Abstract
Aims
Crates used to transport live poultry can be contaminated with Campylobacter, despite periodic sanitization, and are potential vectors for transmission between flocks. We investigated the microbial contamination of standard and silver ion containing crates in normal use and the genetic structure of associated Campylobacter populations.
Methods and Results
Bacteria from crates were enumerated by appropriate culture techniques, and multilocus sequence typing (MLST) was used to determine the genetic structure of Campylobacters isolated from standard and silver ion containing crates. Compared to standard crates, counts of bacteria, including Campylobacter, were consistently lower on silver ion containing crates throughout the decontamination process. In total, 16 different sequence types were identified from 89 Campylobacter jejuni isolates from crates. These were attributed to putative source population (chicken, cattle, sheep, the environment, wild bird) using the population genetic model, structure. Most (89%) were attributed to chicken, with 22% attribution to live chicken and 78% to retail poultry meat. MLST revealed a progressive shift in allele frequencies through the crate decontamination process. Campylobacter on crates survived for at least 3 h after sanitization, a period of time equivalent to the journey from the processing plant to the majority of farms in the catchment, showing the potential for involvement of crates in transmission.
Conclusions
Inclusion of a silver ion biocide in poultry transportation crates to levels demonstrating acceptable antibacterial activity in vitro reduces the level of bacterial contamination during normal crate use compared to standard crates. Molecular analysis of Campylobacter isolates indicated a change in genetic structure of the population with respect to the poultry-processing plant sanitization practice.
Significance and Impact of the Study
The application of a sustainable antimicrobial to components of poultry processing may contribute to reducing the levels of Campylobacter circulating in poultry.
Keywords: antimicrobial, Campylobacter, chicken transportation crates, genetic diversity, MLST
Introduction
Campylobacter is the most common zoonotic cause of bacterial gastroenteritis in the industrialized world with the financial burden estimated at £500 million in the United Kingdom (Humphrey et al. 2007) and $8 billion in the United States (Buzby and Roberts 1997). Human campylobacteriosis occurs after consumption of contaminated meat, water and milk (Friedman et al. 2000; Kapperud et al. 2003; Neimann et al. 2003), and poultry meat is implicated as the major source of disease, accounting for 40–80% of cases (Corry and Atabay 2001; Vellinga and Van Loock 2002; Neimann et al. 2003; Friedman et al. 2004; Wilson et al. 2008; Sheppard et al. 2009a). The widespread occurrence of Campylobacter jejuni and Campylobacter coli in the digestive tract of farmed chickens and the automated methods used to slaughter and present these birds to retail outlets help to explain the contribution of poultry meat as a source of human campylobacteriosis. Poultry-processing plants have also been implicated as a source of contamination as flocks that are free of Campylobacter at slaughter may produce Campylobacter-positive meat after processing (Johnsen et al. 2006). The crates used to transport live birds from farms to processing have been shown to harbour Campylobacter spp. (Slader et al. 2002; Hansson et al. 2005; Allen et al. 2008; Peyrat et al. 2008). Because these crates circulate between unrelated flocks, they may present a contamination risk for uninfected birds (Rivoal et al. 1999; Newell et al. 2001; Miwa et al. 2003; Posch et al. 2006; Van Worth et al. 2006; Allen et al. 2007). The cleaning of equipment involved in poultry processing, including transportation crates, is ineffective (Wedderkopp et al. 2001), and better decontamination of these crates may be expected to reduce the numbers of pathogens circulating between flocks.
Molecular biological studies have enhanced understanding of the distribution and survival of Campylobacter genotypes in the environment and at various stages in the food chain (Berndtson et al. 1996a; Herman et al. 2003; Lindmark et al. 2006; Allen et al. 2007; Klein et al. 2007; Lienau et al. 2007). Multilocus sequence typing (MLST) has revealed sufficient genetic variation within Camp. jejuni and Camp. coli to identify host- and source-associated genotypes (Manning et al. 2003; McCarthy et al. 2007; Sheppard et al. 2009b; Colles et al. 2010). This allowed the development of source attribution studies to infer the origin of disease. By comparison of sequence types (STs) from clinical isolates with those from reference datasets, chicken-associated lineages are consistently found to be major sources of human disease (Wilson et al. 2008; Mullner et al. 2009; Sheppard et al. 2009b; Strachan et al. 2009). Variation in Campylobacter genotypes throughout poultry processing could provide insight into the contamination of retail meat, improving the understanding of the transmission routes to human infection. This would act as a guide for the implementation of effective intervention measures to reduce the burden of human disease (Baker et al. 2006).
In this study, we investigated the incidence and genetic structure of Campylobacter populations contaminating crates used to transport live chickens. Standard and silver ion containing crates were examined throughout the decontamination process, providing information on the epidemiology and transmission of Campylobacter through the poultry-processing plant.
Materials and methods
Environmental site and samples
Samples were collected from two types of polyethylene chicken transportation crate (1165 mm length, 765 mm width, 255 height, 10 kg weight; Anglia Autoflow, Diss, UK), one standard crate and the other incorporating antimicrobial silver ions (BioCote Ltd, Wolverhampton, UK). The silver ions were incorporated into the crate polymer as a 1% (w/w) masterbatch addition during crate moulding to produce a final concentration of Ag+ of 16 ppm. This concentration of Ag+ had previously been empirically determined using similar polymers and the same silver ion additive in products unrelated to this study to achieve acceptable antimicrobial efficacy (data not shown). Ten silver ion-treated crates entered the chicken processing system during early February 2009 and were used continuously for the transportation of live chickens from farms to the UK processing factory for at least 4 weeks before sampling commenced.
Samples were collected on site from the ten silver ion containing crates and ten standard crates at various stages through the crate decontamination process in March 2009 and included swabs from crates and wash water. Six Amies charcoal transport swabs (TCS, Heywood, UK) were collected from each of the ten treated and untreated crates, swabbing an area of 25 cm2 on the inside surface of empty crates avoiding visible organic material. Of these six swabs, three were later processed in the laboratory for aerobic colony counts and three processed for Campylobacter isolation. Swabs were collected at defined points throughout the decontamination process. Swabbing points were as follows: (i) before the water wash (immediately following live bird removal); (ii) after the water wash; (iii) after peracetic acid sanitization; (iv) 1 h after sanitization; (v) 2 h after sanitization; (vi) 3 h after sanitization (representing the approximate maximum journey time from processing plant to next farm for bird collection. This 3-h point also represented the microbiological state of crates before live birds were placed in them). Crates are not normally stored after sanitization, rather loaded immediately for the collection of more live birds. Samples of wash water (100 ml) were collected in sterile, screw-topped containers at hourly intervals throughout the sampling programme from the wash tank. After washing and sanitization, crates were stored in metal-framed modules (routinely used for transporting crates) for 3 h. On the day of sampling, it was noted that crates stored in the modules and exposed to the open air had dried considerably after washing, but not completely. Collected swabs were labelled and stored in chilled containers prior to transportation to the laboratory.
