially deacetylates H4K16ac12, we hypothesized that this histone modification could be altered upon autophagy induction.
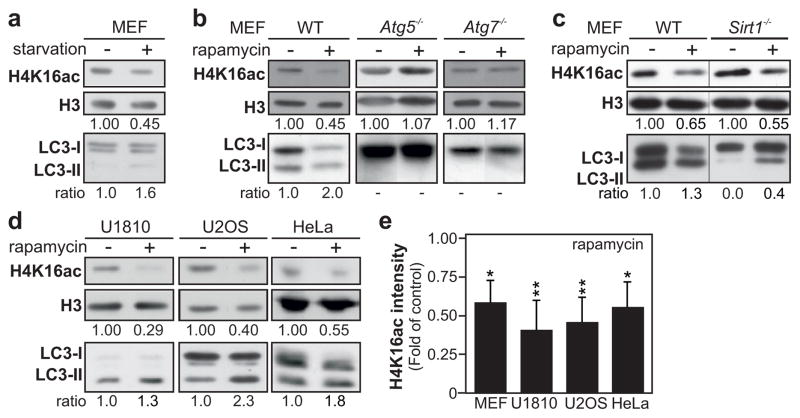
We induced autophagy in mouse embryonic fibroblast (MEF) cells by amino acid starvation and observed a decrease in acetylation of H4K16 (Fig. 1a). To elucidate whether the observed effect on H4K16ac was linked to the role of SIRT1 during starvation-induced autophagy, or if the deacetylation of H4K16 is a general feature of the autophagic process, treatments with rapamycin or Torin1, respectively allosteric and catalytic inhibitor of the kinase mechanistic target of rapamycin (MTOR), were used to induce SIRT1-independent autophagy (Supplementary Figs 2a and 3a). The global level of H4K16ac was robustly reduced in MEF cells after those treatments (Fig. 1b and Supplementary Fig. 4a). Interestingly, in the Sirt1 null MEF cells, both rapamycin and torin-1 treatments, but not amino acid starvation, induced the downregulation of H4K16ac, confirming that SIRT1 is not required for the repression of this histone modification (Fig. 1c and Supplementary Figs 2b and 4b).
Figure 1. Autophagy is associated with reduced acetylation of histone H4 lysine 16.
(a) Starvation (3 h)-induced autophagy results in a downregulation of H4K16ac in histone extracts of MEF cells. (b) Upon rapamycin treatment (300 nM) LC3 conversion and downregulation of H4K16ac are observed in WT MEF cells but not in the autophagy-deficient Atg5−/− and Atg7−/− MEF cells. (c) Rapamycin treatment increased the LC3-II/LC3-I ratio and promoted H4K16ac decrease in Sirt1−/− and WT MEF cells. (d) Rapamycin-induced autophagy led to downregulation of H4K16ac at 48 h in histone extracts of HeLa and U20S cells, and after 6 h in U1810 cells. (e) Quantification of H4K16 acetylation by immunoblotting is depicted for rapamycin-treated cells. Data are expressed as mean ± SEM (n=3–5); *Pvalue<0.05; **Pvalue<0.01.
The downregulation of H4K16ac upon autophagy induction occurred in various human cancer cell types, i.e. U1810, HeLa and U2OS cells (Fig. 1d–e and Supplementary Fig. 4c–d) and was even found to occur in yeast (Supplementary Fig. 5). Rapamycin treatment did not affect total histone H4 levels (Supplementary Fig. 6). The changes in H4K16 acetylation status were linked to the occurrence of autophagy as established by an increased lipidation of the autophagic marker LC3, resulting in an increased ratio of the lipidated form (LC3-II) to the unlipidated form (LC3-I), referred to later as LC3 conversion (Fig. 1a–d and Supplementary Fig. 3a–b). Similarly, in yeast this treatment resulted in increased lipidation of Atg8 (yeast homolog of LC3), and both cleavage and vacuolar localization of GFP-Atg8 (Supplementary Fig. 7a–c).
