Abstract
Clinical mutations in patients diagnosed with Type 2A von Willebrand disease (vWD) have been identified that break the single disulfide bond linking N- and C-termini in the vWF A1 domain. We have modeled the effect of these mutations on the disulfide-bonded structure of A1 by reducing and carboxy-amidating these cysteines. Solution biophysical studies show that loss of this disulfide bond induces a molten globule conformational state lacking global tertiary structure but retaining residual secondary structure. The conformational dependence of platelet adhesion to these native and molten globule states of A1 is quantitatively compared using real-time high-speed video microscopy analysis of platelet translocation dynamics under shear flow in a parallel plate micro-fluidic flow chamber. While normal platelets translocating on surface-captured native A1 domain retain the catch-bond character of pause times that increase as a function of shear rate at low shear and decrease as a function of shear rate at high shear, platelets that interact with A1 lacking the disulfide bond remain stably attached and do not translocate. Based on these findings, we propose that the shear stress-sensitive regulation of the A1-GPIb interaction is due to folding the tertiary structure of this domain. Removal of the tertiary structure by disrupting the disulfide bond destroys this regulatory mechanism resulting in high-strength interactions between platelets and vWF A1 that are dependent only on residual secondary structure elements present in the molten globule conformation.
Keywords: Von Willebrand Factor, Von Willebrand Disease Type 2, Platelet Adhesiveness, Shear Stress, Rheology, Disulfide Bond, Protein Folding, Thermodynamics, Molten Globule
2 Introduction
Type 2A Von Willebrand Disease is clinically identified by a loss of high and intermediate von Willebrand factor multimers resulting in a loss of platelet-dependent function. The majority of missense mutations that cause type 2A disease occur in the A2 domain and are grouped by their effects on cellular retention of VWF or the susceptibility of VWF to proteolysis by ADAMTS13. However, a few mutations also classified as 2A occur in the A1 domain. The most puzzling are those that abolish the single disulfide bond in the A1 domain which is required to maintain the native structure of the domain. These point mutations identified in VWD patients result in the substitution of Cysteine 1272 for Arginine, Glycine, Phenylalanine and Serine and Cysteine 1458 for Tyrosine [1, 2, 3, 4]. Patients with C1272S and C1272F substitutions have been reported to be heterozygous so that one of the alleles is normal and although not reported, it is likely that the other known mutations are also heterozygous in VWD patients [1, 2]. Phenotypically, these patients display the distinctive clinical features of abnormal bleeding, decreased VWF ristocetin cofactor activity relative to VWF antigen, and diminished ristocetin-induced platelet agglutination (RIPA). Platelet counts are generally normal, but thrombocytopenia can occur [3].
Early studies resulted in disparate conclusions regarding the effect of the C1272 − C1458 disulfide bond on the structure and function of the A1 domain. Initially, two independent studies using recombinant A1 domain demonstrated 1) a requirement of the disulfide bond for effective inhibition of RIPA using fixed platelets in the presence of purified plasma vWF and 2) the aggregation of washed platelets in the absence of VWF induced by the recombinant domain [5, 6]. However, a subsequent report demonstrated that a reduced and alkylated dispase-digested fragment of VWF containing residues V1262 − K1491 of the A1 domain displayed dose-dependent inhibition of shear-induced platelet agglutination (SIPA) and was more effective at inhibiting the binding of purified VWF and LJ-Ib1 (an antibody against GPIbα) to GPIbα than the disulfide-intact fragment [7, 8]. To further complicate the issue, studies on recombinant multimeric VWF containing mutations C1272R, C1272G and C1458G illustrated that binding of VWF to fixed platelets was dependent on the specific cysteine that was mutated [9]. Mutating C1272 with either Arginine or Glycine caused a spontaneous interaction with platelets whereas C1458G resulted in a complete loss of function. Multimer gels also showed low to intermediate molecular weight multimers for C1272R and C1272G and a complete loss of all multimeric species for C1458G.
Studies from the Ruggeri lab illustrated an increase in GPIbα binding affinity of 1–2 orders of magnitude upon reducing and alkylating the disulfide bond. On the basis of fluorescence and circular dichroism, this result was interpreted to be a ”loosening of native tertiary structure” [10]. Furthermore, it was later demonstrated in flow chamber studies that a greater number of platelets adhered to glass surface-immobilized reduced/alkylated A1 domain at low shear than for disulfide-intact A1. At high shear this observation was reversed suggesting that fluid shear in laminar flow could switch the conformation of A1 from a low to a high affinity state that was similar to reduced/alkylated A1 with respect to platelet adhesion at low shear [11]. These two key observations combined with our identification of a thermodynamically stable intermediate conformation in the three-state unfolding pathway of the A1 domain led to current understanding of how gain and loss of function mutations in the A1 domain affect the linkage between conformation and GPIbα binding affinity and the effect of shear stress on this linkage [12, 13, 14]. This mechanism appears to involve a delicate balance in the thermodynamic equilibrium between the low affinity native and high affinity intermediate state resulting in differential rates of dissociation of GPIbα from the A1 domain that modulate the efficiency of platelet adherence to VWF as a function of shear stress [12, 15]. Type 2 mutations affect the efficacy of platelet adherence to VWF and change the affinity of GPIbα binding by altering this conformational equilibrium in favor of one or the other thermodynamic state [12, 13].
In principle, type 2A mutations that break the disulfide bond in A1 could cause severe problems in hemostasis that lead to microvasular thrombosis because of the enhanced affinity of this intermediate conformation for platelet GPIbα. Here, we provide a rigorous functional and biophysical comparison of these two thermodynamic conformational states using native disulfide-intact A1 and reduced and carboxyamidated (RCAM) A1 as a model of the intermediate state [12]. Using a parallel-plate micro-fluidic flow chamber that immobilizes the A1 domains via an N-terminally fused 6xHistidine-Tag engineered into their sequence, we find that disulfide-intact A1 exhibits catch-bond characteristics in the platelet pause times with a biexponential survival function that quantifies a unique shear dependence of the dissociation rate constants. In contrast, the enhanced affinity of RCAM A1 for platelet GPIbα abolishes platelet rolling on surface immobilized RCAM A1 resulting in net zero translocation velocities under shear flow. Biophysical, thermodynamic and spectroscopic comparisons between A1 and RCAM A1 illustrate that the intermediate conformation exists as a molten globule state that lacks tertiary structure while retaining residual secondary structure. This observation implies that folding inhibits the binding potential of A1 and that stable platelet attachment is primarily dependent upon secondary structure elements that are retained in the high affinity intermediate conformation. The implications of these findings pertaining to type 2A von Willebrand disease, VWF multimerization and protective mechanisms for VWF clearance are discussed.
3 Results
3.1 Platelet adhesion to surface captured disulfide-intact A1 and RCAM A1 under shear flow
The interaction of disulfide-intact A1 and of RCAM A1 with platelets was studied by immobilizing the proteins via the 6xHis-Tag on Cellix biochips in which the internal channel surface was coated with a Cu2+ chelating chemistry. A 5µM concentration of protein was immobilized in the biochip and citrated whole blood was perfused at a shear rate of 800s−1 (Figure 1). This perfusion was followed by TBS buffer and after red blood cells cleared the channel, the shear was decremented to 100s−1 or incremented to 9000s−1 every two minutes at logarithmic intervals. Surface bound platelets were allowed to equilibrate at a given shear rate for 50s and a 60s video at 24 frames per second was recorded and analyzed as described in the methods. The internal channel surface lacking immobilized A1 domain was completely inert to platelets.