Crate decontamination
The standard decontamination procedure for transportation crates on the processing plant used for sampling was as follows: emptied crates passed through an unheated mains-supplied wash tank by conveyer belt taking approx. 10 s. The wash tank was filled with fresh water at ambient temperature at the beginning of each day and not refreshed or treated. There was no physical action on the crates during the washing stage; crates were fully submerged and not dried before progressing to the next stage. Within 20 s of emerging from the wash water, crates entered a short-soak metal housing (approx. 5 s duration without agitation) for spraying with 0·25% peracetic acid (Holchem Laboratories, Preston, UK). On exiting the housing, crates were manually slotted into metal holding modules for return to transportation trailers. We estimated in excess of 1000 empty crates had passed through the wash water tank by the time sampling commenced.
Aerobic colony counts
Swabs were processed for aerobic colony counts by immersing the swab tips in 1 ml sterile phosphate-buffered saline (pH 7·0) and vortexing for 20 s. The bacterial suspensions were serially diluted to a factor of 10−6, and 0·1 ml volumes of each dilution were inoculated onto plate count agar (Oxoid, Basingstoke, UK) and incubated for up to 48 h at 37°C. Mean colony counts were calculated from the cultures of the three swabs collected from individual crates.
Campylobacter isolation, identification and enumeration
Tips of swabs collected for Campylobacter isolation were broken off into sterile bijous filled with Bolton broth containing a selective supplement (Oxoid), vortexed for 20 s and incubated for 4 h at 37°C, followed by further incubation at 42°C. After 24 h, 0·1 ml of the inoculated broth was subcultured onto modified CCDA (mCCDA) (Oxoid) and incubated for 48 h at 42°C in 10% CO2. Presumptive Campylobacter colonies were Gram-stained and tested by the oxidase reaction (HPA 2005). For Campylobacter enumeration, 0·1 ml of Bolton broth was withdrawn from the bijou containing the swab tip after 24-h incubation and serially diluted in fresh Bolton broth to a dilution of 1 : 10 000. A volume of 0·1 ml of each dilution was subcultured onto modified CCDA, spread with a sterile glass spreader and incubated for 48 h at 42°C in 10% CO2. Campylobacter-like colonies were identified by Gram stain and oxidase reaction. Colonies were counted at the appropriate dilution and converted to colony forming units (CFUs) per swab.
DNA extraction and PCR sequencing
Single Campylobacter colonies were subcultured on mCCDA plates to obtain pure cultures for DNA sequencing. Half of the colonial growth from a single plate was harvested from each isolate and suspended in 0·15 ml of PBS in a sealable 1·5-ml Eppendorf tube. Cell suspensions were heated at 100°C for 10 min to obtain a final DNA concentration of 10–30 ng μl−1. DNA concentrations were measured by UV spectroscopy (Nanodrop 1000, Thermo Scientific, Cramlington, UK).
PCR, sequencing and bioinformatics
DNA template arrays were created in 96-well Thermo-fast®, polyethylene plates (Abgene, Epsom, UK), and seven-locus MLST was carried out by standard methods using published primers (Dingle et al. 2005; Miller et al. 2005). Each 25 μl PCR comprised molecular grade water (Sigma-Aldrich, Poole, UK), 2·5 μl 10× PCR buffer (Qiagen, Crawley, UK), 0·25 μmol l−1 each of forward and reverse primer, 0·2 mmol l−1 dNTP mix (Invitrogen, Paisley, UK), 0·025 units μl−1 (0·125 μl) Taq polymerase (Qiagen) and 2 μl of template DNA. The PCR thermal cycle began with a 15 min denaturation step at 95°C, followed by 35 cycles of 94°C (30 s), 50°C (30 s) and 72°C (1 min), with a final extension at 72°C (5 min). Five microlitres of PCR products was visualized with ultraviolet transillumination following electrophoresis at 200 V (10 min) on a 1% (w/v) agarose gel containing ethidium bromide (10 mg ml−1) in 1× TBE buffer (1 mmol l−1 EDTA, 40 mmol l−1 Tris-acetate). The amplification products were purified by precipitation with 20% polyethylene glycol–2·5 mol l−1 NaCl (Embley 1991) and stored at −20°C. Nucleotide sequencing PCRs were performed in both directions, using the same primers (f or r), diluted (1:15) in water. Reactions were carried out in 10 μl volumes containing 2 μl of PEG-precipitated DNA resuspended in water, 1·0 μl 5× buffer, 0·02 μl BigDye Terminator v3·1 mix (Applied Biosystems, Warrington, UK) and 0·25 μmol l−1 of either the forward or the reverse primer. Cycling parameters were as follows: 30 cycles of 96°C (10 s), 50°C (5 s) and 60°C (2 min). Unincorporated dye terminators were removed by precipitation of the termination products with 95% ethanol, and the reaction products were separated and detected with an ABI Prism 3730 automated DNA sequencer (Applied Biosystems). Forward and reverse sequences were assembled from the resultant chromatograms using the staden suite of computer programs (Staden 1996). The consensus sequences were queried against the Campylobacter database to give an allele number. The combination of alleles for the seven housekeeping genes gave the STs that were assigned into genetically related clonal complexes, based on sharing four or more alleles with the central genotype. A database was developed in the framework provided by the existing Campylobacter profiles database, [http://pubmlst.org/Campylobacter/] which covers the species Camp. jejuni and Camp. coli and is based on mlstdbNet software (Jolley et al. 2004).