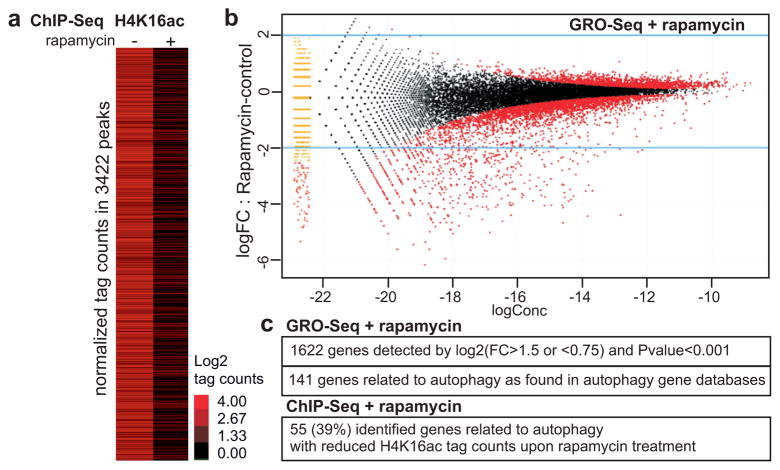
Dynamic histone modifications play a pivotal role in cell regulatory events16 and the H4K16 residue is of particular interest, as its acetylation influences higher order chromatin structure17 and plays an important role in transcription18. Chromatin immunoprecipitation targeting H4K16ac, followed by high-throughput sequencing (ChIP-Seq) was performed to elucidate the genome-wide occurrence of this histone mark in U1810 cells undergoing autophagy (Fig. 2a). H4K16ac ChIP-Seq data analysis revealed 3422 called peaks in untreated U1810 cells, which subsequently showed reduced H4K16ac occupancy after 8 h rapamycin treatment. To gain insight into the role of this induced H4K16 deacetylation in the regulation of gene expression during autophagy, we performed a global run-on-sequencing (GRO-Seq) assay19,20 to generate a genome-wide view of the location, orientation, and density of nascent transcripts engaged by RNA polymerases at high resolution in rapamycin-treated versus untreated U1810 cells (Fig. 2b and Supplementary Fig. 8a). This approach unveiled a significant alteration of the U1810 transcriptome with the identification of 1622 significantly (FC>1.5 or <0.75 and Pvalue<0.001) up- or downregulated genes (Fig. 2b–c and Supplementary Fig. 8a). A large fraction of the identified genes (141 genes; 8.7%) were related to autophagy (Fig. 2c and Supplementary Fig. 8b). There is an overall coincidence across the autophagy-related genes between the alteration of the GRO-Seq signal and the absence of H4K16 acetylation. Indeed, 55 genes, i.e. 39% of the autophagy-related genes (including genes belonging to the autophagic core machinery) identified by GRO-Seq analysis, exhibited reduced H4K16ac tag counts upon rapamycin treatment (Fig. 2c and Supplementary Fig. 9a–b).
Figure 2. Deacetylation of H4K16 by rapamycin treatment is associated with transcriptional regulation of autophagy-related genes.
(a) Heat map of H4K16Ac ChIP-Seq performed in U1810 cells without or with 8 h rapamycin treatment. Data are shown as log2 values of tag counts in the 3422 regions defined as peaks in the control sample. (b) De novo detection of transcripts using GRO-Seq analysis was performed in 8 h rapamycin-treated U1810 cells and compared to untreated U1810 cells. GRO-seq data visualized as ‘MA’ plots (log ratio versus abundance). The plot shows GRO-Seq gene expression for pair-wise comparison between rapamycin-treated vs control cells. The red points denote the differentially expressed genes. (c) Autophagy-related genes identified as regulated by rapamycin in the GRO-Seq data analysis and in the ChIP-Seq data analysis.