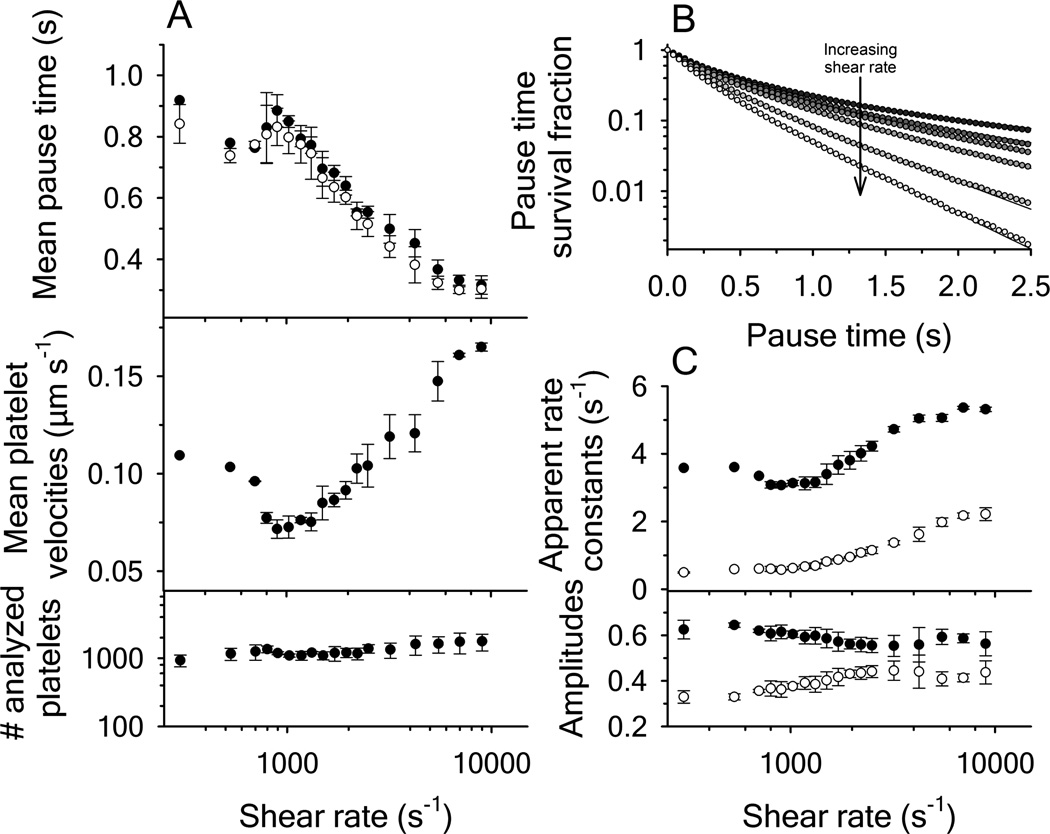
Figure 1.
Platelet translocation on disulfide-intact A1 domain as a function of shear rate. (A, upper panel) Mean pause times derived from the statistical average of platelet distributions (filled circles) and from biexponential fitting of the pause time survival fraction using equation (1) (open circles), mean platelet velocities (center panel) and the amount of platelets analyzed (lower panel). (B) Survival fraction decay functions of the platelet pause times with increasing shear rates. Rates are: 800, 1320, 1500, 1950, 2500, 4250 and 9000 s−1 from top to bottom. Curves were fit to a biexponential functions. (C) Apparent rate constants k1 (black circles) and k2 (open circles) and the fractional amplitudes associated with k1 (black circles) and k2 (open circles). Error bars represent the mean standard deviation from four independent experiments.
At each shear rate, platelet mean pause times and mean translocation velocities were calculated from the X-Y trajectories of moving platelets as described in the methods. Approximately 1000 platelet trajectories were analyzed at each shear rate (Figure 1A). The mean pause times were also calculated from the pause time survival fraction obtained from the cumulative integral of the pause time distribution (Figure 1B) [16]. The survival fraction exponentially decreases with faster decay rates as shear increases. Fitting the survival fraction with a bi-exponential decay function results in the apparent rate constants and amplitudes as a function of shear rate (Figure 1C). The mean pause times were calculated from the survival fraction using equation (1) and were found to agree with taking the statistical mean of all platelet pause time distributions (Figure 1A).
| (1) |
Mean platelet pause times increased from ≃ 0.75s at a shear rate of 530s−1 to ≃ 0.9s at 1025s−1 and then decreased again upon further increase of the shear rate. The mean platelet translocation velocities mirrored the pause times and were minimal at a shear rate of 1025s−1 and increased at lower and higher shear rates. While performing the flow experiments, it was observed that platelets began to detach from surface immobilized disulfide-intact A1 at shear rates ≤ 300s−1.
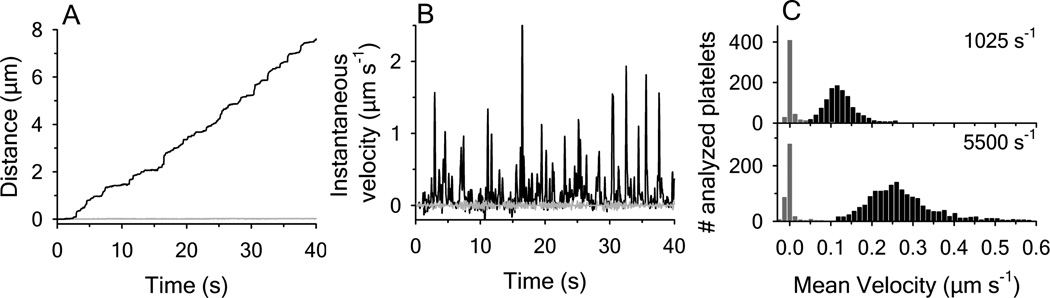
In contrast to disulfide-intact A1, the interactions between surface-immobilized RCAM A1 and platelet GPIbα resulted in captured platelets that did not translocate across the surface at all applied shear rates. Pause time analysis was not possible to do for RCAM A1 because the maximum was limited by the length of the recorded movies. However, we did compare the distance traveled and instantaneous velocities. Figure 2A illustrates the traveled distance for a single platelet translocating on disulfide-intact A1 with a platelet on RCAM A1 over a 40s time frame. The traveled distance was converted into instantaneous velocities in Figure 2B. While the interaction of a platelet with A1 caused many translocation events yielding nonzero velocities, the velocity of a platelet translocating on RCAM A1 was statistically zero. This is also shown by the histograms given in Figure 2C which report velocity distributions of all analyzed platelet translocations at shear rates of 1025 and 5500s−1. Furthermore, platelets remained firmly adhered to RCAM A1 at all shear rates investigated up to 9000s−1 and remained attached to RCAM A1 even after removal of shear stress by stopping the flow. However, the total number of platelets interacting with RCAM A1 was less, by approximately half, than with disulfide-intact A1 indicating that the on-rate for bond formation is reduced for RCAM A1 relative to disulfide-intact A1. Representative movies of platelets translocating on disulfide-intact A1 and RCAM A1 at 1025s−1 are provided in the supporting information.
Figure 2.
Comparison of platelet translocations on disulfide-intact A1 with RCAM A1. (A) Traveled distance of a single platelet as a function of time at 1025 s−1; disulfide-intact A1 (black line), RCAM A1 (grey line). (B) Comparison of the instantaneous velocities obtained for a single platelet interacting with disufide intact A1 (black line) and RCAM A1 (grey line) at 1025 s−1. (C) Histograms of the mean velocity of platelets translocating on disulfide-intact A1 (black bars) and RCAM A1 (gray bars) at shear rates of 1025 and 5500 s−1. Total number of platelets analyzed were 526 and 422 for RCAM A1 at 1025 and 5500 s−1 respectively; and 1132 and 1687 for disulfide-intact A1 at 1025 and 5500 s−1 respectively. Data are representative of four independent experiments.