Published data
Data from this study were compared with data from published studies comprising a comparison dataset for analysis containing, in total, 598 cattle, 1312 chicken (195 from live bird faeces and 1117 from poultry meat), 250 sheep, 91 environmental (sand and environmental waters) and 609 wild bird (1 jackdaw, 1 crow, 2 swan, 8 duck, 43 gull, 67 pigeon, 180 goose, 284 starling and 23 unknown birds) Camp. jejuni isolates (McCarthy et al. 2007; Colles et al. 2008a; Sheppard et al. 2009a). The details of these datasets are given in Table 1.
Table 1.
Multilocus sequence typing and isolate data from published sources
| Source | Number of isolates | Number of STs | Number of clonal complexes | Date | References |
|---|---|---|---|---|---|
| Chicken live (faeces) | 195 | 25 | 12 | 2005–2006 | Wedderkopp et al. 2001; Sheppard et al. 2009b |
| Chicken dead (meat) | 1117 | 224 | 28 | 2004–2006 | Rivoal et al. 1999; Wedderkopp et al. 2001 |
| Wild bird | 609 | 159 | 22 | 2002–2006 | Rivoal et al. 1999; Colles et al. 2008b |
| Cattle | 598 | 130 | 19 | 1981–2003 & 2005/2006 | Rivoal et al. 1999; Lienau et al. 2007 |
| Sheep | 250 | 72 | 12 | 1982–2003 & 2005/2006 | Rivoal et al. 1999; Lienau et al. 2007 |
| Environment | 91 | 71 | 17 | 2003, 2005 & 2005/2006 | Rivoal et al. 1999; Wittwer et al. 2005; Wilson et al. 2008 |
All isolates from UK sources.
DNA sequence analysis
The similarity of the isolate genotypes from crates to published datasets of Campylobacter, from chickens and other host species, was assessed by probabilistic assignment of genotypes to host species based on MLST alleles. This analysis used the no-admixture model of structure, a Bayesian model-based clustering method designed to infer population structure and assign individuals to populations using multilocus genotype data (Pritchard et al. 2000; McCarthy et al. 2007). Differences in genotype frequency between populations allow probabilistic assignment of isolates to populations, even if there is some sharing of genotypes between those populations. MLST data were analysed as allelic profiles, and 10 000 iterations were performed following a 1000 iteration burn-in. In each analysis, an external set of chicken crate-derived isolates was assigned to host based on genotype information using a training set with isolates from known host species and environmental origin as outlined in the following sections. These were distinguished from the test data using the usepopinfo flag of structure. Two separate analyses were carried out in which the STs from isolates sampled from poultry transport crates were assigned to host population using (i) 2860 isolates from chicken, cattle, sheep, the environment and wild birds and (ii) 1312 chicken only isolates from chicken faeces and meat.
In vitro antimicrobial efficacy determination
Antimicrobial efficacy of silver ion-treated crates was determined by standard methodology. This involved subjecting triplicate 5 × 5 cm sample coupons of silver ion-treated polyethylene, cut from manufactured chicken crates, to the ISO 22196 protocol. Test bacterial strains were methicillin-resistant Staphylococcus aureus (MRSA) NCTC 11939 and Escherichia coli ATCC 8739.
Live-Dead fluorescence staining
A single Campylobacter colony was randomly chosen from each of three mCCDA plates containing Campylobacter isolates. These colonies were isolated from different prewash silver ion-treated crates. They were suspended together in Bolton broth/PBS (1 : 500) to an approximate density of 1·0 × 106 cells per ml. Sample coupons (5 × 5 cm) of silver ion-treated polyethylene cut from manufactured chicken crates were inoculated with 0·1 ml of this cell suspension in triplicate overlaid with sterile polyethylene film and incubated in moistened Petri dishes for up to 24 h at 42°C in 10% CO2. Cells were retrieved from the test surface by immersion in 1 ml of sterile PBS, agitation for 20 s and centrifugation at 13 000 rev min−1 in a bench top micro centrifuge (Eppendorf, Cambridge, UK). Pellets of cells were washed in PBS, resuspended in 25 μl of PBS and stained with the Live-Dead® BacLight™ Bacterial Viability kit according to manufacturer’s recommendations (Invitrogen). The procedure was repeated using untreated polyethylene as a control.
Results
Aerobic colony counts
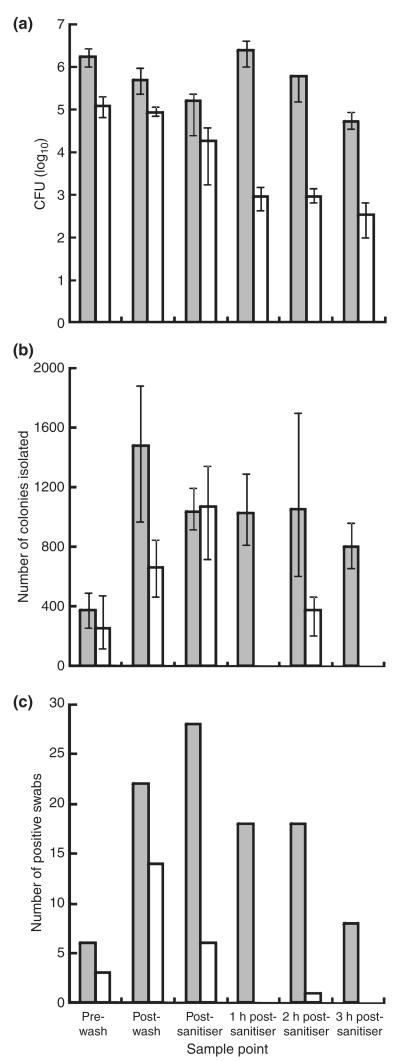
Aerobic colony counts obtained from both standard untreated and silver ion containing chicken transportation crates varied across the processing plant decontamination process. Counts were obtained from swabs taken immediately after live chickens (12 birds per crate) had been removed from the crates and before a water wash and then at key points throughout the decontamination process (Fig. 1a). The mean aerobic colony count (CFU) on standard crates before washing was 1·7 × 106 CFU compared to 1·2 × 105 CFU from silver ion containing crates. Water washing diminished counts for both crate types to 4·9 × 105 CFU (71·2% reduction) on standard crates and 8·7 × 104 CFU (27·5% reduction) on silver ion containing crates. Spray sanitization with 0·25% peracetic acid further reduced counts to 1·6 × 105 CFU (67·5% reduction) for standard crates and 1·9 × 104 CFU (78·2% reduction) for silver ion containing crates. Standard crates exhibited a 15-fold increase in aerobic colony count after 1 h post-sanitization (2·4 × 106 CFU), whilst silver ion containing crates exhibited a reduction in count to 9·2 × 102 CFU (95·2% reduction). After a further 1 and 2 h, the standard crate counts had reduced to 6·1 × 105 CFU (74·6% reduction) and 5·4 × 104 CFU (91·2% reduction), respectively. The silver ion containing crate counts increased to 9·3 × 102 CFU (1·1% increase) and then decreased to 3·5 × 102 CFU (62·4% reduction) after the further 1 and 2 h, respectively. The mean count of aerobic colonies from wash water was 3·0 × 1010 CFU ml−1.
