Abstract
Expression of invasion genes in Salmonella pathogenicity island 1 (SPI-1) is mainly driven by the transcriptional activator HilA. Transcription of hilA is subject to complex control and is stimulated by the SPI-1-encoded HilC and HilD proteins. The C-terminal domain of RpoA contributes to hilA activation by HilC/D under certain inducing conditions.
Salmonella enterica serovar Typhimurium is a facultative intracellular pathogen that causes gastroenteritis in humans and cattle or a systemic disease in mice similar to typhoid fever (reviewed in reference 27). Its ability to invade host intestinal cells is crucial for onset of localized disease following oral ingestion and depends on the production of the type III secretion system encoded in Salmonella pathogenicity island 1 (SPI-1) and of the effector proteins injected by that secretory machine (reviewed in references 11 and 25). Expression of these invasion genes is influenced by many environmental signals, including osmolarity, oxygen level, growth phase, and the presence of fatty acids and bile salts. These cues act through a complex contribution of many regulatory proteins which somehow influence expression of the major SPI-1-encoded transcriptional regulator HilA. The HilA protein has a helix-loop-helix DNA activation domain similar to that in the OmpR/ToxR family (3, 4). It activates expression of the invasion genes present in SPI-1 and elsewhere by a direct action at their promoters and also indirectly by increasing the level of another activator protein, InvF (6). Transcription of hilA is subject to direct or indirect negative control by multiple nucleoid binding proteins, including Hha, H-NS, HU, and Fis (10, 23), and other proteins, including HilE, Pag, and Ams (5). hilA transcription is elevated in response to two SPI1-encoded AraC/XylS family transcription factors, HilC and HilD (8, 20, 22), and to the RtsA protein, which is encoded on another SPI (9). The HilC and HilD proteins bind to their own gene promoters as well as to at least two regions in the hilA promoter, called sites A1 (positions −242 to −182, relative to the start of transcription) and A2 (positions −85 to −61) (18, 24). Although HilC and HilD bind to the same DNA regions, they have different requirements with respect to the sequence and length of their DNA target (18).
Various models for HilC/D action at the hilA promoter have been proposed from analysis of hilA-lacZ reporters. The expression of 5′ truncations of the hilA upstream region suggested that negative elements acted upstream of the promoter and that their repressive effect might be overcome by the HilC/D proteins (22, 23). In contrast, Boddicker et al. (5) found that the HilD protein was required to activate hilA-lacZ transcription even when several negative regulators, Hha, Ams, HilE, and/or Pag, were deleted in combination. Because activation by AraC family members often requires interaction with the C-terminal domain of the α subunit (α-CTD) of RNA polymerase (RNAP) (examples in references 7, 12, and 15), they also showed that the α-CTD L289F variant resulted in greatly decreased hilA-lacZ expression in vivo. Thus, these different studies suggested that HilD could act at the hilA promoter either as a direct activator or indirectly by relieving the action of repressors.
Investigation of the effect of HilC/D on in vitro transcription at the hilA promoter might help resolve these apparently conflicting models by testing whether these proteins affect promoter activity in the absence of other regulatory proteins. We show here that the hilA promoter is active in the absence of repressive proteins, but both HilC/D proteins can further stimulate this activity. The importance of the HilC/D-binding sites in the promoter is determined. The effect of alanine substitutions in α-CTD confirms a role of this domain in activation of hilA during in vivo and in vitro expression, but dependence on this domain was found only under certain inducing conditions.
In vitro transcription of the hilA promoter.
