Abstract
During the course of screening for exoprotease-deficient mutants among Bacillus subtilis gene disruptants, a strain showing such a phenotype was identified. The locus responsible for this phenotype was the previously unknown gene ybaL, which we renamed salA. The predicted gene product encoded by salA belongs to the Mrp family, which is widely conserved among archaea, prokaryotes, and eukaryotes. Disruption of salA resulted in a decrease in the expression of a lacZ fusion of the aprE gene encoding the major extracellular alkaline protease. The decrease was recovered by the cloned salA gene on a plasmid, demonstrating that the gene is involved in aprE expression. Determination of the cis-acting region of SalA on the upstream region of aprE, together with epistatic analyses with scoC, abrB, and spo0A mutations that also affect aprE expression, suggested that salA deficiency affects aprE-lacZ expression through the negative regulator ScoC. Northern and reverse transcription-PCR analyses revealed enhanced levels of scoC transcripts in the salA mutant cells in the transition and early stationary phases. Concomitant with these observations, larger amounts of the ScoC protein were detected in the mutant cells by Western analysis. From these results we conclude that SalA negatively regulates scoC expression. It was also found that the expression of a salA-lacZ fusion was increased by salA deficiency, suggesting that salA is autoregulated.
Bacillus subtilis aprE, which encodes the major extracellular alkaline protease, is one of the genes whose expression is induced postexponentially in response to stationary-phase stresses, and the transcription of aprE is strictly regulated by complex and redundant systems (43). Spo0A-AbrB and DegS-DegU are the major factors that control aprE expression. Spo0A is a master regulator of early developmental responses, such as sporulation initiation, and regulates transcription of many genes negatively and positively (11, 43). During transition from growing to stationary phase, Spo0A is activated by phosphorylation, causing the transcriptional repression of abrB, which encodes a global transcription factor regulating many target genes (44, 45). This event is critical for a B. subtilis cell to adapt to stationary-phase stresses. For example, the repression of abrB by phosphorylated Spo0A results in the activation of spo0E, spo0H, spoVG, and aprE expression (5, 6, 41, 45) and the inactivation of scoC and rbs operon expression (32, 42).
ScoC (Hpr) was identified as a repressor for the expression of aprE and nprE, the gene encoding the major extracellular neutral protease (4, 14, 33). It was further demonstrated that ScoC regulates the expression of sinI and two oligopeptide transporter operons (20, 36), and a consensus DNA sequence, 5′-RATAnTATY-3′, has been proposed for the binding of the ScoC protein (16). The sinI gene, located directly upstream of sinR, encodes a protein that binds SinR and thereby inhibits its regulatory activity (1, 7, 9). SinR binds the upstream regions of the target genes, including aprE, amyE, sacB, spoIIA, spoIIG, spoIIE, and spo0A (7, 8, 9, 23, 24, 25). The sinI and sinR genes are, however, in different transcription units and subject to different regulation. Thus, the expression of sinI but not sinR is regulated by Spo0A, AbrB, and ScoC (Fig. 1A) (1, 9, 36). Consequently, ScoC and SinR form a regulatory unit repressing aprE expression, in which SinR is activated through the inhibition of sinI transcription by ScoC (Fig. 1A). Furthermore, SinR and Spo0A are in a transcriptional circuit, i.e., Spo0A activates sinI expression, SinI inhibits SinR and SinR inhibits spo0A expression (24, 25, 36). Recently, it was shown by DNA microarray analysis that many genes are under the control of ScoC (2).
FIG. 1.
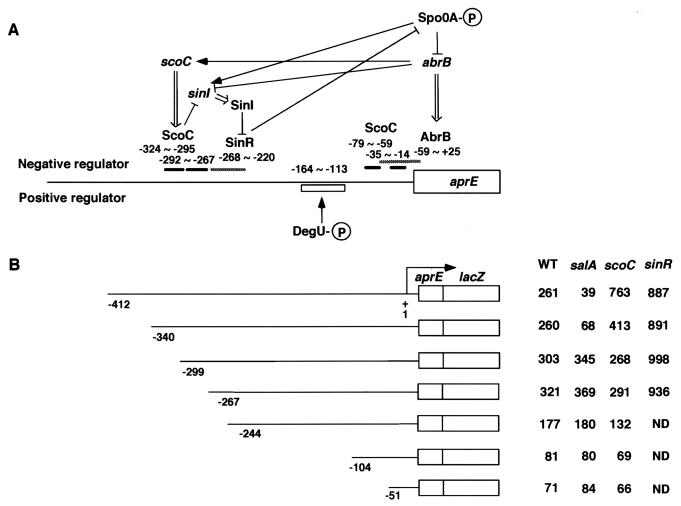
Regulation of aprE by multiple regulators, and determination of the cis-acting region of SalA on the control region of aprE. (A) Schematic representation of aprE regulation by trans-acting regulators. The horizontal line indicates the DNA region upstream of aprE. The thick and hatched bars above the horizontal line show the regions protected by binding of ScoC, SinR, and AbrB (7, 16, 45). The box below the horizontal line shows the cis-acting site for DegU as deduced by analyses using deletions of the upstream region and the degU32(Hy) mutation (13). The numbers indicate the nucleotides relative to the transcriptional start site of aprE. The arrows and T bars indicate stimulation and inhibition, respectively, and the double-lined arrows show translation of the gene. The P's in the circles depict the phosphate group incorporated in DegU and Spo0A. (B) The lines indicate the regions containing the control region of aprE, and the boxes denote translational fusions between the 8th codon of aprE and the15th codon of lacZ (13). The numbers below the lines indicate the nucleotide numbers relative to the transcription start site of aprE, and those in the four columns show β-galactosidase activities expressed in Miller units. ND, not determined. Strains OAM145, OAM146, OAM147, OAM218, OAM148, OAM149, and OAM150 were used as wild-type strains (WT); OAM151, OAM152, OAM153, OAM 219, OAM154, OAM155, and OAM156 were used as salA mutants; OAM157, OAM158, OAM159, OAM 220, OAM160, OAM161, and OAM162 were used as scoC mutants; and OAM221, OAM222, OAM223, and OAM224 were used as sinR mutants. Cells were grown in Schaeffer's sporulation medium, and β-galactosidase activities were determined as described in Materials and Methods. Three independent experiments were done in which samples were withdrawn at hourly intervals in stationary phase (T1 through T5). The highest values observed between T2 and T3 from the three experiments were averaged and shown. The standard deviations were within 15% in all the experiments.
Expression of aprE is also subject to positive regulation. DegU, the response regulator of the DegS-DegU two-component system, serves as a positive factor when it is phosphorylated (21). In addition, five more positive factors (DegQ, DegR, SenS, TenA, and ProB) are known to enhance aprE expression (30, 39).
These elaborate and redundant regulatory factors influencing the expression of aprE presumably constitute a network that responds to and integrates both internal and external signals and leads to the fine-tuning of aprE expression to cope with changing environments.