3.2 Spectroscopic properties of A1 and RCAM A1
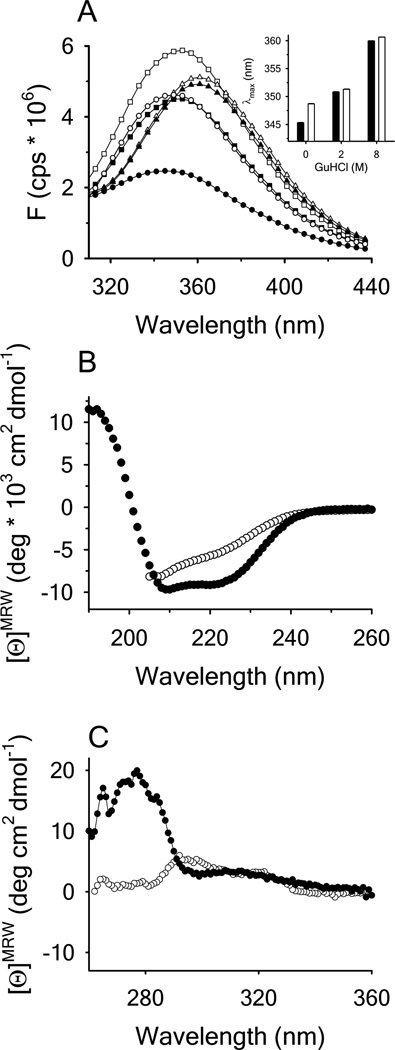
Fluorescence and circular dichroism spectra of A1 and of RCAM A1 were measured to compare the basic spectroscopic properties of the two protein variants (Figure 3). The fluorescence spectrum of native A1 has a relatively low intensity with a λmax of ~ 345nm. In presence of 2M GdnHCl the fluorescence intensity increases as the interior of the protein becomes solvent accessible and a red shift of the spectrum to ~ 351nm occurs. 8M GdnHCl decreased the fluorescence intensity and resulted in an additional redshift to ~ 360nm. The fluorescence spectrum of RCAM A1 in presence of 0.5M GdnHCl showed an increased fluorescence intensity and a slightly more red shifted λmax in comparison to A1. Increasing the GdnHCl concentration to 2M and 8M resulted in an increased intensity and a λmax comparable to that of the disulfide-intact A1 domain. Far UV CD spectra of the A1 domain are dominated by a high α-helical content whereas RCAM A1 had a reduced α-helical content. Nevertheless, RCAM A1 is still retains significant secondary structure content in solution; the distribution of which is summarized in Supplemental Table 1. In the near UV, the CD of disulfide-intact A1 show a well-defined spectral banding pattern between 260 and 300nm indicating significant tertiary structure content that is absent in RCAM A1.
Figure 3.
(A) Fluorescence emission spectra of disulfide-intact A1 (closed symbols) and RCAM A1 (open symbols) in the presence of 0M (circles), 2M (squares) and 8M GdnHCl (triangles) with excitation λ = 280nm. Inset: Wavelength of maximum fluorescence intensity (λmax) for disulfide-intact A1 (black bars) and RCAM A1 (white bars). (B) Far UV CD spectra of disulfide-intact A1 (closed circles) in buffer and RCAM A1 (open circles) in 0.5M GdnHCl. (C) Near UV CD spectra for disulfide-intact A1 (closed circles) and RCAM A1 in buffer (open circles).
3.3 Chemical denaturation of A1 and RCAM A1
The thermodynamic stability of A1 and of RCAM A1 was determined from GdnHCl and urea induced unfolding measured via CD and FL at 25°C (Figure 4). Three states, native, intermediate and unfolded, were observed by CD for disulfide-intact A1 in GdnHCl, but urea did not completely unfold the intermediate state. In contrast, the ellipticity for RCAM A1 in the absence of denaturant is comparable to that of the A1 domain in the intermediate state. Complete unfolding of A1 and of RCAM A1 occurs in GdnHCl but not in urea. However, in presence of urea, a small expansion of RCAM A1 compared to A1 in its intermediate state is observed. The obtained parameters for the transitions are summarized in Supplemental Table 2.
Figure 4.
GdnHCl denaturation (A) and urea denaturation (B) of disulfide-intact A1 (closed circles) and RCAM A1 (open circles) monitored by circular dichroism at λ = 222nm.
3.4 Thermal denaturation of A1 and of RCAM A1
The unfolding of disulfide-intact A1 and RCAM A1 was also monitored via CD at 222nm, intrinsic protein fluorescence at 280nm=359nm and ANS fluorescence at 350nm=495nm as a function of temperature. As thermal unfolding of the A1 domain is not reversible, the thermal transitions were analyzed using a two state irreversible model described in detail previously and all three spectroscopic methods resulted in comparable unfolding enthalpies and transition temperatures for A1, Supplemental Table 3 [17]. Figure 5 illustrates that all three methods resulted in a cooperative unfolding transition for disulfide-intact A1 that was absent for RCAM A1.
Figure 5.
Thermal denaturation. (A) disulfide-intact A1 in 1M urea (closed circles) and RCAM A1 in 1M urea (closed squares) monitored by circular dichroism at λ = 222nm. (B) disulfide-intact A1 in buffer (closed circles), 9M urea (open circles) and RCAM A1 in buffer (closed squares) monitored by intrinsic protein fluorescence emission at λem = 359nm with excitation at λex = 280nm. (C) disulfide-intact A1 in buffer (closed circles) and RCAM A1 in buffer (closed squares) monitored by ANS fluorescence emission at λ = 495nm with excitation at λex = 350nm. Inset: Spectra of A1 and RCAM A1 in the presence of ANS at 20°C before (closed circles and squares) and after the thermal scan (open circles and squares).
The circular dichroic ellipticity of disulfide-intact A1 changed from ~ −9.4 to ~ −5.5 upon thermal denaturation, Figure 5A. Ellipticity of the thermally denatured state was comparable to that of RCAM A1 in presence of 1M urea, ~ −5.0. The ellipticity of RCAM A1 was constant over the full range of temperature. RCAM A1 has a slight tendency for aggregation followed by precipitation even in presence of 1M urea at high temperature. A thermal scan of RCAM A1 in absence of urea was significantly affected by protein aggregation beginning at ~ 40°C. The high tyrosine and tryptophan fluorescence intensity of RCAM A1 relative to disulfide-intact A1 indicated that most of the fluorophors were solvent exposed even at 20°C, Figure 5B. Thermal unfolding of RCAM A1 was comparable to disulfide-intact A1 in presence of 9M urea.
Thermal denaturation in the presence of ANS was comparable to CD and intrinsic fluorescence, Figure 5C. Spectra of the ANS fluorescence measured before thermal denaturation resulted a low initial ANS fluorescence for the disulfide-intact A1 domain indicating a relatively low exposure of hydrophobic surfaces. In contrast, the ANS spectrum of RCAM A1 resulted in a significantly enhanced fluorescence intensity due to exposed hydrophobic surfaces. After each thermalscan, the protein samples were equilibrated at 20°C and spectra were measured. The ANS fluorescence increased for disulfide-intact A1 and decreased slightly for RCAM A1 probably as a result of high temperature protein aggregation during the thermalscan.
3.5 Acrylamide quenching of tryptophan fluorescence
The spectroscopic characterization of A1 and of RCAM A1 showed that RCAM A1 corresponds well to the intermediate state of A1 which still contains a high amount of secondary structure, but lacks well ordered tertiary structure or a hydrophobic core. This was also confirmed via acrylamide induced tryotophan fluorescence quenching performed with an excitation wavelength of 295nm. The extent of the acrylamide quenching and hence the accessibility of the tryptophan residue was determined via the Stern Volmer equation and the resulting Stern-Volmer-constant.
Initially a quenching experiment was performed with disulfide-intact A1 at 20°C and at 1°C in order to distinguish between static and collisional quenching. Since the frequency of collisions between two molecules decreases at lower temperatures, the efficiency of a collisional quencher is reduced at lower temperatures and this results in a decrease Stern-Volmer constant. Clear evidence for collisional quenching could be obtained for the A1-domain since the Stern-Volmer constant decreased from 2.6 ± 0.03 to 2.2 ± 0.03 M−1, Figure 6A, ruling out a static quenching mechanism.