The in vitro transcription of the hilA promoter by Escherichia coli RNA polymerase and purified His-tagged versions of HilC and HilD was carried out as described previously (18). Plasmids philA-242+90txn, philA-100+90txn, and philA-55+90txn were used as DNA templates. These plasmids carry hilA promoter sequences in which the upstream ends are at positions −242, −100, and −55, respectively, and the downstream ends are at +90. These fragments were amplified by PCR using pINO3 plasmid DNA (18) as template and Vent polymerase, as specified by the manufacturer (New England Biolabs). Sequences of the PCR primers are available upon request; they introduce EcoRI and HindIII sites at the upstream and downstream ends, respectively. Each fragment was ligated into EcoRI-HindIII-digested plasmid pSR (17), in which the strong Rho-independent rrnB transcription terminator is downstream of the HindIII site. The sequences of all DNA inserts were verified by automated sequencing at the University of Virginia Biomolecular Research Facility.
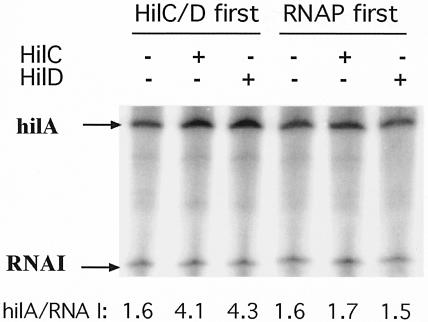
The effect of the order of addition of the transcription factors HilC/D and RNAP in single-round transcription of the hilA promoter was examined (Fig. 1). Supercoiled philA-242+90txn plasmid DNA was incubated first with saturating levels of either HilC/D or RNAP for 6 min prior to addition of the other components, RNAP or HilC/D, respectively. After another 6 min, nucleoside triphosphates (NTPs) were added together with heparin to final concentrations of 1 nM DNA template; 50 nM HilC or HilD; 0.25 U of E. coli RNAP (U.S. Biochemicals [USB]); 50 μg of heparin (Sigma)/ml; ATP, CTP, and GTP at 200 μM; and 40 μM [α-32P]UTP (2.5 Ci/nmol) in a final volume of 25 μl. After 15 min, the reaction was terminated and the products were resolved and quantified by PhosphorImager and ImageQuant analysis (Molecular Dynamics) as previously described (18). The amount of the hilA transcript of the expected size (22) was expressed relative to the level of the RNA I transcript produced from the plasmid vector in each transcription reaction. Each experiment was repeated at least three times and representative results are shown.
FIG. 1.
Effect of order of addition of transcription factors on in vitro transcription of the hilA promoter. Plasmid philA-242+90txn DNA (1 nM) was first incubated with purified His-tagged HilC or HilD protein (50 nM) or with E. coli RNAP (0.25 U; USB) for 6 min, as indicated. RNAP, HilC/D, or buffer, respectively, was then added. After another 6 min, NTPs were added together with heparin and single-round runoff transcription was allowed to proceed for 15 min. Amounts of the hilA and RNA I transcripts were determined by PhosphorImager quantitation, and their ratios are given at the bottom of each lane.
The hilA promoter was efficiently transcribed by RNAP alone in the absence of other proteins (Fig. 1). This high basal activity of the hilA promoter was consistent with its 9-of-12-position match to the promoter consensus, namely, TTTACA-N16-TAAGAT. Addition of either HilC or HilD protein stimulated the production of the hilA transcript by two- to threefold when they were incubated with the DNA before (Fig. 1) or together (data not shown) with RNAP. In replicate experiments, the average degree (± standard deviation) of stimulation by HilC, HilD, or both was 2.77 ± 0.96, 2.35 ± 0.87, and 2.75 ± 1.05, respectively. Both proteins were almost equally effective in vitro, with HilC slightly more active. No stimulation occurred when the HilC/D proteins were added 6 min after RNAP (Fig. 1). No transcript was seen in the absence of RNAP, and the level of transcription and the extent of stimulation by the HilC/D proteins were greatly diminished when a linear DNA template was used (data not shown). The low basal activity of the promoter in vivo indicates the contribution of negative factors which are not present in the in vitro system. However, the finding that HilC/D proteins are not required for transcription from the hilA promoter is in contrast to the strong requirement for HilD for hilA-lac expression in cells lacking some of the negative factors (5).
