Abstract
The TtgGHI efflux pump of Pseudomonas putida extrudes a variety of antibiotics and solvents. We show that the ttgGHI operon is transcribed in vitro and in vivo from a single promoter and not from two overlapping promoters as previously proposed. The expression of this promoter is controlled by the TtgV repressor, whose operator expands through four helical turns that overlap the −10 region of the promoter. We also show that TtgV is released from its operator on binding of effectors such as aliphatic alcohols. Mutational analysis of the ttgGHI promoter revealed that substitutions at −13, −12, and −8 yielded promoters that were unable to drive transcription whereas certain mutations at −9, −11, and −6 to −3 increased expression in vivo. The cause of the increased expression was either a decrease in the affinity of the TtgV protein for its operator or an increase in the affinity of RNA polymerase for the mutant promoters.
Multidrug efflux pumps are widely distributed among prokaryotic and eukaryotic organisms (6, 16-18, 25, 29). In bacteria, resistance to a number of antibiotics, superoxide-generating agents, dyes, and organic solvents is mediated by different families of multidrug transporters. Toluene, styrene, and toxic solvents with log Pow values between 1.5 and 3.5 (i.e., the logarithm of the partition coefficient of the target compound in a mixture of octanol and water) partition preferentially in the cell membrane, leading to the disorganization of the inner membrane, which causes cell death. In solvent-tolerant gram-negative bacteria, these organic solvents are removed by pumps which belong to the resistance-nodulation-division (RND) family, so that the solvents are kept below their toxicity threshold (19).
RND efflux pumps are made of three components: an inner membrane transporter (13, 30), an outer membrane channel (8), and a lipoprotein anchored in the inner membrane that extends in the periplasm (31) and that is probably involved in the correct assembly of the other two elements and is needed for optimal functioning of the efflux pump. These efflux pumps are energized by a proton motive force (15).
Pseudomonas putida DOT-T1E is able to thrive in liquid medium containing toluene (21). This strain exhibits an innate high resistance to solvents, which increases when bacteria are preexposed to sublethal concentrations of toluene (2, 20, 23). This is achieved through the cooperative efflux of the solvent by three RND pumps called TtgABC, TtgDEF, and TtgGHI (12, 23). The expression of ttgABC is constitutive, and its level does not vary significantly in the presence of toluene (2). The ttgDEF operon is not expressed at all in the absence of solvents, and its expression is higher in the presence of aromatic hydrocarbons (23). The ttgGHI operon is expressed at a basal level in the absence of solvents, and its expression increases about threefold in response to toluene. The TtgGHI efflux pump plays a pivotal role in the innate and induced tolerance to solvents in this strain (12, 23), and a ttgH knockout mutant is extremely sensitive to solvent shocks (22).
We suggested that ttgGHI expression occurred from two overlapping tandem promoters (23) and that it was controlled by the ttgV gene product (22). In the present study we show that expression from the ttgGHI operon takes place from a single promoter and that solvents such as 1-hexanol trigger the release of the repressor from the operator site, allowing transcription.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture medium.
The bacterial strains, cloning vectors, and plasmids constructed in this study are listed in Table 1. Bacterial strains were grown in Luria-Bertani (LB) medium at 30°C as described previously (22). Liquid cultures were shaken on an orbital platform operating at 200 rpm. Escherichia coli DH5α was used as the host strain to construct and maintain different plasmids (1). P. putida cultures were supplemented with solvents at 3 mM when indicated. When required, cultures were supplemented with the following antibiotics: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; rifampin, 20 μg/ml; and tetracycline, 20 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| P. putida DOT-T1E | Rifr | 21 |
| E. coli DH5α | recA1 | 1 |
| Plasmids | ||
| pANA96 | Tcr, ttgG promoter cloned in pMP220 | 22 |
| pET28b(+):ttgV | Apr, vector use to produce TtgV | 22 |
| pGG1 | pUC18 bearing an 8-kb BamHI fragment with ttgGHI and ttgVW | 23 |
| pMP220 | Tcr, promoterless lacZ expression vector | 26 |
| pMExb | Mutant ttgG promoter cloned in pMP220 | This study |
| pET103-P ttgG | Apr, promoter of ttgG cloned upstream of the T7 terminator in pET103 | This study |
Apr, Kmr, Rifr, and Tcr stand for resistance to ampicillin, kanamycin, rifampin, and tetracycline, respectively.
x indicates the plasmid number bearing different mutant promoters fused to ′lacZ.