In this paper, we report a new locus, salA, whose disruption resulted in a reduction of aprE expression. It was shown that salA encodes a negative regulator of scoC, which explains the phenotype of the salA disruption. It was also suggested that salA was under negative autoregulation.
MATERIALS AND METHODS
Bacterial strains and culture media.
All the B. subtilis strains used in this study are listed in Table 1. The DNA sequences upstream of the aprE-coding regions in constructs SG35.18, SG 35.21, SG 35.20, SG35.8Δ34, and SG35.8Δ6 (13) were confirmed by sequence determination. B. subtilis and Escherichia coli cells were grown in Schaeffer's sporulation and 2× SG (52) media and in Luria-Bertani (LB) medium, respectively. LBCG is a modified version of LB medium in which casein and gelatin were added (46). The concentrations of the antibiotics added to the medium were 1 μg/ml for erythromycin, 10 μg/ml for kanamycin and tetracycline, 100 μg/ml for spectinomycin and ampicillin, and 5 μg/ml for chloramphenicol and phleomycin.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotypea | Reference or source |
|---|---|---|
| CU741 | trpC2 leuC7 | S. A. Zhaler |
| OM210 | trpC2 leuC7 salA::pMutin-salA (Emr) | pMutin-salA × CU741 |
| OM211 | trpC2 leuC7 salA::pMutin-salA (Emr) (lacZ::Tcr) | pLacZ::Tc × OM210 |
| OM212 | trpC2 leuC7 salA::pUKM-salA (Kmr) | pUKM-salA × CU741 |
| OM213 | trpC2 leuC7 scoC::pMutin-scoC (Emr) | pMutin-socC × CU741 |
| OM214 | trpC2 leuC7 scoC::pMutin-scoC (Emr) (lacZ::Tcr) | pLacZ::Tc × OM213 |
| OM215 | trpC2 leuC7 sinR::Pmr | pPHL2-sinR × CU741 |
| KK101 | trpC2 pheA1 spo0A::Spr | 18 |
| TT734 | trpC2 leuC7 abrB::Kmr | pGCB2 × CU741 |
| BG4224 | trpC2 his-1 thr-5 amyE::aprE-lacZ(−412 [SG35.18], Cmr) | 13 |
| BG4226 | trpC2 his-1 thr-5 amyE::aprE-lacZ(−340 [SG35.21], Cmr) | 13 |
| BG4225 | trpC2 his-1 thr-5 amyE::aprE-lacZ(−244 [SG35.20], Cmr) | 13 |
| BG4202 | trpC2 his-1 thr-5 amyE::aprE-lacZ(−104 [SG35.8Δ34], Cmr) | 13 |
| BG4160 | trpC2 his-1 thr-5 amyE::aprE-lacZ(−51 [SG35.8Δ6], Cmr) | 13 |
| OAM145 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) | BG4224 DNA × CU741 |
| OAM146 | trpC2 leuC7 amyE::aprE-lacZ(−340 [SG35.21], Cmr) | BG4226 DNA × CU741 |
| OAM147 | trpC2 leuC7 amyE::aprE-lacZ(−299, Cmr) | pHTP15 × CU741 |
| OAM218 | trpC2 leuC7 amyE::aprE-lacZ(−267, Cmr) | pTSN35 × CU741 |
| OAM148 | trpC2 leuC7 amyE::aprE-lacZ(−244[SG35.20], Cmr) | BG4225 DNA × CU741 |
| OAM149 | trpC2 leuC7 amyE::aprE-lacZ(−104 [SG35.8Δ34], Cmr) | BG4202 DNA × CU741 |
| OAM150 | trpC2 leuC7 amyE::aprE-lacZ(−51 [SG35.8Δ6], Cmr) | BG4160 DNA × CU741 |
| OAM151 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) salA::Emr (lacZ::Tcr) | OM211 DNA × OAM145 |
| OAM152 | trpC2 leuC7 amyE::aprE-lacZ(−340 [SG35.21], Cmr) salA::Emr (lacZ::Tcr) | OM211 DNA × OAM146 |
| OAM153 | trpC2 leuC7 amyE::aprE-lacZ(−299, Cmr) salA::Emr (lacZ::Tcr) | OM211 DNA × OAM147 |
| OAM219 | trpC2 leuC7 amyE::aprE-lacZ(−267, Cmr) salA::Emr (lacZ::Tcr) | OM211 DNA × OAM218 |
| OAM154 | trpC2 leuC7 amyE::aprE-lacZ(−244 [SG35.20], Cmr) salA::Emr (lacZ::Tcr) | OM211 DNA × OAM148 |
| OAM155 | trpC2 leuC7 amyE::aprE-lacZ(−104 [SG35.8Δ34], Cmr) salA::Emr (lacZ::Tcr) | OM211 DNA × OAM149 |
| OAM156 | trpC2 leuC7 amyE::aprE-lacZ(−51 [SG35.8Δ6], Cmr) salA::Emr (lacZ::Tcr) | OM211 DNA × OAM150 |
| OAM157 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) scoC::Emr (lacZ::Tcr) | OM214 DNA × OAM145 |
| OAM158 | trpC2 leuC7 amyE::aprE-lacZ(−340 [SG35.21], Cmr) scoC::Emr (lacZ::Tcr) | OM214 DNA × OAM146 |
| OAM159 | trpC2 leuC7 amyE::aprE-lacZ(−299, Cmr) scoC::Emr (lacZ::Tcr) | OM214 DNA × OAM147 |
| OAM220 | trpC2 leuC7 amyE::aprE-lacZ(−267, Cmr) scoC::Emr (lacZ::Tcr) | OM214 DNA × OAM218 |
| OAM160 | trpC2 leuC7 amyE::aprE-lacZ(−244 [SG35.20], Cmr) scoC::Emr (lacZ::Tcr) | OM214 DNA × OAM148 |
| OAM161 | trpC2 leuC7 amyE::aprE-lacZ(−104 [SG35.8Δ34], Cmr) scoC::Emr (lacZ::Tcr) | OM214 DNA × OAM149 |
| OAM162 | trpC2 leuC7 amyE::aprE-lacZ(−51 [SG35.8Δ6], Cmr) scoC::Emr (lacZ::Tcr) | OM214 DNA × OAM150 |
| OAM221 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) sinR::Pmr | OM215 DNA × OAM145 |
| OAM222 | trpC2 leuC7 amyE::aprE-lacZ(−340 [SG35.21], Cmr) sinR::Pmr | OM215 DNA × OAM146 |
| OAM223 | trpC2 leuC7 amyE::aprE-lacZ(−299, Cmr) sinR::Pmr | OM215 DNA × OAM147 |
| OAM224 | trpC2 leuC7 amyE::aprE-lacZ(−267, Cmr) sinR::Pmr | OM215 DNA × OAM218 |
| OAM163 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) salA::Kmr | OM212 DNA × OAM145 |
| OAM164 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) salA::KmrscoC::Emr (lacZ::Tcr) | OM214 DNA × OAM163 |
| OAM167 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) spo0A::Spr | KK101 DNA × OAM145 |
| OAM168 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) abrB::Kmr | TT734 DNA × OAM145 |
| OAM169 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) spo0A::SprabrB::Kmr | OAM167 DNA × OAM145 OAM168 DNA |
| OAM170 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) salA::Emr(lacZ::Tcr) spo0A::Spr | OAM167 DNA × OAM151 |
| OAM171 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) salA::Emr(lacZ::Tcr) abrB::Kmr | OAM168 DNA × OAM151 |
| OAM180 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) salA::Emr(lacZ::Tcr) spo0A::SprabrB::Kmr | OAM169 DNA × OAM151 |
The numbers in parentheses and the letters in brackets indicate the deletion end points upstream of the transcription start point of aprE and promoter constructs, respectively. Emr, erythromycin resistance; Tcr, tetracycline resistance; Pmr, phleomycin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Spr, spectinomycin resistance.