Requirement for HilC/D-binding sites.
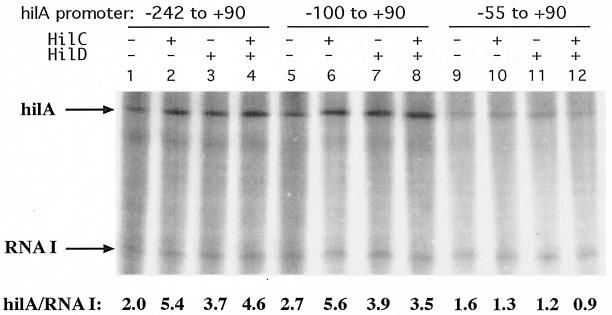
To examine the extent of the promoter region that is required for activation by HilC/D, the in vitro expression from templates carrying different lengths of the upstream region were compared. The basal level of transcription and the degree of stimulation by HilC or HilD were not significantly different for promoter regions which begin at −242, possessing HilC/D-binding sites A1 and A2; at −186 (data not shown); and at −100, possessing only site A2 (Fig. 2). The −55 to +90 template, which lacks site A2, showed a moderate decrease in basal activity and no stimulation by HilC/D. Thus, the presence of site A2 was sufficient for stimulation by HilC/D of in vitro transcription.
FIG. 2.
Effect of upstream DNA on stimulation of hilA transcription by HilC and HilD proteins. Plasmid philA-242+90txn, philA-100+90txn, or philA-55+90txn (1 nM) was first incubated with purified HilC or HilD protein (50 nm) for 6 min, as indicated, before addition of E. coli RNAP (0.25 U; USB). After another 6 min, NTPs were added together with heparin and single-round runoff transcription was allowed to proceed for 15 min. Amounts of the hilA and RNA I transcripts were determined by PhosphorImager quantitation, and their ratios are given.
Comparison to hilA-lacZ expression.
For comparison with the in vitro activities, three promoter fragments carrying the hilA promoter region beginning at positions −242, −100, or −55 and extending to +505 were fused to the lacZ gene in the moderate-copy-number plasmid pRS415P (18, 26). The longer length of the transcribed region was chosen to include the entire hilA leader which has significant impact on in vivo but not in vitro expression (unpublished data). These plasmids were introduced into serovar Typhimurium strain SL1344 and its ΔSPI-1 derivative, SD11. Levels of β-galactosidase were determined after cells were grown under inducing conditions of static culture in Luria-Bertani medium (LB) plus 1% NaCl.
Expression of the full-length promoter fragment (−242 to +505) was stimulated roughly 24-fold by the presence of SPI-1, presumably reflecting the action of HilC and HilD (Table 1). A similar, 14-fold SPI-1-dependent stimulation was seen with the fragment truncated to −100. The roughly twofold difference in SPI-1-independent activity when comparing these two promoter fragments might suggest that some negative factors might bind in the upstream region between −242 and −100. Consistent with the in vitro expression, stimulation was almost absent in the fragment truncated to −55. The low activity remaining in the strains with SPI-1 deleted could reflect stimulation by RtsA (9) or the residual activity of the silenced promoter. The degree of stimulation by the presence of SPI-1-encoded factors seen in vivo was much greater than the two- to threefold stimulation by the HilC/D protein in vitro. These results are consistent with the action of HilC/D both as direct activators and as derepressors when the negative factors are present.
TABLE 1.