In order, to provide further evidence that the downregulation of H4K16ac during autophagy is part of a specific program, three additional histone modifications were examined, i.e. H3K4me3, H4K12ac and H4K8ac. In fact, whereas H4K16ac and H3K4me3 are known to be associated histone marks21–23, K8 and K12 acetylation levels on histone H4 are reported to be independent of H4K16 acetylation13,24. In agreement with the established molecular link between H4K16ac and H3K4me3, rapamycin-induced autophagy was associated with a reduction in H3K4me3 (Supplementary Fig. 10a–b), whereas H4K12ac and H4K18ac were left unaffected (Supplementary Fig. 10c). The joint downregulation of the H4K16ac and the H3K4me3 histone modifications was also observed in wild-type (WT) yeast, but not atg1 Δ, atg5 Δ and atg7 Δ autophagy-deficient yeast or Sas2-overexpressing yeast (Supplementary 10d). Collectively, these genome-wide deep-sequencing analyses indicate that the observed deacetylation of H4K16 during autophagy results in transcriptional regulation of autophagy-related genes.
Since MTOR is involved in a wide variety of signaling pathways, treating cells with rapamycin or Torin1 could cause the observed epigenetic changes by mechanisms unrelated to autophagy. To exclude this possibility, we tested the effect of rapamycin on the H4K16ac histone modification in Atg5- and Atg7-deficient MEF cells. These genes encode two ATG proteins that are essential for the canonical autophagy pathway (Fig. 1b, lower panels). Treatment of Atg7−/− or Atg5−/− MEFs, i.e., autophagy-deficient cells, with rapamycin, did not lead to a similar degree of downregulation of H4K16ac (Fig. 1b, upper panels). Identical effects were observed in yeast (Supplementary Fig. 5).
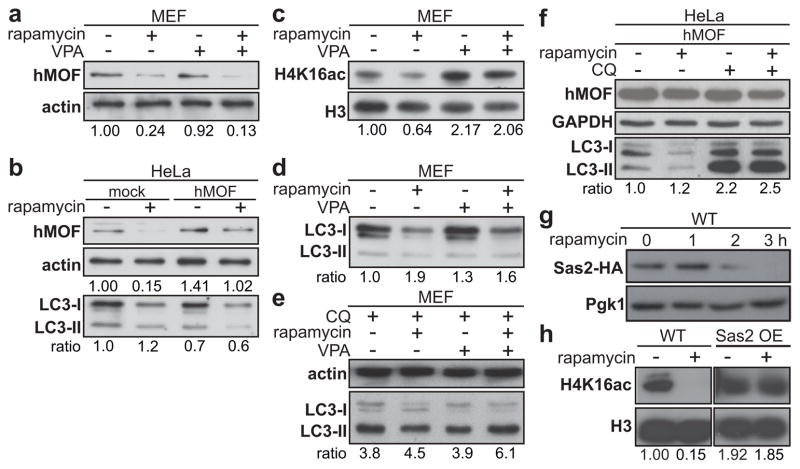
Thus, the process of autophagy, independent of whether its induction required a SIRT1-dependent signaling pathway, was associated with deacetylation of H4K16. Collectively, these data suggest that alteration in another histone modifying enzyme should be responsible for the observed modification in the acetylation status of H4K16. This observation prompted us to examine the status of hMOF during autophagy. Interestingly, while SIRT1 expression was not significantly altered upon rapamycin treatment (Supplementary Fig. 11a), hMOF expression was effectively downregulated in mammalian cells (Fig. 3a–b and Supplementary Figs. 3a–c and 11a). Similarly, robust downregulation of hMOF expression was observed upon Torin1 treatment or under amino acid starvation (Supplementary Fig. 3a–d). Remarkably, the observed downregulation of hMOF levels upon rapamycin, Torin1, or starvation treatment, was abrogated when cells were co-treated with inhibitors of autophagy such as chloroquine (CQ), or 3-methyladenine (3MA) (Supplementary Fig. 3a–c). In yeast cells engineered to express an HA-tagged version of the yeast homolog of hMOF, Sas2, rapamycin treatment induced a nearly complete loss of the HA signal within 3 h (Fig. 3g). Overexpression of Sas2 repressed the downregulation of H4K16ac upon rapamycin treatment in yeast cells (Fig. 3h). Altogether, these results revealed that the downregulation of hMOF is part of the autophagy program.