Materials.
Synthetic oligonucleotides were commercially prepared by Espec Oligo Service (Ibaraki, Japan) (Table 2). Sequence determination was carried out by using a 377 DNA Sequencer (Perkin-Elmer) and a Dye Terminator Cycle Sequencing Kit (Applied Biosystems).
TABLE 2.
Oligonucleotides used in this study
| Oligo-nucleotide | Sequence |
|---|---|
| Sal-d1-F | 5′-GGAAGCTTCGATTTGAAGAGCTGCC-3′ |
| Sal-d1-R | 5′-TTGGATCCAACCTTTTTCCCTAGACG-3′ |
| Sal-d2-F | 5′-CTAGGATCCGGGGCGCTTATAGAAGTCGG-3′ |
| Sal-d2-R | 5′-TGAAAGCTTATAAACAGACGGAGCAAATT-3′ |
| Sal-op-F | 5′-GATAAGCTTGTAAACGAATACACAAAAGGG-3′ |
| Sal-op-R | 5′-GATCTCGAGTTTTATACCTGCACTGACAT-3′ |
| Sal-f-F | 5′-ATGGAATTCAGCTTTTATTACGGGAAATAT-3′ |
| Sal-f-R | 5′-ATGAAGCTTTACTTCATCTTCTCTTATCA-3′ |
| SinR-F | 5′-GATAAGCTTATTGGCCAGCGTATTAAA-3′ |
| SinR-R | 5′-GATGAATTCTCATGTTTCTCATCGAGCA-3′ |
| SinR-R2 | 5′-AAAATCCCAAAAAGAGGAGTAG-3′ |
| HPRCON | 5′-CAGTGAATTCATCATGCTTTGAAAAAATATCACG-3′ |
| SINCON | 5′-CAGTGAATTCCATTGTTCTCACGGAAGCAC-3′ |
| LACZ3 | 5′-CTGACGCAGTCGGCATAACC-3′ |
| ScoC-F | 5′-AGTAAGCTTGAGTGGAACCGCCCTATGA-3′ |
| ScoC-R | 5′-ATCGGATCCGCCGTTTGGAGAACCTTAAA-3′ |
| Sal-m1 | 5′-AAAAGGCGGCGTAGATCTGTCAACTGTGTCAG-3′ |
| Sal-m2 | 5′-CTGACACAGTTGACAGATCTACGCCGCCTTTT-3′ |
| ScoC-1 | 5′-ATCGAGTGGAACCGCCCTATGA-3′ |
| ScoC-2 | 5′-CAGCCGGTTCATCCGCCGCAA-3′ |
Screening for mutants in exoprotease production.
The strains used for screening were the gene disruption mutants constructed by the insertion of pMutin-derived plasmids (18, 49) into B. subtilis genes, which were obtained from Japanese Consortium of Bacillus Functional Genomics. Into the mutants we introduced a multicopy pUBH1 derivative, pLC1, which carries proB and enhances exoprotease production in a DegS-DegU-dependent manner (17, 29). We used this strategy so that the disruption mutants show higher exoprotease activities, and thus any phenotype defective in exoprotease production was easily detected on LBCG plates. The mutant strains carrying pLC1 were spread on LB plates containing erythromycin and kanamycin, and after overnight incubation at 37°C, the colonies formed were transferred by toothpicks onto the LBCG plates containing kanamycin. The halos around the colonies were compared with that formed by B. subtilis 168 carrying pLC1 after more than 24 h.
Plasmid construction.
All the plasmids used in this study are listed in Table 3. To create pMutin-salA and pMutin-scoC, the PCR products amplified by using the oligonucleotide primer pairs Sal-d1-F and Sal-d1-R, and ScoC-F and ScoC-R, respectively, were treated with HindIII and BamHI and cloned between the HindIII and BamHI sites of pMutinIII (49). To construct pUKM-salA, the PCR product amplified by using Sal-d2-F and Sal-d2-R as oligonucleotide primers was treated with BamHI and HindIII and cloned into pUKM504 (28) treated with the same enzymes. To make pPHL2-sinR, the PCR product amplified by using SinR-F and SinR-R as primers was treated with HindIII and EcoRI and cloned into plasmid pPHL2 (31) treated with the same enzymes. To construct ptrp-salA the PCR product amplified by using the primers Sal-f-F and Sal-f-R was treated with EcoRI and HindIII and cloned into ptrpBG1 (38) that had been treated with the same enzymes. Plasmid pTHP15 was constructed by the insertion between the EcoRI and ClaI sites in ptrpBG1 of a PCR fragment, which was obtained by using the primers HPRCON and LACZ3 and DNA from strain BG4224 as a template. This procedure resulted in the same construct as SG35.18 except that the new construct contains the sequence up to −299 with respect to the transcription start site of aprE. Plasmid pTSN35 was constructed in the same procedure as that used for pTHP15 except that primer SINCON was used instead of HPRCON. To make pDG-salA, the PCR product amplified by using Sal-op-F and Sal-op-R as primers was treated with HindIII and SalI and cloned into plasmid pDG148, a shuttle vector capable of replication in both E. coli and B. subtilis (40), which had been treated with the same enzymes. Sequences were confirmed for the entire, cloned DNA regions. To construct pGCB2, the kanamycin resistance gene cassette obtained by the SmaI digestion of pBEST508 (15) was inserted into the blunt-ended BstXI site in the abrB gene cloned on a pBR322-derived plasmid, pGC516R (34).
TABLE 3.