Nucleic acid techniques.
DNA preparation, digestion with restriction enzymes, analysis by agarose gel electrophoresis, isolation of DNA fragments, ligations, and transformations were done by standard procedures (1). Plasmid DNA was sequenced on both strands with specifically designed primers, using an automatic DNA sequencer (ABI-PRISM 3100; Applied Biosystems). P. putida cells were electroporated done as described previously (23). RNA preparation and primer extension were done as described by Marqués et al. (11).
For footprint assays, we used the TtgV protein prepared as described by Rojas et al. (22). DNase I footprint assays were carried out as described previously for identification of the operator to which TtgV binds (22), whereas dimethyl sulfate (DMS) footprint analyses were carried out as described by Ausubel et al. (1).
Electrophoretic mobility shift assay (EMSA).
A 210-bp DNA fragment containing the sequence between the nucleotides before the first start codons of ttgV and ttgG was amplified by PCR from plasmid pGG1 using the following set of primers: 5′-GGAATTCATCTTTCCTCTGCGCGTACG-3′ and 5′-AACTGCAGACGGGGGCTATTGCTGAATCG-3′. Cycling parameters were 4 min at 96°C followed by 30 cycles at 96°C for 1 min, 60°C for 30 s, and 72°C for 30s, and ending with 3 min at 72°C. These fragments were isolated from agarose gels and end labeled with 32P as described previously (22, 27). About 1 nM labeled DNA (∼1.5 × 104 cpm) was incubated with increasing amounts of purified TtgV for 10 min at 30°C in 10 μl of TGED binding buffer (10 mM Tris-HCl [pH 8.0], 5% [vol/vol] glycerol, 0.1 mM EDTA, 1 mM dithiothreitol) containing 20 μg of poly(dI-dC) per ml and 200 μg of bovine serum albumin per ml. Reaction mixtures were electrophoresed in a nondenaturing 4.5% (wt/vol) polyacrylamide gel in Tris-glycine buffer (0.2 M glycine, 0.025 M Tris-HCl [pH 8.6]). The results were analyzed with Molecular Imager FX equipment (Bio-Rad, Madrid, Spain).
Construction of PttgG mutant promoters by PCR.
The PttgG mutant promoters were generated by overlap extension PCR mutagenesis, as described previously (5). The internal oligonucleotide primers used for mutagenesis exhibited one mismatch with the wild-type sequence. After DNA amplification, the resulting DNA was digested with EcoRI and PstI and the 210-bp EcoRI-PstI PttgG mutant was inserted between the EcoRI and PstI sites of pMP220 to yield plasmid pMEx (the x indicates the plasmid number). All mutant PttgG promoters generated in this study were confirmed by DNA sequencing.
Single-round in vitro transcription assays with supercoiled plasmid DNA.
Reactions (20-μl reaction mixtures) were performed with STA buffer (10 mM Tris-acetate [pH 8.0], 8 mM magnesium acetate, 3.5% [wt/vol] polyethylene glycol, 10 mM KCl, 1 mM dithiothreitol) containing 10 or 100 nM σ70-holoenzyme (Epicentre), 20 U of RNAsin (Promega), and around 10 nM supercoiled plasmid DNA template. The reaction mixtures were incubated for 10 min at 30°C before the addition of the following elongation mixture: 0.1 mM each ATP, CTP, GTP, and UTP; 0.3 μCi of [α-32P]UTP (20 μCi/μl); and 100 μg of heparin per ml. After a further 10-min incubation at 30°C, the reactions were stopped by chilling to 4°C and the products were precipitated with 0.25 volume of 10 M ammonium acetate and 2.5 volume of ethanol. The pellets were washed with 80% (vol/vol) ethanol. Dried pellets were resuspended in 8 μl of water plus 4 μl of formamide sequencing dye. Samples were electrophoresed in a 6.5% (wt/vol) polyacrylamide denaturing sequencing gel. The results were analyzed using Molecular Imager GS525 equipment with Quantity One software (Bio-Rad, Madrid, Spain).
RESULTS
In vitro evidence suggests that the ttgGHI promoter is expressed from a single promoter rather than two overlapping promoters.