Effect of HilC/D binding sites A1 and A2 on β-galactosidase expression from hilA-lac fusion strains grown under inducing conditionsa
| Plasmid | SPI genotype | β-Galactosidase activity |
|---|---|---|
| philA-242 + 505L | + | 673 ± 80 |
| Δ | 34 ± 4 | |
| philA-100 + 505L | + | 855 ± 108 |
| Δ | 65 ± 2 | |
| philA-55 + 505L | + | 105 ± 11 |
| Δ | 79 ± 9 | |
| pRS415P (vector) | + | 6 ± 0.4 |
Plasmids philA-242+505L, philA-100+505L, and philA-55+505L were constructed by cloning the appropriate PCR-generated fragments of the hilA promoter into plasmid pRS415P to generate lacZYA transcription fusions. The plasmids were introduced by electroporation into serovar Typhimurium strains SL1344 (SPI1+) and SD 11 (ΔSPI1). Cells were grown overnight in LB plus 1% NaCl in capped, static culture tubes at 37°C to an A650 of around 0.6. β-Galactosidase activity was measured in triplicate by continuous assay in a microplate reader. Activity is expressed as ΔA415 per minute per A650.
Effect of changes in α-CTD on hilA expression.
The α-CTD domain of RNAP is involved in the response to AraC family and other transcription activators (15). Presence of the α-CTD L289F variant decreased hilA expression in vivo (5). To extend these findings on the role of this domain, we tested the response to expression of a series of alanine substitutions in α-CTD. Elevated expression of an RpoA variant allows it to compete with wild-type RpoA for assembly into RNAP holoenzyme. If the variant is altered at a residue important for the response to an activator, this expression results in diminished transcription of the target promoter (14). In preliminary experiments (data not shown), we tested the effect of overexpression of RpoA with the entire CTD beyond residue 256 deleted on hilA-lacZ activity under a variety of growth-inducing conditions. These conditions included combinations of different pHs and levels of NaCl, sodium acetate, or bile salts. The only condition under which we found that overexpression of rpoAΔ265 impaired hilA-lacZ expression was LB, pH 7.2, containing 1.5% bile salts (Sigma). A plasmid library of 69 E. coli rpoA mutants expressing alanine substitutions between residues 255 and 329 under lac control (provided by T. Gaal and R. Gourse) was screened in serovar Typhimurium strain TF79 hha::Km hilA::Tn5lacZY (provided by B. Jones) grown overnight in static capped tubes at 37°C in that medium with ampicillin at 100 μg/ml and 1 mM isopropyl-β-d-thiogalactopyranoside. This strain lacks the potent negative factor Hha and exhibits elevated hilA expression (5). Although the rpoA variants are from E. coli, the sequences of wild-type rpoA of E. coli and Salmonella are identical, and we are not aware of cases where the behavior of RNAP from these species differs.
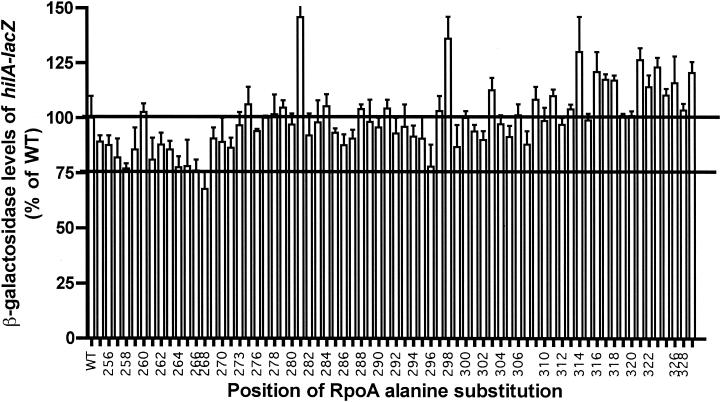
Expression of some rpoA variants affected hilA-lacZ expression. Alanine substitutions at positions 258, 264 to 268, and 296 (group 1) decreased hilA expression by 20 to 30% (Fig. 3). Less-pronounced reduction (10 to 19%) occurred with other substitutions (group 2), including those at residues 257, 259, 261, 263, 271, 286, 287, and 299. A few substitutions resulted in elevated expression. The decrease in hilA expression upon expression of rpoA variants was much less prominent than was seen with other α-CTD-dependent activators, such as CAP, UhpA, and MetR (13, 19, 21). This modest decrease is consistent with the high level of HilC/D-independent transcription that was seen in the in vitro assays.