Plasmids used in this study
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| pUBH1 | Multicopy B. subtilis plasmid; Kmr | 17 |
| pLC1 | pUBH1 carrying proB; causes overexpression of aprE | 29 |
| pMutinIII | Insertional Emr plasmid carrying promoterless lacZ | 49 |
| pMutin-salA | Emr pMutinIII carrying transcriptional fusion salA-lacZ | This study |
| pMutin-scoC | Emr pMutinIII carrying transcriptional fusion scoC-lacZ | This study |
| pUKM504 | Insertional Kmr plasmid | 28 |
| pUKM-salA | Kmr pUKM504 carrying an internal part of salA | This study |
| pPHL2 | pUC19-based Pmr plasmid/31 pPHL2-sinR/pPHL2 carrying an internal part of sinR | This study |
| pLacZ::Tc | Ampr Tcr; carrying lacZ::Tcr | 32a |
| ptrpBG1 | Insertional Ampr Cmr plasmid carrying trpE-lacZ | 38 |
| pTHP15 | ptrpBG1 carrying the aprE promoter region up to −299 | This study |
| pTSN35 | ptrpBG1 carrying the aprE promoter region up to −267 | This study |
| pDG148 | Multicopy B. subtilis and E. coli plasmid; Ampr Kmr | 40 |
| pDG-salA | Ampr Kmr pDG148 carrying salA | This study |
| pDG-salA-m | Ampr Kmr pDG148 carrying mutant salA (G119D and K120L) | This study |
| pBEST508 | Ampr Kmr | 15 |
| pGC516R | Ampr; carrying abrB | 34 |
| PGCB2 | Ampr pGC516R carrying abrB::Kmr | This study |
Ampr, ampicillin resistance. For other abbreviations see Table 1, footnote a.
Site-directed mutagenesis.
To introduce amino acid substitutions into SalA, site-directed mutagenesis of the cloned salA gene on pDG-salA was carried out with the oligonucleotide pairs Sal-m1 and Sal-m2 and a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the procedure described in the instructions. Mutations were confirmed by sequence determination of the entire salA-coding region on the plasmid constructed.
β-Galactosidase and sporulation assays.
Samples were withdrawn at hourly intervals for the measurement of β-galactosidase activities as described previously (32). For a sporulation test, cells were grown in 2× SG medium at 37°C for 24 h (31), and the serial dilutions of the cell culture were plated on LB agar plates before and after heating at 80°C for 10 min.
RNA isolation and Northern blot analysis.
Total RNA was isolated as described previously (53). A DNA probe for detecting scoC was prepared by PCR with a DIG Probe Synthesis Kit (Boehringer Mannheim, Mannheim, Germany) and the primers ScoC-1 and ScoC-2 (Table 2). RNA was denatured with formamide and blotted to a Nylon membrane (Boehringer Mannheim). mRNA for scoC was detected with the DNA probe and a DIG Luminescent Detection Kit (Boehringer Mannheim).
Real-time reverse transcription (RT)-PCR analysis.
cDNA was synthesized and amplified using relevant PCR primers and a Real Time One Step RNA PCR Kit (Takara Biomedicals, Shiga, Japan) according to the procedure recommended by the supplier. The amplification step of DNA was monitored by the Smart Cycler System (Cepheid) with SYBR Green I, and the relative concentrations of mRNAs were estimated by the procedure provided by Cepheid. The PCR products synthesized by this procedure were verified by melting curve analysis and by agarose gel electrophoresis for size determination. The rRNA levels in the RNA samples were also determined to confirm that the same amounts of RNA were used for the PCR.
Western blot analysis.
Western blotting and the subsequent detection of digoxigenin-labeled protein bands were done according to the procedure described previously (12). Polyclonal antibody against a synthetic 13-amino-acid peptide, NH2-ADEPAEELEPVNS-COOH, derived from a C-terminal part of ScoC was raised in rabbits and prepared by Sawady technology Inc. (Tokyo, Japan).
RESULTS
Disruption of salA resulted in a decrease of aprE-lacZ expression.
We screened for exoprotease-deficient strains among the B. subtilis gene disruption mutants by the halo assay (see Materials and Methods). As a result, we found a strain carrying a disruption at the ybaL locus to produce a significantly smaller halo around the colony (data not shown). We renamed ybaL salA, which stands for a regulator of scoC, aprE, and ybaL (see below). The salA gene was originally reported as a locus (rec223) that gave rise to a mitomycin C-sensitive phenotype and caused a defect in DNA-mediated transformation when present on a multicopy plasmid (10, 35). Although we compared the sensitivity of the wild-type CU741 and salA strains by inoculating them in liquid LB medium or on LB plates containing 1.6-fold serial dilutions of mitomycin C, we could not detect any difference in sensitivity between the strains, nor could we reproduce the multicopy effect of salA on transformation (data not shown).
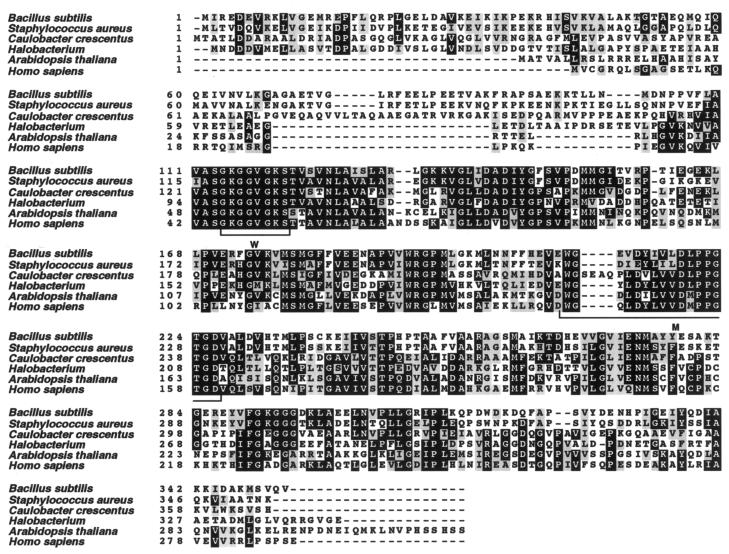
The salA gene is predicted to encode a protein of the Mrp family that is conserved among eukaryotes, prokaryotes, and archaea and carries an ATP-binding motif (Fig. 2) (3, 50). It should be noted that the term Mrp was defined by its genetic location in E. coli (3) but not by the function. To confirm the phenotype of low exoprotease production, we examined the expression of an aprE-lacZ translational fusion placed at the amyE locus in a salA disruption mutant. Although the halo is formed mainly by the exocellular neutral protease, the nprE gene product (17), we used the aprE-lacZ fusion to study the effect of salA on aprE expression, since the regulation of aprE expression has been better studied than that of nprE. The region upstream of the aprE-coding sequence in the strain used extends as far as −412 relative to the transcriptional start site. As shown in Fig. 3, the β-galactosidase activity derived from aprE-lacZ in the mutant was about 1/10 the level at the peak value detected in the wild-type cells. In this experiment, we used the wild-type and salA strains carrying a multicopy plasmid, pDG148, so that they serve as the control strains for the next experiment. The presence or absence of the vector plasmid had no effect on aprE expression (data not shown).
FIG. 2.