Figure 1.
Quantification of aerobic colony count (a) and Campylobacter colony count (b) of 25-cm2 sample areas, and the number of Campylobacter-positive swabs (c) from standard  and silver ion containing
and silver ion containing  chicken transportation crates obtained after removal of live birds at the processing plant and across the decontamination process.
chicken transportation crates obtained after removal of live birds at the processing plant and across the decontamination process.
Campylobacter colony counts
The number of Campylobacter cells present on crates was quantified by determining the number of Campylobacter colonies isolated from swabs (Fig. 1b). The mean Campylobacter colony count from prewash standard crates was 4·7 × 103 CFU compared to 3·2 × 103 CFU for silver ion containing crates. Both crate types yielded increased counts after washing, suggesting the wash water contaminated the crates with Campylobacter; 1·9 × 104 CFU and 8·3 × 103 CFU from standard and silver ion containing crates, respectively. Wash water samples yielded a mean count of 2·3 × 104 CFU of Campylobacter. Sanitization reduced the counts of Campylobacter from standard crates (1·3 × 104 CFU) but increased the counts from silver ion containing crates (1·3 × 104 CFU). Counts of 1·3 × 104 CFU, 1·3 × 104 CFU and 1·0 × 104 CFU were then obtained from standard crates at 1-, 2- and 3-h postsanitization sampling points, respectively. Corresponding counts from silver ion containing crates were 0 CFU, 4·7 × 102 CFU and 0 CFU. The mean Campylobacter count from wash water was 2·3 × 104 CFU ml−1.
The number of Campylobacter-positive swabs from standard crates increased through processing from 6/30 (prewash) to 22/30 (postwash), increasing further after sanitization (28/30) (Fig. 1c). The number of Campylobacter-positive swabs decreased over the 3-h postsanitization period on the standard crates (1 h: 18/30, 2 h: 18/30 and 3 h: 8/30). Campylobacter-positive swabs also increased on silver ion containing crates during washing from 3/30 (prewash) to 14/30 (postwash). The number of Campylobacter-positive swabs then decreased (6/30) after sanitization. The 3-h period after sanitization produced a single Campylobacter-positive swab 2 h postsanitization (1/30).
In vitro antimicrobial efficacy determination
Antimicrobial efficacy was confirmed in vitro using the ISO 22196 protocol. A mean loss of cultivability, 99·35% from an inoculum of 1·5 × 104 E. coli cells and 99·63% from an inoculum of 2·5 × 104 MRSA cells, was observed after 24 h exposure to silver ion-treated (BioCote®) crate test samples compared to untreated crate material. Nine replicates were tested per bacterial species (data not shown).
Live-Dead fluorescence staining
The Live-Dead® BacLight™ Bacterial Viability kit employs the SYTO® 9 and propidium iodide stains (the green and red-fluorescent nucleic acid stains, respectively). A combination of these stains enables the differentiation of live bacterial cells with intact cell membranes (green) from cells with damaged membranes (red). Campylobacter cells were harvested from Bolton broth culture during logarithmic growth and stained with the Live-Dead® kit, fluoresced bright green (Fig. 2a). Using this technique, Campylobacter cells exposed to silver ion containing crate material showed a loss of viability over time (Fig. 2b–d). In contrast, Campylobacter cells harvested from the same culture exposed to untreated crate material for the same incubation times retained more viability (Fig. 2e) compared to cells exposed to silver ion containing material.
Figure 2.
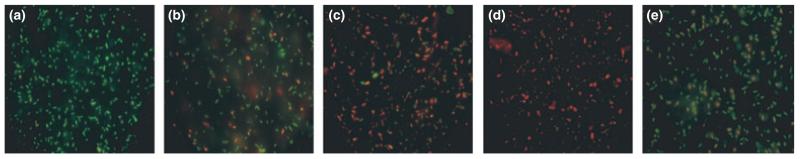
Live-Dead-stained Campylobacter cells exposed to silver ion-treated crate material and standard crate material. Cells stained during logarithmic phase growth in Bolton broth (a) fluoresce green indicating viability. Cells photographed 1 h (b), 2 h (c) and 4 h (d) after exposure to silver ion containing crate material show an increasing red/orange fluorescence indicating a progressive reduction in viability. Live-Dead-stained Campylobacter cells exposed to untreated crate material show less reduction in viability over a 4-h period (e) compared to d.
Genetic diversity
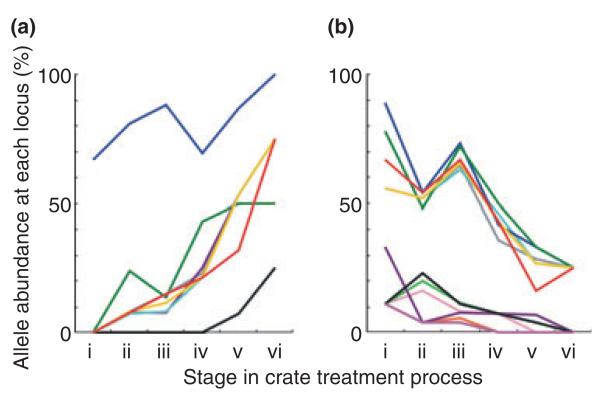
MLST was completed for 89 isolates, all of which gave genetic profiles characteristic of Camp. jejuni (Table 2). The 89 isolates were retrieved from the following samples: sample point i standard crate 6; sample point i silver ion containing crate 3; sample point ii standard crate 13; sample point ii silver ion containing crate 9; sample point iii standard crate 20; sample point iii silver ion containing crate 4; sample point iv standard crate 13; sample point v standard crate 14; sample point vi standard crate 4; wash water 3. There were a total of 16 STs. The four most common STs accounted for 80% of genotypes. There were four clonal complexes (ST-45, ST-354, ST-574, ST-661) defined as groups of STs that share four or more alleles in common with the central genotype. The ST-45 complex is known from multiple hosts (McCarthy et al. 2007; Colles et al. 2008a; Sheppard et al. 2009b), and the other clonal complexes have been found in chicken samples (Colles et al. 2008a; Sheppard et al. 2009b). They contrast with those most commonly associated with cattle (for example, ST-61, ST-206 and ST-403) and wild birds (for example, ST-177, ST-179, ST-682, ST-1034, ST-1275) (Colles et al. 2008b). The variation in MLST allele frequency is related to the change in dominant STs but provides additional information about the progressive shift in abundance and persistence of particular lineages throughout the crate decontamination process (Fig. 3).