Figure 6.
(A) Acrylamide quenching of tryptophan fluorescence in disulfide-intact A1 at 20°C (closed circles) and 1°C (closed squares), RCAM A1 at 20°C (open symbols) and 1µM NATA at 20°C (grey circles) in buffer. (B) Resulting Stern-Volmer quenching constants derived from the slope of F0/F as a function of acrylamide molarity at the indicated concentrations of GdnHCl. Disulfide-intact A1 (black bars), RCAM A1 (white bars) and NATA in buffer (grey bar).
In further quenching experiments the accessibility of the tryptophan residue was determined for disulfide-intact A1 and RCAM A1 in 0, 2 and 7M GdnHCl (Figure 6)B. In the presence of 2M GdnHCl, the Stern-Volmer constants for both protein variants increased relative to the absence of GdnHCl. In 7M GdnHCl, these quenching constants for both disulfide-intact A1 and RCAM A1 were comparable to the quenching of NATA in water. The Stern Volmer constant of RCAM A1 in buffer was 5.6 ± 0.4 M−1, comparable to disulfide-intact A1 in 2M GdnHCl (6.0±0.1 M−1). Increasing GdnHCl concentrations reduced the efficiency of acrylamide quenching which is likely due to favorable interactions between GdnHCl and the tryptophan side chain which compete with collisional interactions between tryptophan and acrylamide [18].
3.6 Analytical Size Exclusion Chromatography
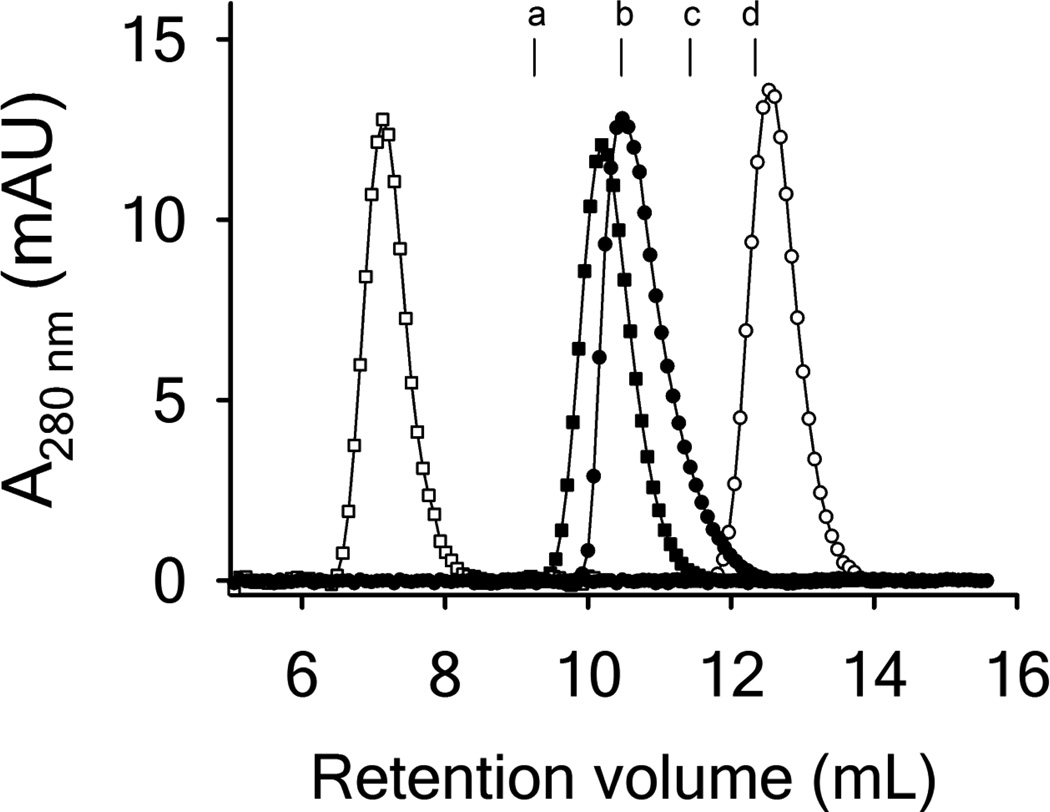
Size exclusion chromatography was performed to determine the apparent hydrodynamic radius of RCAM A1 relative to disulfide intact A1. In agreement with our previously reported results, the elution volume in buffer for RCAM A1 was higher (12.56mL) than for disulfide-intact A1 (10.48mL) [12]. While the elution volume for WT A1 corresponded well to a protein of ~ 28 kDa, RCAM A1 eluted at a volume characteristic for a protein of ~ 12 kDa. This atypical elution behavior indicates a partial unfolded protein that behaves like a significantly smaller protein when partitioning between the mobile and solid phases of the sizing column. However, addition of 0.25M GdnHCl, resulted in a more expanded form of RCAM A1 that eluted at 7.13mL relative to 10.19mL for disulfide intact A1 in 0.25M GdnHCl. Elution of both proteins in the presence of 2M GdnHCl resulted in highly expanded conformations that approached the void volume of the column, 6.8mL. Unfortunately, the low solubility of RCAM A1 precluded any direct measure of the size and shape of the protein due to limited detection by more direct measures such as analytical ultracentrifugation, dynamic light scattering or small angle X-ray scattering.
4 Discussion
Reduction and carboxyamidation of the disulfide bond preferentially stabilizes a conformation that is spectroscopically similar to the urea- and GdnHCl-induced intermediate conformation of disulfide-intact A1 domain in the far UV CD. This unrestrained conformation exhibited an enhanced apparent binding affinity for GPIbα on fixed platelets that enabled a quantitative thermodynamic description of how von Willebrand disease mutations alter the coupling between shear force dependent dissociation from GPIbα and the conformational stability of A1 [12]. Here we show that RCAM A1 arrests platelets resulting in net zero translocation velocities. This is in stark contrast to disulfide-intact A1 where platelets are observed to stick and roll on the surface captured domain. Pause times determined for platelets translocating on disulfide-intact A1 represent the average length of time a platelet is immobile. This time has a complex dependence on the shear rate which is seen to decrease at low shear, increase to a maximum at intermediate shear and then decrease again at higher shear rates. In single molecule experiments designed to measure the lifetime of a bond between two proteins as a function of applied force, this behavior is referred to as a catch-slip interaction where the bond initially becomes weaker at low force, strengthens at intermediate forces, and weakens again at higher forces [19, 20].
In our experiment, the mean pause times for disulfide-intact A1 are longer than previously reported flow chamber measurements [21] and longer than single bond lifetime measurements by AFM [22] indicating that multiple bonds between surface captured A1 and platelet GPIbα may be occurring. Multiple bonds might also explain the complex shear dependence of the apparent rate constants obtained from the pause time survival fraction (Figure 1B & C). Although others have reported rolling velocities of platelets or GPIbα coated microspheres on immobilized A1 at ≤ 100s−1 [22], our experience with the low shear regime is that the interaction is so weak that platelets dissociate from the surface captured domain at less than 300s−1. We attribute these differences to the method of immobilization in which our protein is captured by the 6xHis-tag rather than by nonspecific interactions between the A1 domain and glass or plastic surfaces. This method of surface capture by specific chelation chemistry preserves the structure of the domain and allows one to distinguish the effects of different A1 domain conformations on platelet adhesion.