Figure 3. Rapamycin-induced hMOF downregulation promotes deacetylation of H4K16.
Rapamycin treatment (48 h) promoted the downregulation of the H4K16 histone acetyltransferase hMOF expression level in MEF (a) and transfected HeLa cells (b). VPA (1mM) treatment counteracted rapamycin-induced H4K16ac downregulation (c) and decreased the LC3 ratio (d). (e) Cotreatment with chloroquine (CQ, 10 μM) showed that the decrease in LC3 ratio was a result of an increase in autophagic flux. (f) Inhibition of autophagy by CQ after hMOF overexpression shows that hMOF does not inhibit autophagic flux. (g) The yeast homolog of hMOF, Sas2, tagged with 3xHA showed a complete disappearance of HA signal upon autophagy induction after 3 h. (h) Overexpression of Sas2 repressed the downregulation of H4K16ac upon rapamycin treatment.
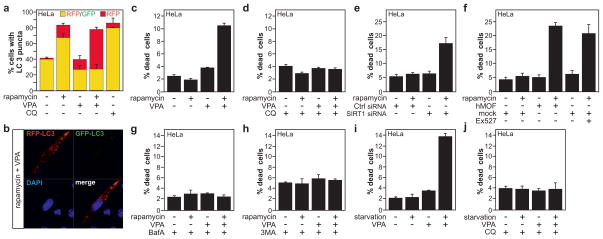
The observed dramatic changes in levels of H4K16 acetylation and associated transcriptional gene regulation suggested that there may be a functional role for this epigenetic change during autophagy. It has only recently become clear how histone modifications can play a regulatory role in apoptosis and how they can influence the decision between life and death (reviewed in 25). A similar regulatory role for histone modifications could be present during autophagy. Shifting the equilibrium of hMOF and SIRT1 expression in favor of SIRT1, leads to a decrease in acetylation of H4K1613. Treatment with valproic acid (VPA) increased the acetylation status of H4K16 (Fig. 3c and Supplementary Fig. 11b–d) by reducing SIRT1 levels13. Treatment with VPA was not only able to reverse rapamycin-induced downregulation of the H4K16ac histone modification (Fig. 3c), but also promoted LC3 conversion and turnover (Fig. 3d and Supplementary Fig. 11e). Importantly, administration of chloroquine to inhibit lysosomal activity enhanced the rapamycin-induced increase in LC3-II levels. Co-treatment with VPA, led to a further increase in the LC3-II levels, confirming the increased autophagic flux in those cells (Fig. 3e and Supplementary Fig. 11f). Combined treatment with bafilomycin A1 (Baf A), which prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes led to similar results (Supplementary Fig. 12). The increase in the autophagic flux in those cells was further confirmed, making use of a tandem reporter construct, mRFP-GFP-LC326 (Fig. 4a–b and Supplementary Figs 13a, 14 and 15). Rapamycin treatment resulted in an increase of yellow color-labeled LC3 puncta in U1810 and HeLa cells. In contrast, a remarkable increase in punctate red fluorescent signals was detected upon VPA and rapamycin-treatment in both cell types (Figure 4a–b and Supplementary Fig. 13a and 14). Similarly, hMOF overexpression in HeLa and U1810 cells correlated with increased autophagic flux upon rapamycin treatment as illustrated by LC3 immunobloting (Fig. 3b,f) and mRFP-GFP-LC3 autophagic flux assay (Supplementary Fig. 15).
Figure 4. Inhibition of H4K16ac downregulation upon autophagy induction results in cell death.