Table 2.
Multilocus sequence typing data for isolates from poultry transport crates
| Allele nucleotide sequence number |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| ST | Frequency* | aspA | glnA | gltA | glyA | pgm | tkt | uncA | Clonal complex |
| 464 | 41 | 24 | 2 | 2 | 2 | 10 | 3 | 1 | |
| 814 | 14 | 2 | 75 | 4 | 48 | 141 | 34 | 1 | 661 |
| 4225 | 11 | 2 | 25 | 5 | 10 | 149 | 12 | 35 | |
| 45 | 6 | 4 | 7 | 10 | 4 | 1 | 7 | 1 | 45 |
| 354 | 5 | 8 | 10 | 2 | 2 | 11 | 12 | 6 | 354 |
| 574 | 2 | 7 | 53 | 2 | 10 | 11 | 3 | 3 | 574 |
| 418 | 1 | 4 | 7 | 2 | 4 | 1 | 7 | 1 | 45 |
| 4226 | 1 | 2 | 75 | 2 | 48 | 141 | 34 | 1 | 661 |
| 4227 | 1 | 24 | 2 | 10 | 2 | 10 | 3 | 1 | |
| 4228 | 1 | 24 | 2 | 2 | 2 | 10 | 34 | 1 | |
| 4229 | 1 | 43 | 25 | 5 | 10 | 149 | 12 | 35 | |
| 4230 | 1 | 43 | 75 | 4 | 48 | 141 | 34 | 1 | 661 |
| 4231 | 1 | 75 | 2 | 14 | 2 | 10 | 3 | 1 | |
| 4232 | 1 | 151 | 75 | 10 | 48 | 141 | 34 | 1 | 661 |
| 4233 | 1 | 249 | 75 | 4 | 48 | 141 | 34 | 1 | 661 |
| 4384 | 1 | 7 | 53 | 2 | 10 | 11 | 3 | 1 | 574 |
The number of times this ST was recorded in the dataset.
Figure 3.
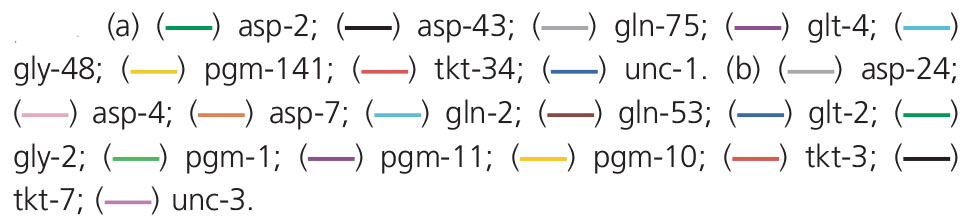
Frequency of alleles that showed an increasing (a) and decreasing (b) trend throughout the crate decontamination process at stages: prewater wash (i); postwash (ii); postsanitization in 0·25% peracetic acid (iii); 1 h postsanitization (iv); 2 h postsanitization (v); 3 h postsanitization (vi). The abundance of alleles is expressed as a percentage of the total number of alleles at that locus at that sample point.
Probabilistic assignment to host
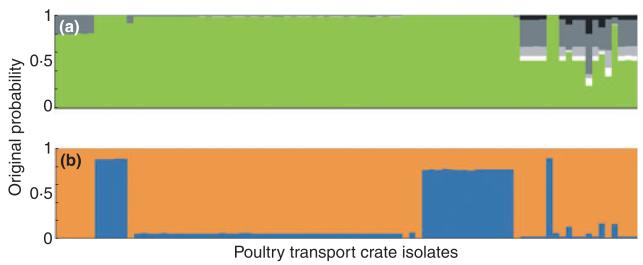
The assignment probability for putative origin was calculated for each chicken transport crate isolate individually (Fig. 4), and the percentage of all isolates attributed to each source was calculated as the sum of these probabilities. In the first STRUCTURE analysis, the crate isolates were attributed to origin populations in the following percentages: 89% to chicken, 1% to cattle, 2% to sheep, 7% to the environment and 1% to wild bird. The attributions in the second analysis using chicken only reference isolates assigned 22% to live chicken (faeces) and 78% to dead chicken (carcass swabs and retail poultry).
Figure 4.
Assignment of Campylobacter jejuni STs from chicken transport crates (n = 89) to origin population using the Bayesian clustering algorithm structure. Each isolate is represented by a vertical bar, showing the estimated probability that it originates from each of the putative sources indicated by different colours. Two independent analyses were carried out assigning tray genotypes to (a) chickens, cattle, sheep, the environment and wild birds, and (b) live chicken (faecal samples) or dead chicken (carcass swabs or retail meat). An equal area of each colour would be expected in the absence of genetic differentiation by host species (a) or food chain stage (b). (a)  wild bird;
wild bird;  environment;
environment;  sheep;
sheep;  cattle;
cattle;  chicken. (b)
chicken. (b)  dead chicken;
dead chicken;  live chicken.
live chicken.
Discussion
The occurrence of Camp. jejuni on poultry transport crates reflected the high prevalence of Campylobacter in chicken flocks (Heuer et al. 2001; Petersen et al. 2001; Newell and Fearnley 2003; Wittwer et al. 2005; Bull et al. 2006; Lindmark et al. 2006; Van Overbeke et al. 2006). The level of contamination and the diversity of genotypes identified by MLST (16 STs) underline the potential importance of transport crates in the transmission of Campylobacter (Slader et al. 2002; Hansson et al. 2005; Allen et al. 2008; Peyrat et al. 2008).