Alignment of amino acids among Mrp family proteins. The numbers indicate the positions of amino acids starting from the N-terminal end of each protein. The amino acids marked by black and gray boxes indicate identical and similar amino acids, respectively. Accession numbers in PIR; B. subtilis, A69743; Staphylococcus aureus, A90012; Caulobacter crescentus, F87508; Halobacterium, H84268; Arabidopsis thaliana, T06147; Homo sapiens, AK022722. Multiple alignment was carried out by CLUSTAL W (47), and the figure was made by BOXSHADE. The brackets marked by W and M under the alignment indicate the conserved Walker-motif and the MRP signature, respectively (51; BSORF Web site [http://bacillus.genome.ad.jp]).
FIG. 3.
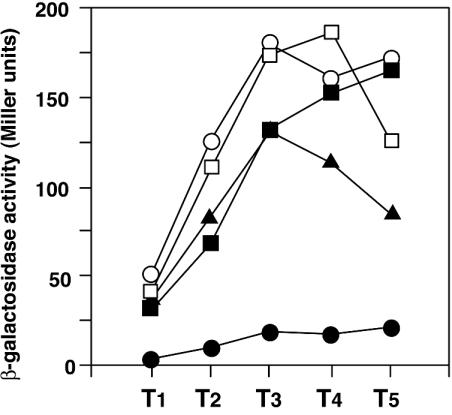
Inhibition of aprE-lacZ expression by salA disruption, and recovery by intact or mutant salA on plasmid pDG148. Cells were grown in Schaeffer's sporulation medium, and β-galactosidase activities were determined as described in Materials and Methods. Data are from one of two experiments. Numbers on the x axis represent the growth time in hours relative to the end of the vegetative growth (T0). ○, OAM145 (WT) carrying pDG148; •, OAM151 (salA) carrying pDG148; □, OAM145 (WT) carrying pDG-salA; ▪, OAM151 (salA) carrying pDG-salA; ▴, OAM151 (salA) carrying pDG-salA-m.
When plasmid pDG-salA carrying the intact salA gene was introduced into the salA disruption mutant, the level of aprE-lacZ expression was recovered roughly to that found in the wild-type cells (Fig. 3), indicating that the decrease in aprE-lacZ expression in the mutant was caused by the salA disruption. In pDG-salA, the salA gene is located downstream of the Pspac promoter that is inducible by IPTG (isopropyl-1-thio-β-d-galactopyranoside), but the addition of IPTG was not necessary for complementation. This result shows that the Pspac promoter is leaky, producing enough SalA to stimulate aprE expression from the pDG-salA construct without the addition of IPTG. The level of aprE-lacZ expression in the wild-type strain carrying pDG-salA (Fig. 3) was similar to that found in the wild-type strain carrying the pDG148 vector (Fig. 3). Even in the presence of IPTG, the expression level of aprE-lacZ was similar (data not shown). This was unexpected, since pDG-salA in the wild-type strain would cause a further enhancement in aprE-lacZ expression. We found by RT-PCR that, in the wild-type cells carrying pDG-salA, the induction levels of salA mRNA synthesis by IPTG diminished after exponential growth (19.0-fold at T0 [the growth time in hours relative to the end of vegetative growth], and 7.1-fold at T1), and reached a similar level (1.4-fold) at T2 as that found in the cells grown without IPTG (data not shown). Since the expression of aprE-lacZ reaches the highest level after T2 in our experimental condition (Fig. 3 and 4), a possible enhancing effect of SalA amplified by the addition of IPTG on aprE-lacZ expression could not be seen in the experiments shown in Fig. 3. However, it may also be possible that the expression level of aprE does not respond linearly to the level of SalA and the enhancing effect is seen only below a certain level of SalA. A further study will be necessary to clarify the relationship between the SalA level and aprE expression.
FIG. 4.
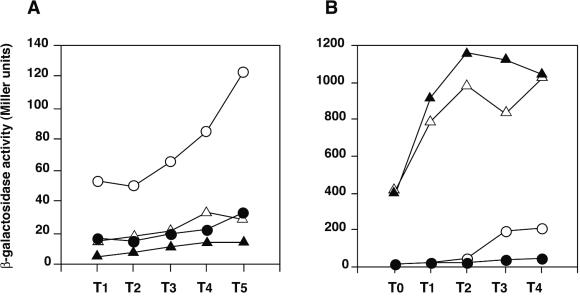
Epistatic relationship between salA and spo0A, abrB, and scoC in aprE-lacZ expression. Cells were grown in Schaeffer's sporulation medium, and β-galactosidase activities were determined as described in Materials and Methods. Data are from one of two experiments. Numbers on the x axis represent the growth time in hours relative to the end of the vegetative growth (T0). (A) ○, OAM169 (spo0A abrB); •, OAM180 (spo0A abrB salA); ▵, OAM167 (spo0A); ▴, OAM170 (spo0A salA). (B) ○, OAM145 (WT); •, OAM151 (salA); ▵, OAM157 (scoC); ▴, OAM164 (scoC salA).
Complementation of salA by pDG-salA was also observed in a strain bearing a translational aprE-lacZ fusion at the original aprE locus constructed by Campbell-type recombination (data not shown).
One characteristic of the Mrp group proteins is that they contain an ATP-binding motif (Fig. 2) (51). Since the SalA protein also carries such a motif, we examined whether it is involved in the regulation of aprE-lacZ expression. We introduced two amino acid substitutions on pDG-salA that are highly conserved among ATP-binding motifs by changing G119 and K120 to D and L, respectively. That an alteration of K in the motif has a dramatic effect on the protein activity has been demonstrated for B. subtilis clpC recently (48). The amino acid changes on plasmid pDG-Sal-m thus constructed, however, did not essentially affect the complementing activity of pDG148-salA in the salA mutant (Fig. 3). The variations in the β-galactosidase activities after T3 might be due to the difference in the stability between the salA mRNAs or the SalA proteins derived from the wild-type and the mutant salA cells. Consequently, we conclude that the putative ATP-binding motif in SalA is not involved in the regulation of aprE at least in a multicopy state.
Determination of the cis-acting region of SalA upstream of aprE.
It has been demonstrated that ScoC, SinR, and AbrB exert their negative effects on aprE expression by binding to the cis elements located upstream of the aprE-coding sequence (Fig. 1A). The positive regulator DegU directed by the degU32(Hy) mutant gene has also been shown to exert a positive effect at a site between −164 and −113 relative to the transcriptional start site (Fig. 1A) (13). If SalA regulates aprE expression through its effect on any of these factors, determination of the cis-acting region of SalA should provide information on the target of SalA. We introduced the salA disruption mutation into strains bearing aprE-lacZ fusions in which sequential deletions upstream of the aprE promoter had been introduced (reference 13 and this study). The β-galactosidase activities determined for these strains are shown in Fig. 1B. The salA mutation caused a decrease in the expression of the aprE-lacZ fusions with the sequences up to −412 and −340 of aprE, whereas it had no negative effect on the five fusions carrying deletions from −299 to −51. The DNA region downstream from −299 contains the SinR and AbrB binding sites, and also the target of DegU (7, 13, 41, 45), suggesting that these factors will not be the primary target of the salA mutation.