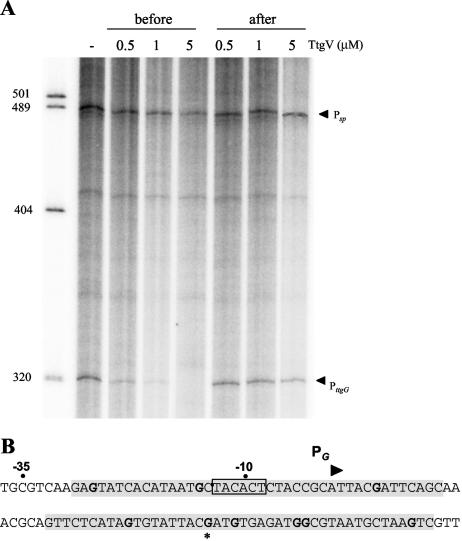
Rojas et al. (23) suggested that in P. putida DOT-T1E, the ttgGHI operon was transcribed from two promoters, PttgG1 and PttgG2, since two potential transcription start points separated by 5 nucleotides were found in vivo regardless of the growth phase and growth in the absence or presence of toluene. To determine whether the two bands represented two promoters or whether the 5-nucleotide-shorter band was a degradation product of the larger one, we carried out independent in vitro transcription assays with two plasmids carrying the ttgGHI promoter region upstream of the early phage T7 terminator. We found that with RNA polymerase in excess, the ttgGHI promoter drove the synthesis of a single mRNA (Fig. 1A). The RNAs produced from these plasmids were 320 and 141 nucleotides. Based on the size, we determined that the transcription start point of the in vitro mRNA corresponded to the longer mRNA obtained in vivo. In further assays, we used the plasmid that yielded the 320-nucleotide mRNA. To confirm that the ttgG promoter was regulated by TtgV, we carried out in vitro transcription assays in the presence of 0.5, 1, and 5 μM TtgV, which was added before or after the RNA polymerase. When TtgV was added before the RNA polymerase, transcription from the ttgGHI promoter decreased to almost undetectable levels for the highest TtgV concentration tested (Fig. 1A). However, when TtgV was added after the RNA polymerase, inhibition of transcription was moderate and the amount of mRNA synthesized was 70 to 80% of that produced in the absence of TtgV. As a control we used a promoter (Psp) of the plasmid that yielded a 488-nucleotide mRNA (Fig. 1A). As expected, the level of mRNA synthesized from the Psp promoter did not change significantly in the presence of TtgV. These results suggested that RNA polymerase may recognize a single promoter upstream from the ttgGHI operon and that modulation of its expression is mediated by TtgV.
FIG. 1.
In vitro transcription from PttgG in pET103-PttgG. (A) Single-round transcription assays were carried out as described in Materials and Methods. The assays were performed for 10 min at 30°C in the absence of TtgV (−) or in the presence of 0.5, 1, or 5 μM TtgV that was added before or after the addition of 100 nM RNA polymerase. The 320- and 488-nucleotide mRNAs synthesized from PttgG and Psp, respectively, are indicated by arrowheads. (B) Binding region of TtgV protein as determined by in vitro DNase I and DMS footprinting assays. The region protected by TtgV deduced from DNase I digestion is shaded. The protected G's (DMS footprinting) are shown in bold. The asterisk identifies a hyperreactive position. The −10 and −35 positions of PttgG are indicated by dots. The +1 position and the direction of transcription are marked with a triangle.
Site-directed mutagenesis of the region upstream from the ttgGHI promoter.
The position of the −10 box in promoters is variable. To define with precision the promoter of the ttgGHI operon, we mutagenized the region of this promoter from positions −3 to −16 (Fig. 1B) by introducing as many single-point mutations as possible using oligonucleotide site-directed mutagenesis (Table 2) and analyzed the mutant promoters for expression in vivo by using fusions of the mutant promoters to ′lacZ, and in vitro by performing in vitro transcription assays.
TABLE 2.