Previous studies have shown that the slaughter process is an important factor in the spread of Campylobacter genotypes in poultry (Berndtson et al. 1996b; Herman et al. 2003; Rosenquist et al. 2003; Allen et al. 2007; Klein et al. 2007; Lienau et al. 2007). Our finding that the ST-45, ST-354, ST-574, ST-661 clonal complexes, commonly associated with chickens (Sheppard et al. 2009a), dominate among the Camp. jejuni on the transport crates is consistent with the crates being involved in transmission. Furthermore, the persistence of these genotypes on the crates through the decontamination process provides a potential source of Campylobacter for uncontaminated flocks as the crates are reused.
There was evidence of differential survival of STs through the crate cleaning process. It is known that some STs are found at a significantly lower frequency on chicken after slaughter compared to the levels in live chickens (Newell et al. 2001; Johnsen et al. 2006). In this study, STs 418 and 4227 were only found on crates before washing and other STs dominated at later processing stages. The reasons for the variation in ST frequency throughout the crate decontamination process are poorly understood. A recent study reported variation in Campylobacter spp. adhering to an inert surface, the initial stage of biofilm formation (Sulaeman et al. 2010). The ability of Campylobacter to form biofilms may then impact on the observed survival of STs. The potential for differential survival of Camp. jejuni strains has also been suggested with the identification of environmentally adapted lineages, in particular ST-45 (Sopwith et al. 2008). This ST was one of those that was more abundant throughout the crate decontamination process, which is consistent with the idea that it is adapted to survival outside a host (Sopwith et al. 2008). The dynamics of genotypic changes were investigated by characterizing the abundance of alleles at various stages throughout crate decontamination. Trends in allele abundance indicate that some alleles decline and others increase at stages through the process. Although these are not necessarily determinants of phenotypes directly related to survival outside the host, this does provide insight into the trends in genotypic variation. It may therefore be possible to identify genotypes that could be important in Campylobacter transmission between flocks, on the basis of their constituent MLST alleles.
A total of 60% of the STs (9/15), accounting for 20% of the isolates (19/89), from the poultry transport crates, had not been previously described from any other source (http://pubmlst.org/campylobacter). However, comparison with isolates from chicken, other farm animal sources and the environment, by probabilistically assigning the putative origin, indicated an 89% similarity between the STs from transport crates and those from chicken (faeces/meat). Taking this analysis further, using chicken only training datasets, 78% of STs isolated from the transport crates were more similar to those from retail meat compared to live chickens. The similarity between the STs isolated during this study and previously recognized STs suggests parameters such as sampling/enrichment methods, times/seasons and temperatures did not influence significantly ST findings. The overlap between the genotypes from transport crates and those from retail meat suggests a common source of contamination.
This study demonstrated the survival of Campylobacter on crates for at least 3 h after on-site sanitization, a period of time equivalent to the crates’ journey from the processing plant to the majority of the farms in the catchment. It has previously been suggested that storage of crates for a period of not less than 2 weeks before contact with the next flock would dramatically reduce the circulation of pathogens, including Campylobacter, between flocks (Berrang et al. 2004). The crate decontamination process operated at our study’s processing plant was shown to be ineffective. Indeed, the wash water contained sufficient bacteria including Campylobacter to increase the levels of contamination on crates passing through it. However, mass storage of crates as an additional intervention would be impractical and expensive. Compared to standard crates, silver ion containing crates were consistently shown in our study to be less contaminated with bacteria, including Campylobacter, under normal conditions. It is possible that bacteria are rendered non-culturable by the action of a silver ion biocide but retain pathogenicity. However, the action of ionic silver on bacteria is lethal, so use of these treated crates may contribute to the interruption of Campylobacter transmission through poultry processing and the application of antimicrobials at key points may have the potential to reduce the prevalence of Campylobacter in the poultry environment. A potentially useful exercise would be to enhance wash/sanitization conditions for silver ion containing crates to determine the optimum combination of these antimicrobial strategies.
In conclusion, this study demonstrates the potential involvement of poultry transport crates in the transmission of Campylobacter. The genotypes colonizing them, characteristic of those from poultry, change through the slaughter process but current crate decontamination practices remove only certain genotypes. This study provides information that will enable targeted intervention by the poultry industry to help reduce the circulation of Campylobacter through the food chain.
Acknowledgements
SKS and MCJM are funded by Wellcome Trust fellowships. Some of the comparison data were produced with funding from DEFRA (contract number: OX0615).