Spectroscopic studies designed to interrogate the globular structure and stability of RCAM A1 confirm that breaking the disulfide linkage abolishes the native state structure. This loss of native structure is evident in 1) the increased wavelength of maximal fluorescence emission; 2) loss of circular dichroism in both the near and far UV; 3) the absence of an N ⇌ I far UV CD transition in the chemical denaturation; 4) the absence of a thermal unfolding transition monitored by CD, FL and ANS; 5) increased intrinsic protein tyrosine and tryptophan fluorescence; 6) increased ANS binding and fluorescence; and 7) an increased Stern-Volmer constant due to collisional acrylamide quenching of solvent exposed tryptophan. Taken together, all of these spectroscopic observables demonstrate that RCAM A1 adopts a molten globule state that is devoid of tertiary structure contacts with exposed fluorophores and hydrophobic residues and only residual secondary structure content remains. Furthermore, ANS is commonly used as a molten globule indicator dye because it has a stronger affinity for proteins with prominent secondary structure but loosely packed tertiary structure elements compared with its affinity for native and random coil proteins [23, 24]. It is commonly observed that molten globule proteins exhibit larger hydrodynamic radii than their native state conformations [25]. While ANS is indicator of a molten globule state due to exposed hydrophobic surfaces, the SEC data show that RCAM A1 has a greater elution time than disulfide-intact A1 suggesting that the macromolecular dimensions of RCAM A1 are smaller than native A1. Near UV CD demonstrates that the tertiary structure of RCAM A1 is largely diminished if not completely absent. We attribute the atypical SEC elution of RCAM A1 relative to native A1 to be due to the presence of significant amounts of supersecondary structure that could alter both the size as well as the shape of the protein in buffer. Small amounts of GdnHCl disrupt these structures due to stacking interactions that cause a greater expansion of RCAM A1 relative to native A1 (Figure 7) [26].
Figure 7.
Size exclusion chromatograms for native disulfide-intact A1 (closed symbols) and RCAM A1 (open symbols) in buffer (circles) and 0.25M GdnHCl (squares). The column was equilibrated with PGA buffer and calibrated with several proteins as indicated by their peak maxima (a - Ovoalbumin from chicken (44.0kDa), b - bovine Carboanhydrase (29kDa), c - Myoglobin from horse (17.0kDa), d cytochrome c from horse (12.4kDa). Void volume = 6.85mL with blue dextran (2000kDa). Included volume 17.54mL with vitamin B12 (1.35kDa).
The co-crystal structure of the disulfide-intact A1 domain in complex with GPIbα shows two hydrogen-bonded contact interactions between 1) the A1-β3 strand and the β-hairpin switch of GPIbα and 2) α1β2 and β3α2 loops of A1 and the N-terminal β-hairpin of GPIbα flanking a highly solvated electrostatic surface area [27]. Aside from the electrostatics, these contact interactions are weak in the presence of shear due to the very few hydrogen bonds that make up the interaction. Furthermore, the GPIbα β-hairpin switch must transition from a coil conformation in the unbound state in order to accommodate binding [27, 28].
The finding that RCAM A1 binds platelets tighter than disulfide-intact A1 is intriguing and counter intuitive. How can loss of structure result in gain of function? Our interpretation is that relaxing the domain structure by reducing the disulfide bond allows the protein to reorganize and form the secondary and supersecondary structural elements that are naturally expressed in the amino acid sequence. These secondary elements are stable in and of themselves and predetermined with a high affinity for the platelet GPIb receptor. Within this context, folding of the A1 domain, by linking the N- and C-terminal cysteines is inhibitory for the A1 GPIbα binding interaction, but not completely. Those secondary elements required for binding remain intact in the disulfide-linked domain and the process of folding and disulfide-bonding induces strain on these elements that reduces GPIb affinity. This results in transient platelet interactions and the lack of stable adhesion to surface-captured disulfide-intact A1, observed as stick and roll translocations in the flow chamber. Releasing these secondary structure elements from the confines of the tertiary fold enables high affinity and high strength bonds with platelets. To support this idea, a small cyclic helical peptide was recently co-crystallized with GPIbα that inhibits the VWF-GPIbα interaction through binding between the concave surface of the leucine-rich repeats and the extended regulatory β-hairpin loop [29]. Although it is not yet clear which secondary structure elements of the molten globule conformation interact with GPIbα, it is plausible that helical regions of A1 could interact in a similar manner as the inhibitor complex. It is therefore probable that the structural basis for the high affinity interaction between GPIbα and the molten globule conformation of A1 could be completely different than the native co-crystal complex portrays.
It is also conceivable that RCAM A1 interacts with other domains of the GPIb-IX-V complex or other receptors on platelet membranes. The VWF A1 domain is already known to bind heparin and collagen in addition to GPIbα and the equilibrium between the native conformation and a dynamic molten globule increases the probability of additional multivalency with respect to new protein ligands. Indeed, specific binding interactions can occur in molten globules. For example, small ligands bind to periplasmic binding proteins in the acid-induced molten globule state without conversion to the native state [30]. Disulfide intact VWF A1 domain also adopts a molten globule conformation at low pH like many other proteins including ribonuclease A, human α-lactalbumin and retinol binding protein [10, 31]. Similar to the A1 domain, α-lactalbumin forms a compact molten globule in the absence of disulfide bonds [32]. Furthermore, the secondary hemostatic chemokine platelet factor 4 (PF4) that is released from activated platelets to promote coagulation though interactions with heparin, also adopts a molten globule conformation in the absence of disulfide bonds [33]. In contrast to RCAM A1, which gains affinity for platelets in the molten globule conformation, PF4 looses affinity for heparin upon reduction of the disulfide bonds.
The high affinity molten globule state adopted by the A1 domain in the disulfide reduced form presents a considerable challenge for primary hemostasis in patients that harbor mutations that break the disulfide bond. Clearly there are both intracellular and extracellular mechanisms involved that reduce the probability of systemic thrombocytopenia. In the case reported for the patient with C1272F, plasma VWF multimer gel analysis showed an absence of high and intermediate molecular weight multimers [1]. However, only high molecular weight multimers were absent in a patient with a C1272S substitution, indicating that VWF can be secreted into the blood as low molecular weight forms [2]. Multimer studies on recombinant VWF containing C1272R and C1272G show an absence of high molecular weight multimers and an absence of both high and intermediate molecular weight species in C1458G VWF [9]. Therefore, cross-linking the disulfide to induce the tertiary folding of A1 is required for efficient multimerization. A free cysteine in the A1 domain, due to the mutation of its partner, has the potential to pair with other cysteines in VWF resulting in incorrect linkages that could alter the quaternary structure required for multimerization. Similarly, other cysteine mutations in the D3 and B domains also result in defective multimerization that induces intracellular retention of VWF causing severe Von Willebrand Disease [34, 35].
Since the single A1 domain with the reduced disulfide is sufficient to arrest translocation of platelets under shear flow, small molecular weight multimers containing an A1 domain cysteine mutation that are secreted could still pose a potential risk for microvascular thrombosis. At the level of the single A1 domain, loss of the disulfide bond is comparable to a severe type 2B gain of function phenotype. In severe cases, Type 2B VWD is also associated with reduced high molecular weight multimers and thrombocytopenia. Because of this functional similarity, ADAMTS13 could have a role in the clearance of affected VWF multimers. Increased levels of VWF pro-peptide relative to mature VWF in 2B VWD indicate an accelerated clearance of mutant VWF from the blood [36, 37]. It has been recently demonstrated in mice that type 2B mutations are associated with an increase in satellite bands in low molecular weight multimers and a decrease in the circulatory half-life of VWF and VWF-platelet complexes [36, 38]. Recombinant type 2B VWF proteolysis by recombinant ADAMTS13 is also enhanced relative to normal VWF [39]. However, evidence to the contrary demonstrates that ADAMTS13 does not proteolyze the A2 domain in recombinant C1272S VWF either in the absence or presence of urea [40]. Furthermore, the kinetics of normal murine VWF clearance from plasma in ADAMTS13 defficient mice was not significantly different from mice expressing ADAMTS13 [36, 41, 42]. Similar to other cysteine mutations [43], it is plausible that A1 domain cysteine mutations could also result in increased clearance of VWF independent of ADAMTS13 due to a disrupted structural integrity of the multimers. Heterozygosity could present some measure of protection since not all monomeric units of multimeric VWF would lack the disulfide bond in the A1 domain. However, it is questionable whether low molecular weight multimers present in patient plasma contain molten globule A1 domains with a cysteine mutation or if they only contain normal disulfide linked A1 domain due to the compensation of the unaffected allele. This would be a reasonable possibility because part of the function of the endoplasmic reticulum (ER) is the retention of misfolded proteins for degradation [44], although not all misfolded proteins are retained in the ER [45].