Gaur et al. (7) reported that the negative effect of multicopy sinR on aprE expression was seen in a construct carrying the region up to −340 but not −244. As shown in Fig. 1B, similar levels of stimulatory effect by sinR deficiency were observed in the constructs carrying deletions up to −412 through −267, indicating that the target of SinR resides in a region downstream from −267.
The stimulation of aprE expression by scoC deficiency was observed with the construct up to −340 but not with those up to −299 (Fig. 1). The results are in concert with the previous results in which ScoC binds to two regions from −324 to −267 (Fig. 1A) (16). On the other hand, the results are somewhat different from those observed previously in which a small but not a full ScoC effect was still observed with a fusion up to −244 (13). The discrepancy may have arisen by a strain difference. We note that aprE-lacZ expression was rather reduced in the fusions with only the proximal ScoC-binding site (Fig. 1B). Similar results were observed previously (13).
The deletion analyses shown here indicate that the target site of SalA is different from that of SinR and resides in a region around or upstream of −299 where the target site of ScoC is located.
Epistatic analyses of salA and other regulators, spo0A, abrB, and scoC.
Spo0A activates aprE expression in two ways: one is through the repression of abrB and the other through the transcriptional activation of sinI (Fig. 1A). We examined whether the decrease in aprE-lacZ expression by salA deficiency was still observed in a strain lacking the Spo0A-AbrB pathway. As shown in Fig. 4A, the salA mutation caused a decrease in aprE-lacZ expression in both the spo0A and spo0A abrB backgrounds. In a separate experiment, we observed that the salA mutation did not affect the sporulation efficiency of the cells grown in 2× SG medium (data not shown). These results demonstrate that the negative effect of SalA on aprE-lacZ expression is not through the regulatory function of Spo0A.
Since the salA mutation resulted in a decrease of aprE-lacZ expression directed from a region upstream from −299, scoC was the next candidate of the target of SalA. If this is the case, then the expression levels of aprE-lacZ in scoC and scoC salA mutants would be similar. The results depicted in Fig. 4B show that the inhibition of aprE-lacZ expression by salA deficiency was overcome by scoC mutation. However, the small difference in the β-galactosidase levels in the scoC and scoC salA mutants was reproducible. These results suggest that the regulatory pathway of SalA in aprE expression overlaps but does not completely coincide with that of ScoC.
Enhanced and prolonged transcription of scoC in the salA mutant in early stationary phase.
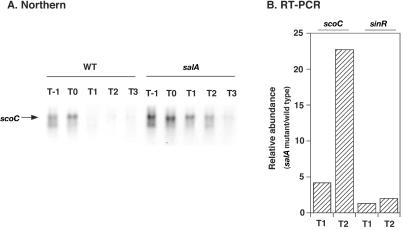
The results on the cis-acting region of SalA and the epistatic analyses may indicate that the salA disruption caused an increase in scoC expression or ScoC activity in the cell, resulting in repression of aprE-lacZ. To test this possibility, we first carried out Northern analysis on scoC using a scoC-specific probe and RNAs obtained from both the wild-type and salA strains. The results depicted in Fig. 5A show that the level of the scoC transcript in the wild-type strain was the highest during transition phase, followed by a decrease in early stationary phase. A similar profile of β-galactosidase activities from a scoC-lacZ transcriptional fusion in strain OM213 (salA+) was observed (data not shown). It was shown previously that the expression of scoC-lacZ declined only slightly during stationary phase (33). The discrepancy may be due to the different stability of the transcripts derived from scoC itself and different scoC-lacZ fusions. In the salA mutant, the scoC transcript levels at T−1 and T0 were significantly higher compared with those in the wild-type cell (Fig. 5A), and the difference was more apparent at T1 and T2. These results show that scoC transcription in the salA mutant was at a higher level and prolonged.
FIG. 5.
Abundance of scoC transcripts in a salA-deficient mutant compared with that in the wild-type strain. Strains used were CU741 and OM210 for the wild-type (WT) and salA strains, respectively. The procedures for RNA isolation and Northern and RT-PCR analyses are described in Materials and Methods. The RNA samples for the Northern and RT-PCR analyses were prepared separately. (A) Northern analysis. Cells were grown in Schaeffer's sporulation medium, and samples (20 ml) were withdrawn from T−1 to T4. Twenty micrograms of the isolated RNAs was applied to each lane, and scoC transcripts were detected with a digoxigenin-labeled PCR probe. In a separate experiment we have confirmed that there was no detectable band corresponding to the scoC transcript in the RNA samples obtained from strain OM213 (scoC). (B) RT-PCR analysis. Levels of mRNAs for scoC and sinR were quantified at T1 and T2 by using 1 μg each of the RNA samples. The oligonucleotide pairs ScoC-1 and ScoC-2 and SinR-F and SinR-R2 were used to detect sinR and scoC mRNAs, respectively. Relative abundance of mRNA at each stage was determined by dividing the mRNA levels in the mutant cells by those estimated in the wild-type cells.
We further quantified the observed differences in the mRNA levels by RT-PCR using RNAs prepared separately. In the salA cells, the scoC mRNA levels were 4- and 22-fold higher at T1 and T2, respectively, than those in the wild-type cells (Fig. 5B). These results are in agreement with those of the Northern analysis. The sinR mRNA contents determined by RT-PCR are also shown in Fig. 5B. The sinR transcript level in the salA mutant was no more than twofold compared to that in the wild-type strain at T2, which supports the notion that sinR is not the primary target of SalA. Similar results were obtained by Northern analysis on sinR mRNAs in the wild-type and salA mutant cells (data not shown).
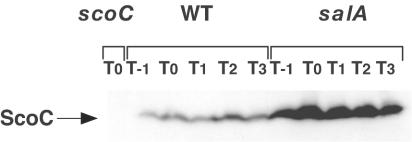
We next examined the ScoC protein levels by Western analysis using an antibody raised against a C-terminal 13-amino-acid peptide of ScoC. As shown in Fig. 6, the ScoC antibody bound to a protein band with a molecular size corresponding to that of ScoC. The protein band was absent in a cell extract from the scoC mutant, indicating that the band detected is indeed that of ScoC. Unexpectedly, the ScoC levels in the wild-type cells were found to be similar among the cell extracts obtained from different stages of stationary phase (Fig. 6). This is in contrast to the results of the Northern experiments described above, and raised an interesting question of how the expression of aprE in the stationary-phase cells is enabled in the presence of the negative regulator, ScoC. In the salA strain, the ScoC levels were higher as expected from the results of the Northern blot analysis (Fig. 5A). On the basis of these results, we conclude that the elevated and prolonged transcription of scoC leads to the production of larger amounts of ScoC in the salA cells.
FIG. 6.