β-Galactosidase expression with fusions of the wild-type ttgG promoter or single-point ttgG mutant promoters to ′lacZa
| Location and base changed | β-Galactosidase activityb
|
|
|---|---|---|
| −Toluene | +Toluene | |
| Wild type | 290 | 700 |
| T-16→A | 280 | 725 |
| T-13→A | 5 | 10 |
| T-13→G | 10 | 10 |
| T-13→C | 2 | 2 |
| A-12→C | 10 | 8 |
| A-12→G | 7 | 10 |
| A-12→T | 10 | 15 |
| C-11→A | 570 | 610 |
| C-11→T | 2,755 | 3,360 |
| A-10→C | 130 | 235 |
| A-10→G | 70 | 150 |
| C-9→A | 1,690 | 2,700 |
| C-9→G | 68 | 180 |
| C-9→T | 175 | 390 |
| T-8→A | 10 | 30 |
| T-8→G | 10 | 22 |
| T-8→C | 3 | 4 |
| C-7→G | 15 | 30 |
| C-7→T | 180 | 465 |
| T-6→A | 15 | 40 |
| T-6→C | 455 | 1,100 |
| T-6→G | 460 | 1,080 |
| A-5→C | 265 | 710 |
| C-4→A | 1,410 | 1,560 |
| C-4→G | 1,580 | 1,825 |
| C-3→G | 1,570 | 1,820 |
P. putida DOT-T1E bearing pANA96 (PttgG::′lacZ) or pMEx bearing a fusion of the indicated mutant PttgG promoter to ′lacZ were grown on Luria-Bertani medium with tetracycline in the absence or presence of 3 mM toluene. After 4 h of incubation, the β-galactosidase activity (Miller units) in permeabilized cells was determined in triplicate (3).
Values are the average of three to five independent duplicate assays. Standard deviations were below 10% of the given values.
The transcriptional fusions to ′lacZ revealed that point mutations at positions −13, −12, and −8 produced mutants with negligible activity in comparison to the wild-type promoter (Table 2). Mutations at −10 (A→C or G), −9 (C→G or T), and −7 (C→G or T) also produced mutant promoters with lower activity than the wild-type promoter (the activities ranged between 23 and 62% of that of the wild type). We also found that certain mutant promoters had higher activity than the wild type (between 1.9- and 9.3-fold that of the wild-type): C-11→T, C-9→A, T-6→C or G, C-4→A or G, and C-3→G (Table 2). Mutations at −14 and −16 had no significant effect on the level of expression of the promoter. Primer extension assays confirmed the results of the β-galactosidase assays; namely, when we found no β-galactosidase activity, we found no mRNA, and when we found lower or higher β-galactosidase activity than those in the wild type, the mRNA levels estimated by primer extension correlated with the results of the enzymatic assay (data not shown).
In vitro transcription assays with mutant promoters revealed that whereas mutant promoters altered at −13, −12, −8 and other mutants altered at −7 and −6 were not transcribed in vitro, promoters altered at any of the other positions were transcribed (data not shown).
Given that mutations at positions −13, −12, and −8 yielded no β-galactosidase activity (or very low levels of activity) and did not drive the expression of mRNA in vivo and in vitro, we considered that these positions delimited the −10 hexamer with the 5′-TACACT-3′ sequence. This sequence exhibits four of six identities to the −10 consensus of the promoters recognized by σ70 (5′-TATAAT-3′) (9).
Why do certain point mutations in the ttgG promoter result in an increase in expression?
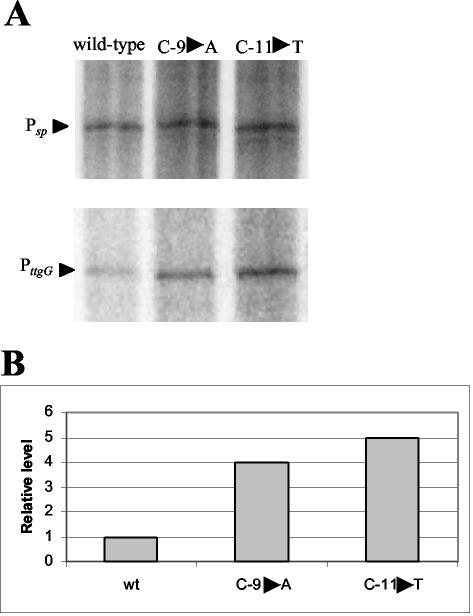
DNase I footprint assays showed that TtgV covered four DNA helical turns that overlap the ttgGHI promoter (22). The increase in expression of certain mutant promoters (i.e., C-3→G, C-4→G or A, C-9→A, and C-11→T) in the ttgG promoter may have resulted from either an improved −10 box for RNA polymerase or a decrease in the affinity of TtgV for its target sequences. We tested whether the mutant promoters that exhibited a higher activity than the wild type in vivo (i.e., C-4→A or G, C-3→G, C-9→A, and C-11→T) also exhibited increased expression in vitro with respect to the wild-type promoter. These assays were done with limiting amounts of RNA polymerase. The level of transcripts obtained in vitro from mutant promoters C-4→A or G and C-3→G was similar to that obtained from the wild-type promoters (see Fig. 3 for the C-4→A mutant), whereas with mutant promoters C-9→A and C-11→T, the level of expression was higher (Fig. 2). We suggest that the increased expression with C-9→A and C-11→T may have resulted from a promoter whose sequence, TATACT in C-11→T and TACAAT in C-9→A, was more similar to the consensus sequence since only one mismatch was found in each mutant promoter.