In conclusion, loss of the A1 domain disulfide bond due to genetic mutation induces a type 2A VWD phenotype because of the loss of mid- to high molecular weight multimeric forms of VWF. We have assessed the functional consequence of a disulfide loss in the single A1 domain and found that it has properties of an extreme gain of function similar to type 2B VWD. This enhanced platelet interaction results from the spontaneous conversion of A1 to a molten globule conformation that retains secondary structural elements which enable high affinity interactions with platelet GPIb. Which secondary structural elements in the A1 domain are involved in this interaction is still under investigation. Despite a severe gain of function at the single A1 domain level, VWD patients afficted with A1 domain cysteine mutations do not show evidence of enhanced platelet-VWF interactions characteristic of type 2B VWD. This is likely a result of a number of possible protective checkpoints occurring throughout the biosynthetic, secretory and clearance pathways of VWF that depend on the proper folding and linkage of the disulfide in the A1 domain.
5 Materials & Methods
5.1 Materials
Tris HCl, GdnHCl, Tween 20 and Na2HPO4 were obtained from Fisher Scientific; sodium acetate and EDTA from ICN Biomedicals; acrylamide from Bio-rad and urea, glycine, NaCl, iodoacetamide, dithiothreitol, N-acetyl-tryptophanamide (NATA) and 8-Anilino-1-naphthalenesulfonic acid (ANS) from Sigma-Aldrich. All chemicals were of analytical grade or higher purity.
5.2 Protein expression, purification, isolation and synthesis
The VWF A1 domain (amino acids Q1238 − P1471) was expressed and purified as described previously [17]. The reduction of A1 in buffer containing 2M GdnHCl with 6mM DTT followed by the carboxyamidation with 12mM iodoacetamide was performed as described previously [12]. A1 was stored on ice in 150mM NaCl, 25mM Tris HCl, pH=7.4 (TBS). RCAM A1 was stored on ice in TBS + 2M GdnHCl. All functional studies were performed in TBS. For all thermodynamic and spectroscopic studies, proteins were dialyzed overnight against PGA Buffer, 10mM sodium acetate, 10mM Na2HPO4, 10mM Glycine, 150mM NaCl, 1mM EDTA, pH=8 before use.
5.3 Concentration determination
Protein, NATA, ANS and peptide concentrations were determined photometrically on a Shimadzu UV2101PC spectrophotometer using the Edelhoch method as modified by Pace [46] from absorption at λ = 280nm minus twice the absorption at λ = 333nm for correction of light scattering. Extinction coefficients for A1 and RCAM A1 (ε = 15350 L/mol/cm) were calculated from eight tyrosines and one tryptophan. The extinction coefficient for NATA is 5630L/mol/cm and the concentration of ANS-solutions was determined at λ = 350nm using an extinction coefficient of 4950L/mol/cm [47].
5.4 Parallel plate flow chamber studies
The determination and analysis of platelet pause times and instantaneous translocation velocities over surface captured A1 and RCAM A1 was performed as previously described [17]. Here, we used the Cellix Mirus Evo syringe pump controlled by VenaFluxAssay Software1. Vena8 CGS biochips were custom designed with 800µM width by 80µM height microchannels bonded to glass slides with a Cu2+ chelated PEG surface serviced by Microsurfaces Inc.2 to which the A1 and RCAM A1 in TBS were captured by the 6xHis-Tag at a total concentration of 5µM. Citrated whole blood was obtained from the informed consent of healthy volunteer donors with approval from the Mayo Clinic Institutional Review Board and 100µL was perfused over the surface captured A1 domains at 700s−1. The shear was increased or decreased in a stepwise manner to measure the dynamics of platelet adhesion at high and low shear. After allowing the flow chamber to equilibrate for 50s at each shear rate, 1min movies were recorded at 24 frames per second in phase contrast at 200× magnification with 2×2 pixel binning on a Zeiss Axiocam Mrm camera (6.45µM/px) attached to a Zeiss Axio Observer D1 inverted microscope. Tracking analysis was performed using MediaCybernetics ImagePro Premier3. The distance travelled in the direction of flow was calculated from the coordinate data for each platelet as a function of time using , where x0 is the coordinates of a platelet on the first frame of the movie and xt is the coordinate on subsequent frames of the movie. These trajectories were differentiated into instantaneous velocities using a Savitzky-Golay algorithm [48] with a five data point window size and a second order polynomial implemented into a Mathematica4 notebook written in our lab. Trajectories were retained for statistical analysis if the platelet was present for ≥ 1s and travelled a total distance greater than 1µM. Distance travelled in pixels (px) was converted to (µm) using 6.45µM/px * 2 / 200 = 0.0645 µm/px. Pause times were determined by the amount of time (sec) a platelets velocity was 0 ± 4px/s ≃ 0 ± 0.23µM/s, within the noise. Data are reported as mean velocities and pause times attained by individual platelets averaged over all platelets analyzed. Integral analysis of the the cumulative pause time histogram gives the survival fraction, equation (2).
| (2) |
where #PTtotal is the total number of pause times counted and #PTt is the total number of pause times that are ≤ to time t. A bi-exponential decay function was fit to the fraction survival data to obtain estimates of the dissociation rates and amplitudes at each shear rate investigated.
5.5 CD spectroscopy
Urea and GdnHCl induced unfolding of A1 and of RCAM A1 at 25°C was monitored via CD at 222 nm on an Aviv Biomedical Model 420C circular dichroism spectrometer. A 1mm quartz cuvette and protein concentrations of 5 or 10µM were used. Samples were equilibrated in their urea or GdnHCl containing buffer at 25°C overnight. CD-signal was averaged for 5 to 10min using an integration time of 1s. CD-spectra were measured at 20°C using 0.1 or 1mm quartz cells with a step width of 1nm between 190 and 260nm with an integration time of 20s and a bandwidth of 1nm. CD-spectra in the near UV range (260 – 360nm) were recorded using a 5cm quartz cell with a 120s integration time. Transition curves and protein spectra were corrected for the CD Signal of the corresponding buffer.
CD thermalscans were recorded between 10 and 95°C at 222nm with a protein concentration of 1µM in a 1cm quartz cell under moderate stirring. Prior to each measurement protein samples were equilibrated at 10°C for 10 – 15min in order to obtain a stable baseline. The scanrate for all Thermalscans was 2°C/min. The integration time for each data point was 20s, the bandwidth 1nm. All measured CD data were converted into mean molar ellipticities per amino acid residue.
5.6 Fluorescence spectroscopy
All fluorescence measurements were performed on a Horiba Jobin-Yvon Fluorolog 3 spectrouorometer equipped with a Wavelength Electronics Model LF1-3751 temperature controller. Protein fluorescence emission spectra of 2µM A1 and RCAM A1 were averaged three times between 305 and 440nm with excitation at 280nm. The step width was 1nm and the integration time 1s. ANS fluorescence emission spectra were averaged three times between 400 and 620nm with excitation at 350nm. Prior to the measurements 1µM A1 or RCAM A1 was incubated for one hour at 20°C with 100µM ANS. All fluorescence spectra were corrected for the signal of the corresponding buffer.