Western analysis on ScoC proteins. Strains used were CU741, OM210, and OM213 for the wild-type (WT), salA mutant, and scoC mutant strains, respectively. Cells grown in Schaeffer's sporulation medium (50 ml) were withdrawn from T−1 to T3, washed and resuspended in 1 ml of TE buffer (10 mM Tris-HCl [pH 8.0], EDTA 1 mM), and disrupted by French press. Equal amounts of the protein were applied on a sodium dodecyl sulfate-14% polyacrylamide gel, and the ScoC protein was detected by ScoC antibody as described in Materials and Methods.
Expression of salA.
To study the expression of salA itself, a salA-lacZ translational fusion was constructed at the amyE locus, and β-galactosidase activities were determined in the salA and wild-type cells. It was found that the expression of salA-lacZ was constitutive, and the β-galactosidase activities at the transition state were twofold higher in the salA disruptant (data not shown), suggesting that salA expression is autoregulated.
DISCUSSION
We have shown in this paper that a new locus, salA (ybaL), negatively regulates scoC expression. It is a monocistronic operon gene located at 13.4° on the B. subtilis chromosome map, and encodes a putative protein belonging to the Mrp family (Fig. 2). Unexpectedly, the putative ATP-binding motif in SalA was unrelated to its activity to regulate aprE expression through ScoC, although the motif is ubiquitous among the Mrp family proteins. The Mrp family proteins are conserved among the three kingdoms (3, 27, 37, 50), but their function is not known so far in any organisms. All we know at present is that NBP35, an Mrp family protein in Saccharomyces cerevisiae, is an essential protein (50). In the case of salA, disruption of the gene did not affect the viability of the host cell (data not shown).
SalA may not be a DNA-binding protein, since no such typical amino acid sequence was predicted in the molecule. This may suggest that the effect of SalA on scoC, and therefore on aprE, expression is indirect. The small difference in the expression levels of aprE-lacZ in scoC and scoC salA mutants (Fig. 4B) leaves open the possibility that some other factor under the regulation of SalA is also involved. Thus, the exact molecular mechanism of transcriptional regulation by SalA remains unknown. As for the transcriptional regulation of scoC, the only known factor that affects scoC expression is AbrB (39, 45). This factor, however, is not involved in the regulation of aprE by salA as shown by the epistatic analyses using spo0A-abrB mutants (Fig. 4A). Thus, the possibility that SalA influences scoC transcription through AbrB is excluded. SalA might affect expression of scoC by associating with some transcription factor or RNA polymerase itself.
Overproduction of ScoC in the wild-type cell results in inhibition of sporulation (33). It was shown that ScoC levels were higher in the salA mutant (Fig. 6), but the sporulation efficiency was not affected in the 2× SG medium. The different observations might be due to different levels of ScoC in the two cells. With respect to sporulation, we note that in the presence of 2% glucose in the 2× SG medium (31) the sporulation efficiency of the salA mutant was reduced to 10% of that observed for the wild-type strain (data not shown). The results imply that catabolite repression becomes more severe as the ScoC concentration is increased. This is in concert with the previous results that catabolite repression of sporulation is rescued by scoC-null mutations (4, 33).
Northern analysis showed that scoC transcripts were not detected after T1 in the wild-type cell (Fig. 5A). In contrast, the Western blot experiment revealed constant levels of the ScoC protein in the cells obtained at T0 and thereafter (Fig. 6). Since aprE-lacZ expression is increased after the end of growth phase (Fig. 3 and 4), there may be some mechanism by which ScoC is stabilized but becomes inactive after entry into stationary phase. There could be several explanations for these observations. (i) ScoC might be inactivated by some effector molecule, since ScoC belongs to the MarR family repressor, which is known to be inactivated by small molecular effectors (26). (ii) ScoC might be sequestered from the aprE promoter by some factor(s). (iii) ScoC binds to the aprE promoter, but its inhibitory function might be counteracted by a positive factor(s). Transcription of aprE is known to be positively regulated by phosphorylated DegU and auxiliary factors including degR, degQ, senS, tenA, and proB (21, 29, 30, 39). One or several of these factors might work as a factor antagonizing the ScoC function in stationary phase, although there is no data concerning this issue at present.
It has been noted that salA (ybaL) expression is induced by disulfide stress (22). It is not known, however, whether this phenomenon is correlated with either scoC or aprE expression.
Acknowledgments
We thank Y. Kijima, S. Kinoshita, H. Kitoh, Y. Tuzuki, T. Inamori, and N. Takahashi for technical assistance and P. Zuber and K. Asai for plasmids. We also thank M. Takeuchi and T. Sato (Tokyo University of Agriculture and Technology), who independently discovered ybaL as an exoprotease-deficient mutant, for discussion.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (C) Genome Biology and a Grant-in-Aid for Scientific Research (B) and (C) from the Ministry of Education, Science, and Sports and Culture of Japan.
REFERENCES
- 1.Bai, U., I. Mandic-Mulec, and I. Smith. 1993. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 7:139-148. [DOI] [PubMed] [Google Scholar]
- 2.Caldwell, R., R. Sapolsky, W. Weyler, R. R. Maile, S. C. Causey, and E. Ferrari. 2001. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J. Bacteriol. 183:7329-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dardel, F., M. Panvert, S. Blanquet, and G. Fayat. 1991. Locations of the metG and Mrp genes on the physical map of Escherichia coli. J. Bacteriol. 173:3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dod, B., and G. Balassa. 1978. Spore control (sco) mutations of Bacillus subtilis. Regulation of extracellular protease synthesis in the spore control mutations scoC. Mol. Gen. Genet. 163:57-63. [Google Scholar]
- 5.Ferrari, E., D. J. Henner, M. Perego, and J. A. Hoch. 1988. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J. Bacteriol. 170:289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari, E., S. M. Howard, and J. A. Hoch. 1986. Effect of stage 0 sporulation mutations on subtilisin expression. J. Bacteriol. 166:173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaur, N. K., J. Oppenheim, and I. Smith. 1991. The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J. Bacteriol. 173:678-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaur, N. K., E. Dubnau, and I. Smith. 1986. Characterization of a cloned Bacillus subtilis gene that inhibits sporulation in multiple copies. J. Bacteriol. 168:860-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaur, N. K., K. Cabane, and I. Smith. 1988. Structure and expression of the Bacillus subtilis sin operon. J. Bacteriol. 170:1046-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gavrilova, E. V., S. L. Mekhedov, A. A. Prozorov, and F. K. Khasanov. 1992. Bacillus subtilis gene rec223: molecular cloning and proposed function of its protein product. Genetika 28:29-39. [PubMed] [Google Scholar]
- 11.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477-508. [DOI] [PubMed] [Google Scholar]
- 12.Hata, M., M. Ogura, and T. Tanaka. 2001. Involvement of stringent factor RelA in expression of the alkaline protease gene aprE in Bacillus subtilis. J. Bacteriol. 183:4648-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henner, D. J., E. Ferrari, M. Perego, and J. A. Hoch. 1988. Location of the targets of the hpr-97, sacU32(Hy), and sacQ36(Hy) mutations in upstream regions of the subtilisin promoter. J. Bacteriol. 170:296-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higerd, T. B., J. A. Hoch, and J. Spizizen. 1972. Hyperprotease-producing mutants of Bacillus subtilis. J. Bacteriol. 112:1026-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itaya, M., K, Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 17:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallio, P. T., J. E. Fagelson, J. A. Hoch, and M. A. Strauch. 1991. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J. Biol. Chem. 266:13411-13417. [PubMed] [Google Scholar]
- 17.Kawamura, F., and R. H. Doi. 1984. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J. Bacteriol. 160:442-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, K., S. D. Ehrlich, et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi, K., K. Shoji, T. Shimizu, K. Nakano, T. Sato, and Y. Kobayashi. 1995. Analysis of a suppressor mutation ssb (kinC) of sur0B20 (spo0A) mutation in Bacillus subtilis reveals that kinC encodes a histidine protein kinase. J. Bacteriol. 177:176-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koide, A., M. Perego, and J. A. Hoch. 1999. ScoC regulates peptide transport and sporulation initiation in Bacillus subtilis. J. Bacteriol. 181:4114-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunst, F., T. Msadek, and G. Rapoport. 1994. Signal transduction network controlling degradative enzyme synthesis and competence in Bacillus subtilis, p. 1-20. In P. J. Piggot, C. P. Moran, Jr., and P. Youngman (ed.), Regulation of bacterial differentiation. American Society for Microbiology, Washington, D.C.