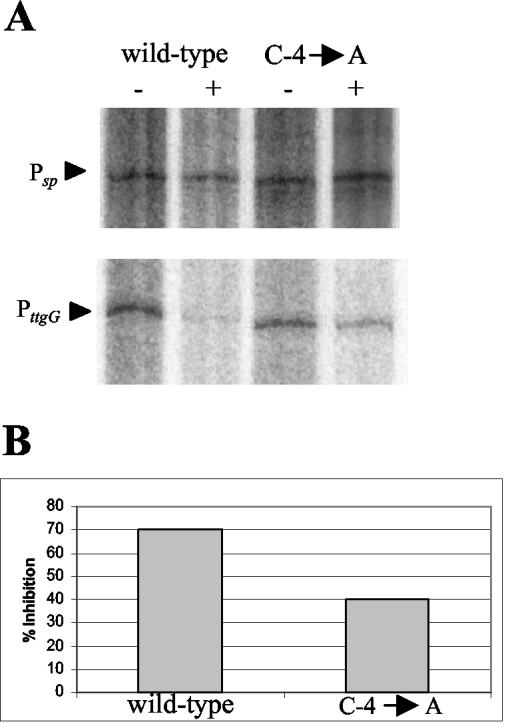
FIG. 3.
In vitro transcription from PttgG and a single-point mutant (C-4→A) in the absence and presence of TtgV. (A) Single-round in vitro transcription assays were carried out as described in the legend to Fig. 1A. The templates contained the wild-type promoter or the mutant (C-4→A) ttgG promoter. The TtgV protein was either not present (−) or added at a concentration of 0.5 μM prior to the addition of RNA polymerase (+). (B) Relative levels of transcription inhibition for each of the promoters tested. The level of transcription was considered 100% for each promoter in the absence of TtgV. The degree of transcription inhibition was calculated from the equation (relative level in the presence of TtgV/relative level in the absence of TtgV) × 100.
FIG. 2.
Single-round transcription in vitro with limiting amounts of RNA polymerase. Conditions are as described in the legend to Fig. 1A, except that 10 nM RNA polymerase was used instead of 100 nM to transcribe the wild-type and the indicated mutant promoters. (A) Section of the gel corresponding to the control Psp promoter, giving rise to a 488-nucleotide mRNA, and the wild-type PttgG or its single-point mutation derivative. (B) Relative levels of transcripts from PttgG. We assigned a relative value of 1 to the amount made from the wild type.
To explain the higher expression in vivo of the other mutant promoters (C-4→A or G, C-3→G), we hypothesized that the affinity of TtgV for its operator might be reduced in the C-4→A or -G and C-3→G promoters and that therefore it competes less effectively with the RNA polymerase for its binding site. To test this hypothesis we performed single-round transcription assays with the wild-type and mutant promoters. While the addition of 0.5 μM TtgV prior to RNA polymerase led to 70% inhibition in the in vitro transcription assay with the wild-type promoter, inhibition with the mutant promoters was only around 40%. Figure 3 shows the results obtained for the mutant promoter C-4→A. Similar results were obtained for the C-3→G mutant (data not shown).