FIG. 3.
Effect of alanine substitutions at positions 259 to 329 in α-CTD on hilA-lacZ expression in vivo. Values of β-galactosidase activity are averages from three independent experiments and are graphed as percentages of the wild-type (WT) RpoA value (96 U). Error bars represent standard deviations. For clarity, the positions where Ala is the native residue were removed and the odd-numbered residue coordinates were deleted from the axis legend.
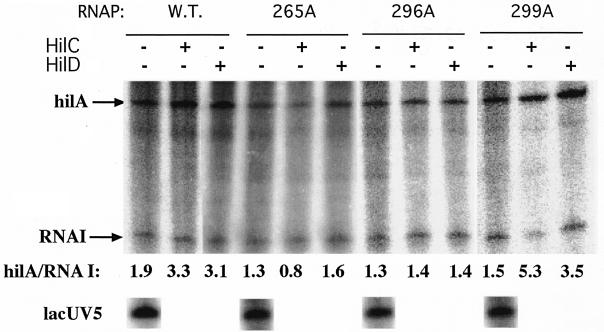
To test whether the decrease in β-galactosidase levels reflects impaired transcription by RNAP with the variant α-CTD, in vitro transcription was carried out with three reconstituted RNAP holoenzymes carrying alanine substitutions at RpoA positions 265 and 296 (group 1) and at 299 (group 2). Purification of these reconstituted RNAPs was described previously (19). The level of transcripts from the α-CTD-independent lacUV5 and RNA I promoters provided a measure of RNAP function. Under conditions where HilC/D stimulated hilA transcription by wild-type RNAP, the RNAP holoenzyme carrying the RpoA R265A and G296A substitutions showed a somewhat decreased basal activity and no stimulation by HilC/D (Fig. 4). These results were consistent with the modest decrease in hilA transcription upon overexpression of the RpoA variants in vivo. The S299A substitution, which was moderately impaired in vivo, allowed stimulation by HilC/D to a comparable degree as the wild-type enzyme. Positions 265 and 296 of RpoA contribute to the DNA-binding surface of α-CTD (14, 16). These results show that α-CTD is involved in activation by HilC/D, probably from their binding site centered at position −73, but is not required for HilC/D-independent expression. It was noteworthy that the only inducing condition where we found a dependence on α-CTD included bile salts as inducer, consistent with the complex control of this promoter and its response to many signals and regulatory proteins.
FIG. 4.
Transcription in vitro of the hilA and lacUV5 promoters by wild-type (W.T.) E. coli RNAP and reconstituted RNAPs carrying the indicated alanine substitutions in RpoA. DNA of plasmid philA-242+90txn (1 nM) was incubated with purified HilC or HilD protein (50 nm) for 6 min. E. coli wild-type or mutant RNAP was then added. After another 6 min, NTPs were added together with heparin and single-round runoff transcription was allowed to proceed for 15 min. Amounts of the hilA and RNA I transcripts were determined by PhosphorImager quantitation. The control promoters lacUV5 and RNA I are independent of α-CTD.
Conclusions.