References
- Allen VM, Bull SA, Corry JE, Domingue G, Jorgensen F, Frost GA, Whyte R, Gonzalez A, et al. Campylobacter spp. contamination of chicken carcasses during processing in relation to flock colonization. Int J Food Microbiol. 2007;113:54–61. doi: 10.1016/j.ijfoodmicro.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Allen VM, Weaver H, Ridley AM, Harris JA, Sharma M, Emery J, Sparks N, Lewis M, et al. Sources and spread of thermophilic Campylobacter spp. during partial depopulation of broiler chicken flocks. J Food Prot. 2008;71:264–270. doi: 10.4315/0362-028x-71.2.264. [DOI] [PubMed] [Google Scholar]
- Baker M, Wilson N, Ikram R, Chambers S, Shoemack P, Cook G. Regulation of chicken contamination is urgently needed to control New Zealand’s serious campylobacteriosis epidemic. N Z Med J. 2006;119:U2264. [PubMed] [Google Scholar]
- Berndtson E, Danielsson-Tham M-L, Engvall A. Campylobacter incidence on a chicken farm and the spread of Campylobacter during the slaughter process. Int J Food Microbiol. 1996a;32:35–47. doi: 10.1016/0168-1605(96)01102-6. [DOI] [PubMed] [Google Scholar]
- Berndtson E, Emanuelson U, Engvall A, Danielsson-Tham M-L. A 1-year epidemiological study of Campylobacters in 18 Swedish chicken farms. Prev Vet Med. 1996b;26:167–185. [Google Scholar]
- Berrang ME, Northcutt JK, Cason JA. Recovery of Campylobacter from broiler feces during extended storage of transport cages. Poult Sci. 2004;83:1213–1217. doi: 10.1093/ps/83.7.1213. [DOI] [PubMed] [Google Scholar]
- Bull SA, Allen VM, Domingue G, Jorgensen F, Frost GA, Ure R, Whyte R, Tinker D, et al. Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Appl Environ Microbiol. 2006;72:645–652. doi: 10.1128/AEM.72.1.645-652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzby JC, Roberts T. Economic costs and trade impacts of microbial foodborne illness. World Health Stat Q. 1997;50:57–66. [PubMed] [Google Scholar]
- Colles FM, Jones TA, McCarthy ND, Sheppard SK, Cody AJ, Dingle KE, Dawkins MS, Maiden MC. Campylobacter infection of broiler chickens in a free-range environment. Environ Microbiol. 2008a;10:2042–2050. doi: 10.1111/j.1462-2920.2008.01623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colles FM, Dingle KE, Cody AJ, Maiden MC. Comparison of Campylobacter populations in wild geese with those in starlings and free-range poultry on the same farm. Appl Environ Microbiol. 2008b;74:3583–3590. doi: 10.1128/AEM.02491-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colles F, McCarthy N, Sheppard SK, Layton R, Maiden MCJ. Comparison of Campylobacter populations isolated from a free-range broiler flock before and after slaughter. Int J Food Microbiol. 2010;132:259–264. doi: 10.1016/j.ijfoodmicro.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry JE, Atabay HI. Poultry as a source of Campylobacter and related organisms. Symp Ser Soc Appl Microbiol. 2001;90:96S–114S. doi: 10.1046/j.1365-2672.2001.01358.x. [DOI] [PubMed] [Google Scholar]
- Dingle KE, Colles FM, Falush D, Maiden MC. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J Clin Microbiol. 2005;43:340–347. doi: 10.1128/JCM.43.1.340-347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM. The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett Appl Microbiol. 1991;13:171–174. doi: 10.1111/j.1472-765x.1991.tb00600.x. [DOI] [PubMed] [Google Scholar]
- Friedman CR, Neiman J, Wegener HC, Tauxe RV. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser MJ, editors. Campylobacter. ASM Press; Washington, D. C: 2000. pp. 121–138. [Google Scholar]
- Friedman CR, Hoekstra RM, Samuel M, Marcus R, Bender J, Shiferaw B, Reddy S, Ahuja SD, et al. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin Infect Dis. 2004;38(Suppl. 3):S285–S296. doi: 10.1086/381598. [DOI] [PubMed] [Google Scholar]
- Hansson I, Ederoth M, Andersson L, Vagsholm I, Olsson Engvall E. Transmission of Campylobacter spp. to chickens during transport to slaughter. J Appl Microbiol. 2005;99:1149–1157. doi: 10.1111/j.1365-2672.2005.02689.x. [DOI] [PubMed] [Google Scholar]
- Health Protection Agency . Oxidase Test. HPA; 2005. Standard Operating Procedure BSOP TP 26. [Google Scholar]
- Herman L, Heyndrickx M, Grijspeerdt K, Vandekerchove D, Rollier I, De Zutter L. Routes for Campylobacter contamination of poultry meat: epidemio-logical study from hatchery to slaughterhouse. Epidemiol Infect. 2003;131:1169–1180. doi: 10.1017/s0950268803001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer OE, Pedersen K, Andersen JS, Madsen M. Prevalence and antimicrobial susceptibility of thermophilic Campylobacter in organic and conventional broiler flocks. Lett Appl Microbiol. 2001;33:269–274. doi: 10.1046/j.1472-765x.2001.00994.x. [DOI] [PubMed] [Google Scholar]
- Humphrey T, O’Brien S, Madsen M. Campylobacters as zoonotic pathogens: a food production perspective. Int J Food Microbiol. 2007;117:237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Johnsen G, Kruse H, Hofshagen M. Genotyping of Campylobacter jejuni from broiler carcasses and slaughterhouse environment by amplified fragment length polymorphism. Poult Sci. 2006;85:2278–2284. doi: 10.1093/ps/85.12.2278. [DOI] [PubMed] [Google Scholar]
- Jolley KA, Chan MS, Maiden MC. mlstdbNet - distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapperud G, Espeland G, Wahl E, Walde E, Herikstad H, Gustavsen S, Tveit I, Natas O, et al. Factors associated with increased and decreased risk of Campylobacter infection: a prospective case-control study in Norway. Am J Epidemiol. 2003;158:234–242. doi: 10.1093/aje/kwg139. [DOI] [PubMed] [Google Scholar]
- Klein G, Beckmann L, Vollmer HM, Bartelt E. Predominant strains of thermophilic Campylobacter spp. in a German poultry slaughterhouse. Int J Food Microbiol. 2007;117:324–328. doi: 10.1016/j.ijfoodmicro.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Lienau JA, Ellerbroek L, Klein G. Tracing flock-related Campylobacter clones from broiler farms through slaughter to retail products by pulsed-field gel electrophoresis. J Food Prot. 2007;70:536–542. doi: 10.4315/0362-028x-70.3.536. [DOI] [PubMed] [Google Scholar]
- Lindmark H, Diedrich IC, Andersson L, Lindqvist R, Engvall EO. Distribution of Campylobacter genotypes on broilers during slaughter. J Food Prot. 2006;69:2902–2907. doi: 10.4315/0362-028x-69.12.2902. [DOI] [PubMed] [Google Scholar]