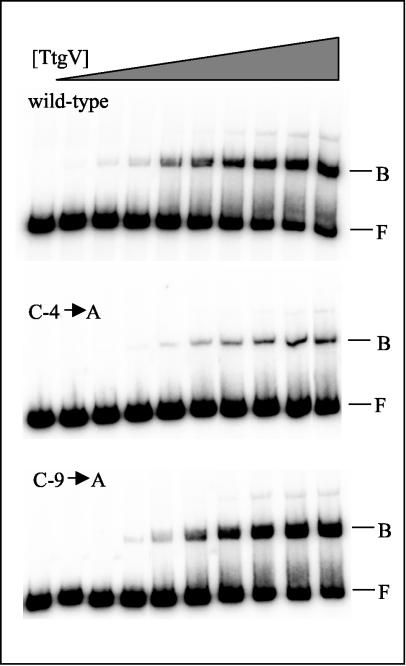
We also carried out EMSAs with the wild-type and mutant promoters. Figure 4 shows the results for the wild-type and mutant promoters C-4→A and C-9→A. We found that the C-4→A mutant promoter was less likely to be bound by TtgV. Similar results were obtained when the mutant promoters C-4→G or C-3→G were used (data not shown). In contrast, when EMSAs were done with the up mutant promoter C-9→A, the results were similar to those obtained with the wild-type promoter (Fig. 4), which supports the notion that increased expression in this mutant promoter could be the result of a higher affinity of the RNA polymerase for the mutant promoter.
FIG. 4.
EMSAs with the wild-type and C-4→A and C-9→A mutant promoters. The promoters were amplified by PCR as 210-bp fragments and labeled at the 5′ ends. About 1 nM DNA containing the ttgG promoter region was incubated with increasing concentrations of TtgV (from left to right, 0, 10, 50, 75, 100, 200, 300, 400, 500, and 700 nM) for 10 min and electrophoresed as indicated in Materials and Methods. B, bound DNA; F, free DNA.
1-Hexanol is an effector for TtgV and releases the repressor from its binding site in PttgG.
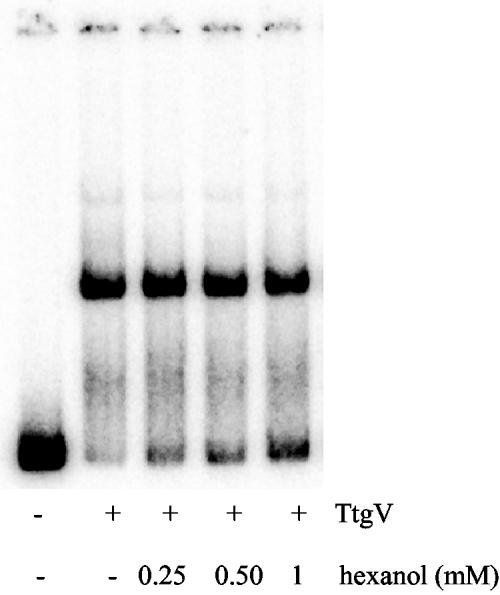
Rojas et al. (22) showed that the TtgGHI efflux pump expels a large number of aromatic hydrocarbons (toluene, styrene, and ethylbenzene), aliphatic alcohols (1-octanol and 1-hexanol), and antibiotics such as ampicillin, carbenicillin, nalidixic acid, and tetracycline. We determined the in vivo effector profile for TtgV by measuring β-galactosidase activity using PttgG::′lacZ fusions in the presence of different compounds. Our assays revealed that antibiotics were not inducers of the expression of the ttgGHI operon, in accordance with the results of Rojas et al. (22). Aromatic compounds such as toluene, styrene, propylbenzene, m-xylene, and indole and aliphatic alcohols, i.e., 1-octanol and 1-hexanol, induced expression of the operon between 2.3- and 3-fold (data not shown). The in vivo assays did not reveal whether this was due to a direct or an indirect effect on the TtgV repressor. When we tested the effect of 1-hexanol on TtgV target binding in EMSA, we observed that hexanol had an in vitro effect on the binding of TtgV to the ttgGHI-ttgV operator site. Figure 5 shows the increase in dissociation of TtgV from the operator containing the DNA fragment in EMSA when increasing concentrations of 1-hexanol were added to the binding region.
FIG. 5.
EMSAs with the wild-type promoter in the presence of 1-hexanol. Conditions are as described in the legend to Fig. 4, except that DNA corresponded to the wild-type ttgG promoter and a fixed concentration of TtgV (500 nM) was used. Samples were incubated for 10 min with the indicated 1-hexanol concentration before electrophoretic separation.