Thermalscans monitoring protein fluorescence were recorded at an emission wavelength of 359nm after excitation at 280nm using a protein concentration of 2µM in a 1cm quartz cell under moderate stirring. ANS-Thermalscans were recorded at 495nm after Excitation at 350nm using 1µM protein and 100µM ANS. Prior to any performed thermalscan samples were equilibrated for 10 – 15min. The scanrate was 2°C/min. At each temperature between 10 or 20°C and 95°C relative fluorescence intensity was collected for 4s and averaged.
Acrylamide quenching of tryptophan was performed using an excitation wavelength of 295nm. After the addition of a certain amount of buffered acrylamide solution the protein solution was equilibrated for ~ 5min and then a fluorescence spectrum between 320 and 400nm was recorded. Stern-Volmer constants were determined from the fluorescence intensity at 359nm using the Stern-Volmer equation as described by Lakowicz [49].
| (3) |
where KD is the Stern-Volmer quenching constant and is the concentration of acrylamide at which F0/F = 2 and 50% of the fluorescence intensity is quenched.
5.7 Analytical size-exclusion chromatography
Analytical size-exclusion chromatography was performed at 4°C using a Superdex 75 column with a total bed volume of 24mL on an Äkta FPLC-system (GE Healthcare). The column was equilibrated with PGA buffer containing 0M, 0.25M and 2M GdnHCl and 500µL of WT A1 (5µM) and RCAM A1 (5µM) were injected followed by elution at 05mL/min flow rate.
Supplementary Material
Acknowlegements
We would like to acknowledge Cellix Limited for the custom design of the biochips used in the flow chamber studies. Also, Jack Aviv for providing a cylindrical cell holder for the Near UV CD. Early in the work, Miguel A. Cruz was helpful in discussions and providing the expression system for the VWF A1 domain, which we gratefully acknowledge. This work was supported by National Heart Lung and Blood Institute of the National Institutes of Health HL109109 to Matthew Auton.
Abbreviations Used
- VWF
Von Willebrand Factor
- VWD
Von Willebrand Disease
- RCAM
Reduced and Carboxy-Amidated
- FL
Fluorescence
- CD
Circular Dichroism
- ANS
8-Anilino-1-naphthalenesulfonic acid
- NATA
N-acetyl-tryptophanamide
- SEC
size exclusion chromatography
- RIPA
ristocetin-induced platelet agglutination
- SIPA
shear-induced platelet agglutination.
Footnotes
Author Contributions: Alexander Tischer performed and analyzed all thermodynamic and spectroscopic studies. Alexander Tischer and Pranathi Madde performed the flow chamber studies and platelet translocation analysis. Luis M. Blancas-Mejia performed the size exclusion chromatography. Matthew Auton designed the research and developed the code for statistical analysis of platelet translocations. Alexander Tischer and Matthew Auton wrote the paper.
References
- 1.Woods AI, Sanchez-Luceros A, Kempfer AC, et al. C1272F: a novel type 2A von Willebrand's disease mutation in A1 domain; its clinical significance. Haemophilia. 2012;18(1):112–116. doi: 10.1111/j.1365-2516.2011.02569.x. [DOI] [PubMed] [Google Scholar]
- 2.Penas N, Perez A, Gonzalez-Boullosa R, Batlle J. C1272S: a new candidate mutation in type 2A von Willebrand disease that disrupts the disulfide loop responsible for the interaction of VWF with platelet GP Ib-IX. Am J Hematol. 2004;75(2):73–77. doi: 10.1002/ajh.10455. [DOI] [PubMed] [Google Scholar]
- 3.Meyer D, Fressinaud E, Gaucher C, et al. Gene defects in 150 unrelated French cases with type 2 von Willebrand disease: from the patient to the gene. INSERM Network on Molecular Abnormalities in von Willebrand Disease. Thromb. Haemost. 1997;78(1):451–456. [PubMed] [Google Scholar]
- 4.Lavergne JM, De Paillette L, Bahnak BR, et al. Defects in type IIA von Willebrand disease: a cysteine 509 to arginine substitution in the mature von Willebrand factor disrupts a disulphide loop involved in the interaction with platelet glycoprotein Ib-IX. Br J Haematol. 1992;82(1):66–72. doi: 10.1111/j.1365-2141.1992.tb04595.x. [DOI] [PubMed] [Google Scholar]
- 5.Azuma H, Hayashi T, Dent JA, Ruggeri ZM, Ware J. Disulfide bond requirements for assembly of the platelet glycoprotein Ib-binding domain of von Willebrand factor. J Biol Chem. 1993;268(4):2821–2827. [PubMed] [Google Scholar]
- 6.Cruz MA, Handin RI, Wise RJ. The interaction of the von Willebrand factor-A1 domain with platelet glycoprotein Ib/IX. The role of glycosylation and disulfide bonding in a monomeric recombinant A1 domain protein. J Biol Chem. 1993;268(28):21238–21245. [PubMed] [Google Scholar]
- 7.Andrews RK, Gorman JJ, Booth WJ, et al. Cross-linking of a monomeric 39/34-kDa dispase fragment of von Willebrand factor (Leu-480/Val-481-Gly-718) to the N-terminal region of the alpha-chain of membrane glycoprotein Ib on intact platelets with bis(sulfosuccinimidyl) suberate. Biochemistry. 1989;28(21):8326–8336. doi: 10.1021/bi00447a010. [DOI] [PubMed] [Google Scholar]
- 8.Miura S, Fujimura Y, Sugimoto M, et al. Structural elements influencing von Willebrand factor (vWF) binding affinity for platelet glycoprotein Ib within a dispase-digested vWF fragment. Blood. 1994;84(5):1553–1558. [PubMed] [Google Scholar]
- 9.Siguret V, Ribba AS, Christophe O, et al. Characterization of recombinant von Willebrand factors mutated on cysteine 509 or 695. Thromb. Haemost. 1996;76(3):453–459. [PubMed] [Google Scholar]
- 10.Miyata S, Goto S, Federici AB, Ware J, Ruggeri ZM. Conformational changes in the A1 domain of von Willebrand factor modulating the interaction with platelet glycoprotein Ibalpha. J Biol Chem. 1996;271(15):9046–9053. doi: 10.1074/jbc.271.15.9046. [DOI] [PubMed] [Google Scholar]
- 11.Miyata S, Ruggeri ZM. Distinct structural attributes regulating von Willebrand factor A1 domain interaction with platelet glycoprotein Ibalpha under flow. J Biol Chem. 1999;274(10):6586–6593. doi: 10.1074/jbc.274.10.6586. [DOI] [PubMed] [Google Scholar]
- 12.Auton M, Zhu C, Cruz MA. The Mechanism of VWF-Mediated Platelet GPIbalpha Binding. Biophys. J. 2010;99(4):1192–1201. doi: 10.1016/j.bpj.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auton M, Sedlak E, Marek J, et al. Changes in thermodynamic stability of von willebrand factor differentially affect the force-dependent binding to platelet GPIbalpha. Biophys. J. 2009;97(2):618–627. doi: 10.1016/j.bpj.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auton M, Cruz MA, Moake J. Conformational Stability and Domain Unfolding of the Von Willebrand Factor A Domains. J. Mol. Biol. 2007;366(3):986–1000. doi: 10.1016/j.jmb.2006.10.067. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Zhang CZ, Zhang X, Springer TA. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 2010;466(7309):992–995. doi: 10.1038/nature09295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas W, Forero M, Yakovenko O, et al. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys. J. 2006;90(3):753–764. doi: 10.1529/biophysj.105.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tischer A, Cruz MA, Auton M. The linker between the D3 and A1 domains of vWF suppresses A1-GPIb catch bonds by site specific binding to the A1 domain. Protein Sci. 2013;22(8):1049–1059. doi: 10.1002/pro.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nozaki Y, Tanford C. The solubility of amino acids, diglycine, and triglycine in aqueous guanidine hydrochloride solutions. J Biol Chem. 1970;245(7):1648–1652. [PubMed] [Google Scholar]