- Manning G, Dowson CG, Bagnall MC, Ahmed IH, West M, Newell DG. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl Environ Microbiol. 2003;69:6370–6379. doi: 10.1128/AEM.69.11.6370-6379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy ND, Colles FM, Dingle KE, Bagnall MC, Manning G, Maiden MC, Falush D. Host-associated genetic import in Campylobacter jejuni. Emerg Infect Dis. 2007;13:267–272. doi: 10.3201/eid1302.060620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, On SL, Wang G, Fontanoz S, Lastovica AJ, Mandrell RE. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J Clin Microbiol. 2005;43:2315–2329. doi: 10.1128/JCM.43.5.2315-2329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa N, Takegahara Y, Terai K, Kato H, Takeuchi T. Campylobacter jejuni contamination on broiler carcasses of C. jejuni-negative flocks during processing in a Japanese slaughterhouse. Int J Food Microbiol. 2003;84:105–109. doi: 10.1016/s0168-1605(02)00398-7. [DOI] [PubMed] [Google Scholar]
- Mullner P, Spencer SE, Wilson DJ, Jones G, Noble AD, Midwinter AC, Collins-Emerson JM, Carter P, et al. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect Genet Evol. 2009;6:1311–1319. doi: 10.1016/j.meegid.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Neimann J, Engberg J, Molbak K, Wegener HC. A case-control study of risk factors for sporadic Campylobacter infections in Denmark. Epidemiol Infect. 2003;130:353–366. doi: 10.1017/s0950268803008355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell DG, Fearnley C. Sources of Campylobacter colonization in broiler chickens. Appl Environ Microbiol. 2003;69:4343–4351. doi: 10.1128/AEM.69.8.4343-4351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell DG, Shreeve JE, Toszeghy M, Domingue G, Bull S, Humphrey T, Mead G. Changes in the carriage of Campylobacter strains by poultry carcasses during processing in abattoirs. Appl Environ Microbiol. 2001;67:2636–2640. doi: 10.1128/AEM.67.6.2636-2640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen L, Nielsen EM, On SL. Serotype and genotype diversity and hatchery transmission of Campylobacter jejuni in commercial poultry flocks. Vet Microbiol. 2001;82:141–154. doi: 10.1016/s0378-1135(01)00382-0. [DOI] [PubMed] [Google Scholar]
- Peyrat MB, Soumet C, Maris P, Sanders P. Recovery of Campylobacter jejuni from surfaces of poultry slaughterhouses after cleaning and disinfection procedures: analysis of a potential source of carcass contamination. Int J Food Microbiol. 2008;124:188–194. doi: 10.1016/j.ijfoodmicro.2008.03.030. [DOI] [PubMed] [Google Scholar]
- Posch J, Feierl G, Wuest G, Sixl W, Schmidt S, Haas D, Reinthaler FF, Marth E. Transmission of Campylobacter spp. in a poultry slaughterhouse and genetic characterization of the isolates by pulsed-field gel electrophoresis. Br Poult Sci. 2006;47:286–293. doi: 10.1080/00071660600753763. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivoal K, Denis M, Salvat G, Colin P, Ermel G. Molecular characterization of the diversity of Campylobacter spp. isolates collected from a poultry slaughterhouse: analysis of cross-contamination. Lett Appl Microbiol. 1999;29:370–374. doi: 10.1046/j.1472-765x.1999.00645.x. [DOI] [PubMed] [Google Scholar]
- Rosenquist H, Nielsen HN, Sommer HM, Norrung B, Christensen BB. Quantitative risk assessment of human campylobacteriosis associated with thermophilic Campylobacter species in chickens. Int J Food Microbiol. 2003;83:87–103. doi: 10.1016/s0168-1605(02)00317-3. [DOI] [PubMed] [Google Scholar]
- Sheppard SK, Dallas JF, Strachan NJ, MacRae M, McCarthy ND, Wilson DJ, Gormley FJ, Falush D, et al. Campylobacter genotyping to determine the source of human infection. Clin Infect Dis. 2009a;48:1072–1078. doi: 10.1086/597402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Dallas JF, MacRae M, McCarthy ND, Sproston EL, Gormley FJ, Strachan NJ, Ogden ID, et al. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int J Food Microbiol. 2009b;134:96–103. doi: 10.1016/j.ijfoodmicro.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slader J, Domingue G, Jorgensen F, McAlpine K, Owen RJ, Bolton FJ, Humphrey TJ. Impact of transport crate reuse and of catching and processing on Campylobacter and Salmonella contamination of broiler chickens. Appl Environ Microbiol. 2002;68:713–719. doi: 10.1128/AEM.68.2.713-719.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopwith W, Birtles A, Matthews M, Fox A, Gee S, Painter M, Regan M, Syed Q, et al. Identification of potential environmentally adapted Campylobacter jejuni strain, United Kingdom. Emerg Infect Dis. 2008;14:1769–1773. doi: 10.3201/eid1411.071678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- Strachan NJ, Gormley FJ, Rotariu O, Ogden ID, Miller G, Dunn GM, Sheppard SK, Dallas JF, et al. Attribution of Campylobacter infections in northeast Scotland to specific sources by use of multilocus sequence typing. J Infect Dis. 2009;199:1205–1208. doi: 10.1086/597417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaeman S, Le Bihan G, Rossero A. Comparison between the biofilm initiation of Campylobacter jejuni and Campylobacter coli strains to an inert surface using BioFilm Ring Test®. J Appl Microbiol. 2010;108:1303–1312. doi: 10.1111/j.1365-2672.2009.04534.x. [DOI] [PubMed] [Google Scholar]
- Van Overbeke I, Duchateau L, De Zutter L, Albers G, Ducatelle R. A comparison survey of organic and conventional broiler chickens for infectious agents affecting health and food safety. Avian Dis. 2006;50:196–200. doi: 10.1637/7448-093005R.1. [DOI] [PubMed] [Google Scholar]
- Van Worth C, McCrea BA, Tonooka KH, Boggs CL, Schrader JS. Diversity of flaA genotypes among Campylobacter jejuni isolated from six niche-market poultry species at farm and processing. J Food Prot. 2006;69:299–307. doi: 10.4315/0362-028x-69.2.299. [DOI] [PubMed] [Google Scholar]
- Vellinga A, Van Loock F. The dioxin crisis as experiment to determine poultry-related Campylobacter enteritis. Emerg Infect Dis. 2002;8:19–22. doi: 10.3201/eid0801.010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedderkopp A, Gradel KO, Jorgensen JC, Madsen M. Pre-harvest surveillance of Campylobacter and Salmonella in Danish broiler flocks: a 2-year study. Int J Food Microbiol. 2001;68:53–59. doi: 10.1016/s0168-1605(01)00463-9. [DOI] [PubMed] [Google Scholar]
- Wilson DJ, Gabriel E, Leatherbarrow AJH, Cheesbrough J, Gee S, Bolton E, Fox A, Fearnhead P, et al. Tracing the source of campylobacteriosis. PLoS Genet. 2008;26:e1000203. doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer M, Keller J, Wassenaar TM, Stephan R, Howald D, Regula G, Bissig-Choisat B. Genetic diversity and antibiotic resistance patterns in a Campylobacter population isolated from poultry farms in Switzerland. Appl Environ Microbiol. 2005;71:2840–2847. doi: 10.1128/AEM.71.6.2840-2847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]