(a) VPA increases the autophagic flux in rapamycin-treated HeLa cells transfected with the mRFP-GFP-LC3 tandem reporter construct which allows distinction between autophagosomes (GFP+/RFP+ yellow puncta) and autolysosomes (GFP−/RFP+ red puncta). (b) Confocal microscopy image of a cell treated with rapamycin and VPA depicting a high ratio of red to green LC3 puncta indicating an increase in autophagic flux. (c) Co-treatment with VPA and rapamycin led to increased cell death. (d) Co-treatment with CQ abrogated VPA+rapamycin-induced cell death. (e–f) Increasing H4K16ac levels by either overexpression of hMOF, inhibition of SIRT1 by siRNA or the chemical inhibitor Ex527 promoted cell death upon rapamycin treatment. Co-treatment with the autophagy inhibitor Bafilomycin A (BafA, 40nM) (g) or 3-methyladenine (3MA, 5 mM) (h) abrogated VPA+rapamycin-induced cell death. Treatment of HeLa cells upon amino-acid starvation with VPA induced cell death (i), which was rescued when cells were co-treated with CQ (j). Data are expressed as mean ± SEM (n=3).
We extended our investigation to the analysis of cell death. We observed a significant increase in cell death in human-derived cell lines co-treated with rapamycin, and VPA or the SIRT1-specific inhibitor Ex52713 as demonstrated by the appearance of condensed or fragmented nuclei (Fig. 4c,f and Supplementary Figs 13b,e, and 16a–b). These results were further confirmed by FACS analysis of the appearance of a sub-G1 hypodiploid DNA peak (Supplementary Fig. 17a–b). In agreement with the observed effect of SIRT1 chemical inhibitors on the outcome of autophagy, we observed a significant increase in cell death in HeLa or U1810 cells upon SIRT1-knockdown and rapamycin treatment (Fig. 4e and Supplementary Fig.13d). To investigate whether the observed cell death upon abrogation of H4K16 deacetylation after rapamycin treatment was a consequence of autophagy induction, we performed an additional set of experiments with CQ, Baf A and 3MA, which inhibit different steps within the autophagic pathway. We noted that co-treatment with these inhibitors of autophagy was able to abrogate both rapamycin+VPA- and rapamycin+Ex527-induced cell death in human cancer cells (Fig. 4d, g and h and Supplementary Fig. 13c and 16c–e). Knock down of ATG7 expression in mammalian cells prevented rapamycin+VPA-induced cell death and thus further strengthens the conclusion about an autophagy-mediated cell death (Supplementary Fig. 18). Worth a notice, the increased cell death upon VPA addition was not limited to rapamycin-induced autophagy, and was also observed in amino acid starved HeLa cells (Fig. 4i–j). Furthermore, we investigated the link between hMOF activity and the outcome of autophagy. In agreement with the discovery that the perturbation of H4K16 acetylation status regulates the outcome of autophagy, we observed a significant increase in cell death in hMOF-overexpressing HeLa or U1810 cells upon rapamycin treatment, mimicking the effect of VPA/rapamycin co-treatment (Fig. 4f and Supplementary Fig. 13e). Collectively, these data indicate that the downregulation of hMOF, the associated reduction in H4K16 acetylation level, and transcriptional regulation of autophagy-related genes are required for the proper progression of the autophagic process, and that disturbance of this epigenetic program results in cell death.
In conclusion, until now, nuclear events have not been considered of primary importance for autophagy, as enucleated cells are still able to accumulate GFP-LC3 puncta in response to autophagic stimuli10. Our data, however, unveil a critical linkage of the induction of autophagy and covalent histone H4K16 modifications linked to altered gene expression, including the regulation of key genes in the autophagy program. Our findings imply a molecular histone switch, where the balancing effects of hMOF and SIRT1 on H4K16 acetylation regulate autophagy (Supplementary Fig. 1). Our results shown here do not oppose the findings about functionality of the autophagic process in enucleated cells, but add a new feedback regulatory network influencing the outcome of autophagy with respect to cell death/survival. The identification of tightly regulated histone modifications associated with the autophagic process offers an attractive conceptual framework both to understand the short-term transcriptional response to stimuli eliciting autophagy, as well as constituting a potential aspect of long-term responses to autophagy.