The in vitro transcription properties of the hilA promoter confirm the complex character of its regulation. This promoter is active in the absence of repressive proteins and is only moderately (two- to threefold) stimulated by the HilC or HilD proteins. Both Hil proteins conferred similar levels of stimulation. Their activation of the promoter may play a secondary role to fine-tune hilA expression, compared to their involvement in the relief of the repression or silencing imposed by several negative factors. In addition, the activators can affect the invasion genes directly in addition to their effect on HilA levels (1, 9). This multiplicity of regulatory effectors complicates analysis of in vivo expression and points out the value of both reporter and biochemical description of this promoter. Our findings show that α-CTD is important for HilC/D stimulation, but the overall effect of overexpression of α-CTD on hilA-lacZ expression was low, consistent with the model that there are multiple modes of regulation and that activation may be less important a contributor than relief of repression. The repressive proteins and the HilC/D and RtsA activators may respond to different environmental signals (2, 9, 23). The reduction in hilA-lac expression by overexpression of the RpoA variant with the CTD deleted was seen only in cells grown in the absence of the repressive Hha protein and in the presence of the inducer bile salts. We cannot conclude how HilC/D proteins participate in hilA derepression, which will require analysis of the effect on transcription of both the positive- and negative-acting proteins. We have found that the hilA promoter has a more complex structure than previously recognized, with additional sites for regulatory intervention (unpublished data). In summary, stimulation of hilA transcription by the AraC/XylS family members HilC and HilD does exhibit the expected dependence on residues in α-CTD, but activation of transcription involves other α-CTD-independent processes as well. The very strong decrease in hilA-lacZ expression in the presence of the RpoA L289F variant is much different from that seen with the alanine substitutions used here and could reflect multiple effects on other regulatory factors.
Acknowledgments
We thank Rick Gourse, Tamas Gaal, Catherine Lee, and Bradley Jones for providing strains and plasmids used in this study.
This work was supported by research grant GM38681 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47:715-729. [DOI] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyetomo, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 68:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 4.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 5.Boddicker, J. D., B. M. Knosp, and B. D. Jones. 2003. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 185:525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egan, S. M. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 184:5529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fierer, J., and D. G. Guiney. 2001. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 107:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finney, A. H., R. J. Blick, K. Murakami, A. Ishihama, and A. M. Stevens. 2002. Role of the C-terminal domain of the alpha subunit of RNA polymerase in LuxR-dependent transcriptional activation of the lux operon during quorum sensing. J. Bacteriol. 184:4520-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritsch, P. S., M. L. Urbanowski, and G. V. Stauffer. 2000. Role of the RNA polymerase α subunits in the MetR-dependent activation of metE and metH: important residues in the C-terminal domain and orientation requirements within RNA polymerase. J. Bacteriol. 182:5539-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaal, T., W. Ross, E. E. Blatter, H. Tang, X. Jia, V. V. Krishnan, N. Assa-Munt, R. H. Ebright, and R. L. Gourse. 1996. DNA-binding determinants of the α subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 10:16-26. [DOI] [PubMed] [Google Scholar]
- 15.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeon, Y. H., T. Negishi, M. Shirakawa, T. Yamazaki, N. Fujita, A. Ishihama, and Y. Kyogoku. 1995. Solution structure of the activator contact domain of the RNA polymerase α subunit. Science 270:1495-1497. [DOI] [PubMed] [Google Scholar]
- 17.Kolb, A., D. Kotlarz, S. Kusano, and A. Ishihama. 1995. Selectivity of the Escherichia coli RNA polymerase E sigma 38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 23:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olekhnovich, I. N., and R. J. Kadner. 1999. RNA polymerase α and σ70 subunits participate in transcription of the Escherichia coli uhpT promoter. J. Bacteriol. 181:7266-7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savery, N. J., G. S. Lloyd, M. Kainz, T. Gaal, W. Ross, R. H. Ebright, R. L. Gourse, and S. J. Busby. 1998. Transcription activation at class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase α subunit. EMBO J. 17:3439-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 23.Schechter, L. M., S. Jain, S. Akbar, and C. A. Lee. 2003. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 71:5432-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40:1289-1299. [DOI] [PubMed] [Google Scholar]
- 25.Scherer, C. A., and S. I. Miller. 2001. Molecular pathogenesis of Salmonellae, p. 265-333. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 26.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, S., R. A. Kingsley, R. L. Santos, H. Andrews-Polymenis, M. Raffatellu, J. Figueiredo, J. Nunes, R. M. Tsolis, L. G. Adams, and A. J. Baumler. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect. Immun. 71:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]