- 19.Thomas W. Catch bonds in adhesion. Annu Rev Biomed Eng. 2008;10:39–57. doi: 10.1146/annurev.bioeng.10.061807.160427. [DOI] [PubMed] [Google Scholar]
- 20.Thomas WE, Vogel V, Sokurenko E. Biophysics of catch bonds. Annu Rev Biophys. 2008;37:399–416. doi: 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- 21.Coburn LA, Damaraju VS, Dozic S, et al. GPIb-vWF rolling under shear stress shows differences between type 2B and 2M von Willebrand disease. Biophys. J. 2011;100(2):304–312. doi: 10.1016/j.bpj.2010.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yago T, Lou J, Wu T, et al. Platelet glycoprotein Ibalpha forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J. Clin. Invest. 2008;118(9):3195–3207. doi: 10.1172/JCI35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin RL, Rose GD. Molten globules, entropy-driven conformational change and protein folding. Current Opinion in Structural Biology. 2013;23(1):4–10. doi: 10.1016/j.sbi.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Semisotnov GV, Rodionova NA, Razgulyaev OI, et al. Study of the molten globule intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991;31(1):119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 25.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim WK, Rösgen J, Englander SW. Urea, but not guanidinium, destabilizes proteins by forming hydrogen bonds to the peptide group. Proc Natl Acad Sci U S A. 2009;106(8):2595–2600. doi: 10.1073/pnas.0812588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huizinga EG, et al. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 28.Uff S, Clemetson JM, Harrison T, Clemetson KJ, Emsley J. Crystal structure of the platelet glycoprotein Ib(alpha) N-terminal domain reveals an unmasking mechanism for receptor activation. J Biol Chem. 2002;277:35657–35663. doi: 10.1074/jbc.M205271200. [DOI] [PubMed] [Google Scholar]
- 29.McEwan PA, Andrews RK, Emsley J. Glycoprotein Ib inhibitor complex structure reveals a combined steric and allosteric mechanism of von Willebrand factor antagonism. Blood. 2009;114:4883–4885. doi: 10.1182/blood-2009-05-224170. [DOI] [PubMed] [Google Scholar]
- 30.Prajapati RS, Indu S, Varadarajan R. Identification and Thermodynamic Characterization of Molten Globule States of Periplasmic Binding Proteins. Biochemistry. 2007;46(36):10339–10352. doi: 10.1021/bi700577m. [DOI] [PubMed] [Google Scholar]
- 31.Vassilenko KS, Uversky VN. Native-like secondary structure of molten globules. Biochim. Biophys. Acta. 2002;1594(1):168–177. doi: 10.1016/s0167-4838(01)00303-x. [DOI] [PubMed] [Google Scholar]
- 32.Redfield C, Schulman BA, Milhollen MA, Kim PS, Dobson CM. Alpha-lactalbumin forms a compact molten globule in the absence of disulfide bonds. Nat Struct Biol. 1999;6(10):948–952. doi: 10.1038/13318. [DOI] [PubMed] [Google Scholar]
- 33.Mayo KH, Barker S, Kuranda MJ, et al. Molten globule monomer to condensed dimer: role of disulfide bonds in platelet factor-4 folding and subunit association. Biochemistry. 1992;31(48):12255–12265. doi: 10.1021/bi00163a040. [DOI] [PubMed] [Google Scholar]
- 34.Tjernberg P, Castaman G, Vos HL, Bertina RM, Eikenboom JC. Homozygous C2362F von Willebrand factor induces intracellular retention of mutant von Willebrand factor resulting in autosomal recessive severe von Willebrand disease. Br J Haematol. 2006;133(4):409–418. doi: 10.1111/j.1365-2141.2006.06055.x. [DOI] [PubMed] [Google Scholar]
- 35.Hommais A, Stepanian A, Fressinaud E, et al. Mutations C1157F and C1234W of von Willebrand factor cause intracellular retention with defective multimerization and secretion. J Thromb Haemost. 2006;4(1):148–157. doi: 10.1111/j.1538-7836.2005.01652.x. [DOI] [PubMed] [Google Scholar]
- 36.Casari C, Lenting PJ, Wohner N. Clearance of von Willebrand factor. Journal of Thrombosis and Haemostasis. 2013 doi: 10.1111/jth.12226. [DOI] [PubMed] [Google Scholar]
- 37.Casonato A, Gallinaro L, Cattini MG, et al. Reduced survival of type 2B von Willebrand factor, irrespective of large multimer representation or thrombocytopenia. Haematologica. 2010;95(8):1366–1372. doi: 10.3324/haematol.2009.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rayes J, Hollestelle MJ, Legendre P, et al. Mutation and ADAMTS13-dependent modulation of disease severity in a mouse model for von Willebrand disease type 2B. Blood. 2010;115(23):4870–4877. doi: 10.1182/blood-2009-11-254193. [DOI] [PubMed] [Google Scholar]
- 39.Rayes J, Hommais A, Legendre P, et al. Effect of von Willebrand disease type 2B and type 2M mutations on the susceptibility of von Willebrand factor to ADAMTS-13 - RAYES - 2007 - Journal of Thrombosis and Haemostasis - Wiley Online Library. J Thromb Haemost. 2007;5(2):321–328. doi: 10.1111/j.1538-7836.2007.02296.x. [DOI] [PubMed] [Google Scholar]
- 40.Hassenpug WA, Budde U, Obser T, et al. Impact of mutations in the von Willebrand factor A2 domain on ADAMTS13-dependent proteolysis. Blood. 2006;107(6):2339–2345. doi: 10.1182/blood-2005-04-1758. [DOI] [PubMed] [Google Scholar]
- 41.Lenting PJ, van Schooten CJM, Denis CV. Clearance mechanisms of von Willebrand factor and factor VIII. J Thromb Haemost. 2007;5:1353–1360. doi: 10.1111/j.1538-7836.2007.02572.x. [DOI] [PubMed] [Google Scholar]
- 42.Badirou I, et al. von Willebrand factor clearance does not involve proteolysis by ADAMTS-13. J Thromb Haemost. 2010;8:2338–2340. doi: 10.1111/j.1538-7836.2010.04012.x. [DOI] [PubMed] [Google Scholar]
- 43.Schooten CJ, Tjernberg P, Westein E, et al. Cysteine-mutations in von Willebrand factor associated with increased clearance. J Thromb Haemost. 2005;3(10):2228–2237. doi: 10.1111/j.1538-7836.2005.01571.x. [DOI] [PubMed] [Google Scholar]
- 44.Lippincott-Schwartz J, Roberts TH, Hirschberg K. Secretory Protein Trafficking and Ogranelle Dynamics in Living Cells. Annu. Rev. Cell Dev. Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kincaid MM, Cooper AA. Misfolded proteins traffic from the endoplasmic reticulum (ER) due to ER export signals. Mol. Biol. Cell. 2007;18(2):455–463. doi: 10.1091/mbc.E06-08-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4(11):2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motono C, Yamagishi A, Oshima T. Urea-Induced Unfolding and Conformational Stability of 3-Isopropylmalate Dehydrogenase from the Thermophile Thermus thermophilusand Its Mesophilic Counterpart from Escherichia coli. Biochemistry. 1999;38(4):1332–1337. doi: 10.1021/bi982380v. [DOI] [PubMed] [Google Scholar]
- 48.Savitzky A, Golay MJE. Smoothing and Differentiation of Data by Simplifies Least Squares Procedures. Anal Chem. 1964;34:1627–1639. [Google Scholar]
- 49.Lakowicz JR. Quenching of Fluorescence. Principles of Fluorescence Spectroscopy. 2006;(8):277–330. [Google Scholar]
- 50.Böhm G, Muhr R, Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.