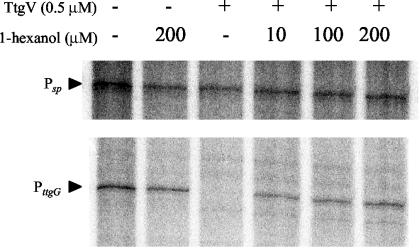
We also tested the effect of this compound in in vitro transcription assays using 20 to 200 μM 1-hexanol. In this range of concentrations, 1-hexanol did not interfere with RNA polymerase activity since the control Psp promoter yielded the expected 488-nucleotide mRNA. We then performed in vitro transcription assays with TtgV added before RNA polymerase and with increasing concentrations of 1-hexanol. We found that the higher the concentration of 1-hexanol, the higher the level of transcription from the wild-type PttgG (Fig. 6).
FIG. 6.
TtgV repression of the transcription of PttgG is alleviated by 1-hexanol. Single-round in vitro transcription assays were performed as described in the legend to Fig. 1A in the absence and presence of 0.5 μM TtgV added before the addition of RNA polymerase. Increasing concentrations of 1-hexanol were added as indicated. Other conditions as are described in Materials and Methods.
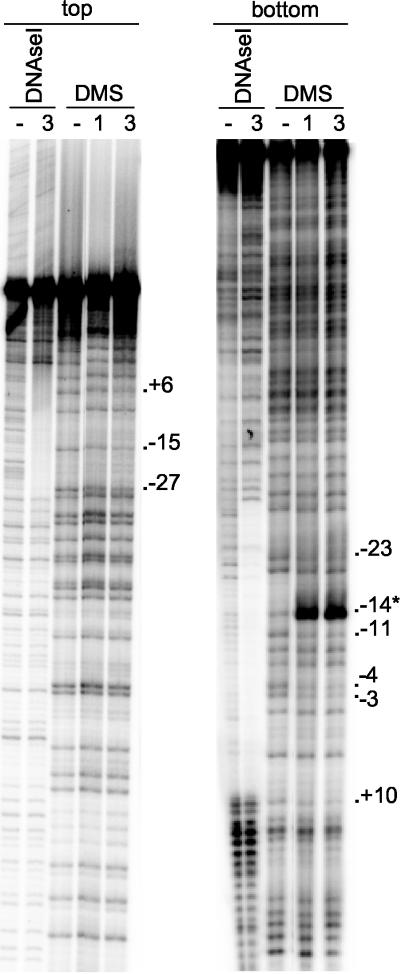
DMS footprint assays.
DNase I footprint assays revealed that TtgV protected the region in the PttgG promoter corresponding to positions +13 to −29 from digestion (22). To characterize the binding region in greater detail, DMS methylation protection assays were carried out. Figure 7 shows that G at position −14 in the bottom strand was hypermethylated in the presence of TtgV. This suggests that binding of TtgV to target DNA triggered local DNA conformational changes. We observed that G's at positions +6, −15, and −27 were protected in the top strand and G's at positions −23, −11, −4, −3, and +10 were protected in the bottom strand, suggesting that they might be contacted by TtgV on binding to its operator. All these positions are in accordance with our previous DNase I footprinting since they are located within the proposed binding site. Furthermore, the importance of G-4 and G-3 in TtgV operator recognition is consistent with our results showing that G-4→A and G-3→C are less prone to be recognized by TtgV.
FIG. 7.
In vitro methylation protection assay of the ttgGHI promoter region. PCR fragments comprising the ttgG promoter region were labeled at one 5′ end and incubated without (−) and with 1 or 3 μM purified TtgV before being subjected to treatment with DMS or DNase I digestion. The positions of the G's that are protected from methylation by the repressor are indicated; the asterisk indicates a hyperreactive position.
DISCUSSION
Our results establish that the ttgGHI operon is probably transcribed from a single promoter rather than from two tandem promoters, as proposed before (23). This assumption is supported by in vitro transcription analysis, which revealed a single transcription start point, and mutational analysis of the region upstream from the +1 position. Mutational studies support the notion that the most probable −10 hexamer of the promoter lies between positions −8 and −13 since a number of mutations in this stretch resulted in mutants that could not be transcribed either in vivo or in vitro. In contrast to these mutants, which lacked activity, we found that certain mutations at −9 and −11 increased promoter activity. This seems to be the result of a better −10 region that is recognized more efficiently by RNA polymerase.