- 22.Leichert, L. I., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louie, P., A. Lee, K. Stansmore, R. Grant, C. Ginther, and T. Leighton. 1992. Roles of rpoD, spoIIF, spoIIJ, spoIIN, and sin in regulation of Bacillus subtilis stage II sporulation-specific transcription. J. Bacteriol. 174:3570-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandic-Mulec, I., N. Gaur, U. Bai, and I. Smith. 1992. Sin, a stage-specific repressor of cellular differentiation. J. Bacteriol. 174:3561-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandic-Mulec, I., L. Doukhan, and I. Smith. 1995. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J. Bacteriol. 177:4619-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, P. F., and M. C. Sulavik. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441-448. [DOI] [PubMed] [Google Scholar]
- 27.Nakashima, H., M. J. Grahovac, R. Mazzarella, H. Fujiwara, J. R. Kitchen, T. A. Threat, and M. S. Ko. 1999. Two novel mouse genes—Nubp2, mapped to the t-complex on chromosome 17, and Nubp1, mapped to chromosome 16—establish a new gene family of nucleotide-binding proteins in eukaryotes. Genomics 60:152-160. [DOI] [PubMed] [Google Scholar]
- 28.Ogura, M., and T. Tanaka. 1996. Transcription of Bacillus subtilis degR is σD-dependent and suppressed by multicopy proB through σD. J. Bacteriol. 178:216-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogura, M., M. Kawata-Mukai, M. Itaya, K. Takio, and T. Tanaka. 1994. Multiple copies of the proB gene enhance degS-dependent extracellular protease production in Bacillus subtilis. J. Bacteriol. 176:5673-5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogura, M., and T. Tanaka. 1997. Expression of alkaline protease gene in Bacillus subtilis mutants that lack positive regulatory genes degR, degQ, senS, tenA and proB. Biosci. Biotechnol. Biochem. 61:372-374. [Google Scholar]
- 31.Ogura, M., Y. Ohshiro, S. Hirao, and T. Tanaka. 1997. A new Bacillus subtilis gene, med, encodes a positive regulator of comK. J. Bacteriol. 179:6244-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogura, M., L. Liu, M. Lacelle, M. M. Nakano, and P. Zuber. 1999. Mutational analysis of ComS: evidence for the interaction of ComS and MecA in the regulation of competence development in Bacillus subtilis. Mol. Microbiol. 32:799-812. [DOI] [PubMed] [Google Scholar]
- 32a.Ogura, M., K. Shimane, K. Asai, N. Ogasawara, and T. Tanaka. 2003. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol. Microbiol. 49:1685-1697. [DOI] [PubMed] [Google Scholar]
- 33.Perego, M., and J. A. Hoch. 1988. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J. Bacteriol. 170:2560-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson, J. B., M. Gocht, M. A. Marahiel, and P. Zuber. 1989. AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation gene and an antibiotic biosynthesis gene. Proc. Natl. Acad. Sci. USA 86:8457-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekiguchi, J., K. Akeo, H. Yamamoto, F. K. Khasanov, J. C. Alonso, and A. Kuroda. 1995. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which affects germination in Bacillus subtilis. J. Bacteriol. 177:5582-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shafikhani, S. H., I. Mandic-Mulec, M. A. Strauch, I. Smith, and T. Leighton. 2002. Postexponential regulation of sin operon expression in Bacillus subtilis. J. Bacteriol. 184:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shahrestanifar, M., D. P. Saha, L. A. Scala, A. Basu, and R. D. Howells. 1994. Cloning of a human cDNA encoding a putative nucleotide-binding protein related to Escherichia coli MinD. Gene 147:281-285. [DOI] [PubMed] [Google Scholar]
- 38.Shimotsu, H., and D. Henner. 1986. Construction of a single copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 39.Smith, I. 1993. Regulatory proteins that control late-growth development, p. 785-800. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C
- 40.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 41.Strauch, M. A. 1995. In vitro binding affinity of the Bacillus subtilis AbrB protein to six different DNA target regions. J. Bacteriol. 177:4532-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strauch, M. A. 1995. AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon. J. Bacteriol. 177:6727-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauch, M. A., and J. A. Hoch. 1993. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol. Microbiol. 7:337-342. [DOI] [PubMed] [Google Scholar]
- 44.Strauch, M., V. Webb, G. Spiegelman, and J. A. Hoch. 1990. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. USA 87:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 8:1615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka, T., and M. Kawata. 1988. Cloning and characterization of Bacillus subtilis iep, which has positive and negative effects on production of extracellular proteases. J. Bacteriol. 170:3593-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turgay, K., M. Persuh, J. Hahn, and D. Dubnau. 2001. Roles of the two ClpC ATP binding sites in the regulation of competence and the stress response. Mol. Microbiol. 42:717-727. [DOI] [PubMed] [Google Scholar]
- 49.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 50.Vitale, G., E. Fabre, and E. C. Hurt. 1996. NBP35 encodes an essential and evolutionary conserved protein in Saccharomyces cerevisiae with homology to a superfamily of bacterial ATPases. Gene 178:97-106. [DOI] [PubMed] [Google Scholar]
- 51.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, L.-F., and R. H. Doi. 1990. Complex character of senS, a novel gene regulating expression of extracellular-protein genes of Bacillus subtilis. J. Bacteriol. 172:1939-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida, K.-I., K. Kobayashi, Y. Miwa, C.-M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]