Online Methods
Cell culture and transfection
Non-small cell lung carcinoma U1810 cells, osteosarcoma U2OS cells, cervical cancer HeLa cells and wild-type, Atg5−/−, Atg7−/− and Sirt1−/− mouse embryonic fibroblasts were cultured using standard procedures13,30. The Atg5−/− and WT MEF cell lines were gifts from Dr. Gerald McInerney, the Atg7−/− MEF cell line was a gift from Dr. Masaaki Komatsu30 and the Sirt1−/− MEF cell line was a gift from Dr. Xiaoling Li. SEY6210 wild-type, atg1Δ, atg5Δ, and atg7Δ yeast cells were grown as described previously31. Plasmids encoding HA-human MOF, Flag-human SIRT1 and mRFP-GFP-LC3 were generous gifts from RG Roeder (The Rockefeller University), L Guarente (Massachusetts Institute of Technology), and G McInerney (Karolinska Institutet) respectively. Transfection of cells was carried out with Lipofectamine and Lipofectamine reagent in HeLa cells and X-tremeGENE HP DNA transfection reagent in U1810 cells.
Histone extracts and immunoblotting
Histone protein extracts were performed as described elsewhere13,27 using TCA precipitation and H2SO4 extraction or using the Histone Purification Mini Kit (Active Motif). Total protein extracts and immunoblotting were performed as reported previously28. GAPDH, beta-actin, histone 3 (H3) and phosphoglycerate kinase 1 (Pgk1) are used as standards for equal loading of protein. Densitometry was done using ImageJ.
Yeast procedures
The GFP-Atg8 processing assay and fluorescence microscopy were carried out as described previously32. If not stated otherwise all experiments were performed at 1h treatment.
Immunofluorescence and confocal microscopy
For confocal microscopy analysis, the adherent mammalian cells were grown on coverslips. Paraformaldehyde-fixed cells were blocked in HEPES, 3% bovine serum albumin, 0.3% Triton X-100 and incubated with primary (4°C, overnight) and secondary (room temperature, 1 h) antibodies. Samples were mounted with Vectashield (Vector Laboratories) and analyzed with Zeiss 510 META confocal laser scanning microscopy (Zeiss)28.
Evaluation of autophagic flux
To monitor the autophagic flux, a tandem reporter construct mRFP-GFP-LC3 was used26. The green fluorescence of this tandem reporter is attenuated in the acidic pH lysosomal environment, by lysosomal hydrolysis, while the mRFP is not. Therefore, the green fluorescent component of the composite yellow fluorescence from this mRFP-GFP-LC3 reporter is lost upon autophagosome fusion with a lysosome, whereas the red fluorescence remains detectable. Thus this probe allows distinguishing between autophagosomes (GFP+/RFP+ yellow puncta) and autolysosomes (GFP−/RFP+ red puncta). At 24 h after plating, the cells were transfected with the mRFP-GFP-LC3 plasmid, alone or in combination with SIRT1 siRNA, control siRNA or hMOF plasmid. The next day, cells were treated for an additional 24 h with the indicated compound(s). Cells were then fixed using 4% paraformaldehyde, and autophagy was determined by quantification of the number of cells with LC3-positive organelles, counting at least 100 cells in triplicate per condition. The presence of autophagic vacuoles expressing endogenous LC3 was also assessed.
Cell death quantification
After treatment, cells were fixed in 4% paraformaldehyde, harvested and cytospins were prepared. Subsequently, DNA was stained with Hoechst 33342 (0.1 mg/ml; Molecular Probes/Invitrogen). The number of dying cells was measured quantitatively by assessing the percentage of cells with fragmented, damaged or condensed nuclei.
Fluorescence-activated cell sorting (FACS) analysis
Quantification of PI (Sigma) and TMRE (Molecular Probes/Invitrogen) staining was performed with a FACSCalibur flow cytometer (Becton Dickinson) using standard procedures28.
ChIP-sequencing
For ChIP-Seq analysis, 5 μg of chromatin was used in two separate IP’s and combined in one elution for each condition. Subsequently, the DNA sequencing library was made using a kit from Illumina (Cat no 1003473) except that Illumina TruSeq adaptors (to enable multiplexing) were used. The library was analyzed by Solexa/Illumina Hi-seq. After pre-filtering the raw data by removing sequenced adapters and low quality reads, the sequence tags were aligned to the human genome (assembly hg19) with the Bowtie alignment tool33. To avoid any PCR-generated spikes we allowed only one read per chromosomal position, thus eliminating PCR bias. From the filtered raw data, 8 million unique reads per sample were used for peak detection. Peak detection was performed using the CisGenome program 34 with a two-sample analysis where sequenced input (1%) was used as a negative control. Peaks were called with a window statistic cutoff of 3 and a log2 fold change of 2. Using the defined chromosomal peak regions from the no-treatment condition, the number of tags were counted in the corresponding rapamycin-treated sample and heat maps were generated using Java Treeview35.
GRO-Sequencing
GRO-Seq experiments were performed as previously reported 19,20. Briefly, cells were washed with cold 1X PBS buffer and swelled in swelling buffer (10 mM Tris-Cl pH 7.5, 2 mM MgCl2, 3 mM CaCl2) for 5 min on ice and harvested. Cells were lysed in lysis buffer (swelling buffer with 0.5% NP-40, 2 u/ml Superase In and 10% glycerol) and finally resuspended in 100 μL of freezing buffer (50 mM Tris-Cl pH 8.3, 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA). For the run-on assay, resuspended nuclei were mixed with an equal volume of reaction buffer (10 mM Tris-Cl pH 8.0, 5 mM MgCl2, 1 mM DTT, 300 mM KCl, 20 units of SUPERase In, 1% sarkosyl, 500 μM ATP, GTP, and Br-UTP, 2 μM CTP) and incubated for 5 min at 30°C. The nuclear-run-on RNA (NRO-RNA) was then extracted with TRIzol LS reagent (Invitrogen) following the manufacturer’s instructions. NRO-RNA was then subjected to base hydrolysis on ice for 40 min and followed by treatment with DNase I and antarctic phosphatase. To purify the Br-UTP-labeled nascent RNA, the NRO-RNA was immunoprecipitated with anti-BrdU argarose beads (Santa Cruz Biotech) in binding buffer (0.5X SSPE, 1 mM EDTA, 0.05% Tween-20). To repair the end, the immunoprecipitated BrU-RNA was resuspended in a 50 μL reaction (45 μL DEPC water, 5.2 μL T4 PNK buffer, 1 μL SUPERase In and 1 μL T4 PNK [NEB]) and incubated at 37°C for 1 h. The RNA was extracted and precipitated using acidic phenol-chloroform. The cDNA synthesis was performed basically as in Ingolia et al., (2009) with a few minor modifications. First, RNA fragments were subjected to a poly-A tailing reaction by poly-A polymerase (NEB) for 30 min at 37°C. Subsequently, reverse transcription was performed using oNTI223 primer (5′-pGATCGTCGGACTGTAGAACTCT; CAAGCAGAAGACGGCATACGATTTTTTTTTTTTTTTTTTTTVN). Second, tailed RNA (8.0 μL) was subjected to reverse transcription using superscript III (Invitrogen). The cDNA products were separated on a 10% polyacrylamide TBE-urea gel and the extended first-strand product (100–500 bp) was excised and recovered by gel extraction. After that, the first-strand cDNA was circularized by CircLigase (Epicentre) and relinearized by Ape1 (NEB). Relinearized single strand cDNA (sscDNA) was separated on a 10% polyacrylamide TBE gel and the product of needed size was excised (~120–320 bp) for gel extraction. Finally, sscDNA template was amplified by PCR using the Phusion High-Fidelity enzyme (NEB) according to the manufacturer’s instructions with two oligonucleotide primers oNTI200 (5′-CAAGCAGAAGACGGCATA) and oNTI201 (5′-AATGATACGGCGACCACCGACAGGT TCAGAGTTCTACAGTCCGACG). DNA was then sequenced on the Illumina HiSeq2000 according to the manufacturer’s instructions, using the small RNA sequencing primer 5′-CGACAGGTTCAGAGTTCTACAGTCCGACGATC.
Database analyses
The following autophagy databases have been used to indentify autophagy-related genes: the human autophagy database available at http://autophagy.lu/ and the autophagy database available at http://tp-apg.genes.nig.ac.jp/autophagy/.
Statistical analyses
Bars and error bars represent mean with SEM. Statistical evaluations were performed by Student’s t-test.
Supplementary Material
Acknowledgments
We thank Drs. G. McInerney, M. Malewicz, S. Orrenius, and T. Panaretakis for valuable discussion and Drs. L. Guarente, V. Kaminskyy, M. Komatsu, G. McInerney, M. Panas, RG. Roeder, and L. Xiaoling for providing us with reagents and cell lines. J.F. is supported by a fellowship from the Karolinska Institutet Foundations, M.A.L-D. is partially supported by a Rackham Predoctoral Fellowship and W.L. is supported by breast cancer research Postdoctoral Fellowship Award (BC110381) from Department of Defense of U.S. This work was supported by NIH grant GM53396 (to D.J.K.) and NIH/NCI and DOD (to M.G.R.), the Swedish Cancer Society, the Swedish Childhood Cancer Foundation (to B.J. and O.H.), and the Swedish Research Council (to B.J.).
Footnotes
Author Contributions
J.F., R.B.S. and B.J. performed experiments in mammalian cells. M.A.L.-D. performed yeast experiments. N.H. performed ChIP-Seq. W.L. performed GRO-Seq. N.H. and Q.M. performed bioinformatical analysis. J.F., O.H., M.G.R., D.J.K., and B.J. designed the study, and analyzed and interpreted the data. The first draft of the paper was written by J.F. and B.J. All authors discussed the results and commented on or edited the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no conflict of interest.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yi C, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 9.Lin SY, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 10.Morselli E, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192:615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morselli E, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 13.Hajji N, et al. Opposing effects of hMOF and SIRT1 on H4K16 acetylation and the sensitivity to the topoisomerase II inhibitor etoposide. Oncogene. 2010;29:2192–2204. doi: 10.1038/onc.2009.505. [DOI] [PubMed] [Google Scholar]
- 14.Taipale M, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith ER, et al. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 18.Kind J, et al. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 19.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruthenburg AJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh H, et al. FOXP3 orchestrates H4K16 acetylation and H3K4 trimethylation for activation of multiple genes by recruiting MOF and causing displacement of PLU-1. Mol Cell. 2011;44:770–784. doi: 10.1016/j.molcel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Grummt I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol. 2005;15:1434–1438. doi: 10.1016/j.cub.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 25.Fullgrabe J, Hajji N, Joseph B. Cracking the death code: apoptosis-related histone modifications. Cell Death Differ. 2010;17:1238–1243. doi: 10.1038/cdd.2010.58. [DOI] [PubMed] [Google Scholar]
- 26.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 27.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 28.Burguillos MA, et al. Caspase signalling controls microglia activation and neurotoxicity. Nature. 2011;472:319–324. doi: 10.1038/nature09788. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Y, Cheong H, Song H, Klionsky DJ. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol. 2008;182:703–713. doi: 10.1083/jcb.200801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.