DNase I footprint analysis revealed that TtgV covers four helical turns that cover the promoter region between positions +13 and −29 (22), which suggests that TtgV represses transcription from the ttgG promoter by competing with the RNA polymerase for promoter binding. To gain insight into the TtgV repression mechanism, we performed in vitro transcription assays. When TtgV was incubated with the ttgG promoter before the addition of the RNA polymerase, transcription from PttgG was repressed but that from a reference promoter, Psp, was unaffected (Fig. 1A). The repression level of ttgG transcription correlated with the dose of TtgV added. However, TtgV did not repress ttgG transcription significantly when it was added after the formation of the RNA polymerase-ttgG promoter open complex (Fig. 1). This indicates that TtgV represses ttgG transcription by physically competing with the RNA polymerase for promoter binding, as in the case of the well-established classic model of repressor action (4, 7, 24, 28). DMS methylation protection assays revealed that in the presence of TtgV, G-14 in the bottom strand becomes hypermethylated. This probably indicates that TtgV binding to the operator region provokes a distortion immediately upstream from the −10 region, which may prevent proper recognition of the promoter by RNA polymerase. DMS footprinting, however, did not help us to discern whether TtgV was able to recognize a relatively highly conserved direct repeat [5′-(A/C)T(G/A)N(C/T)NCA-3′] that appeared in four consecutive helical turns (Fig. 1B) or whether TtgV recognized some of the imperfect inverted repeats in the protected region. Analysis of mutant promoters revealed that mutations C-4→A or G and C-3→G resulted in increased expression in vivo but not in vitro and correlated with a decrease in the affinity of TtgV for the target operator as determined in EMSAs, as well as the protection of the bases at these positions in DMS footprint assays.
The TtgV protein belongs to the IclR family of regulators (10, 14). Members of this family repress the transcription of specific cognate genes in the absence of the target chemical and detach from the operator in response to the presence of a specific signaling molecule. A similar mechanism seems to operate for TtgV: EMSAs revealed that in the presence of increasing concentrations of 1-hexanol, TtgV dissociated from its target operator. This was corroborated by in vitro transcription assays showing that when TtgV was present, there was no mRNA synthesis, but in the presence of 1-hexanol, mRNA levels increased. This suggests that in the presence of 1-hexanol, TtgV was released from its operator site and RNA polymerase was able to access and transcribe the ttgG promoter. These are the first experimental indications of a compound able to directly promote TtgV dissociation from its operator site. Future work on the ttgGHI-ttgV system should reveal more intimate details of the molecular interactions between the regulator, its effectors, and its target DNA.
Acknowledgments
This work was supported by EU project QLK3-CT-2001-00435, grant RGY 0021/2002 from The Human Frontier Science Program, and funds from the Junta de Andalucía to Research Group CVI-191.
We thank C. Meng and A. Felipe for Technical Assistance, M. M. Fandila and C. Lorente for secretarial assistance, and K. Shashok for checking the English in the manuscript.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Duque, E., A. Segura, G. Mosqueda, and J. L. Ramos. 2001. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 39:1100-1106. [DOI] [PubMed] [Google Scholar]
- 3.Gallegos, M. T., S. Marqués, and J. L. Ramos. 1996. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a σ70-dependent promoter or from σ70- and σ54-dependent tandem promoters according to the aromatic compound used for growth. J. Bacteriol. 178:2356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geanacopoulos, M., and S. Adhya. 2002. Genetic analysis of GalR tetramerization in DNA looping during repressosome assembly. J. Biol. Chem. 277:33148-33152. [DOI] [PubMed] [Google Scholar]
- 5.González-Pérez, M. M., J. L. Ramos, M. T. Gallegos, and S. Marqués. 1999. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J. Biol. Chem. 274:2286-2290. [DOI] [PubMed] [Google Scholar]
- 6.Gottesman, M. M., and I. Pastam. 1993. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu. Rev. Biochem. 62:385-427. [DOI] [PubMed] [Google Scholar]
- 7.Heldwein, E. E., and R. G. Brennan. 2001. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409:378-382. [DOI] [PubMed] [Google Scholar]
- 8.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2001. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 9.Lisser, S., and H. Margalit. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maloy, S. R., and W. D. Dunn. 1982. Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J. Bacteriol. 149:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marqués, S., J. L. Ramos, and K. N. Timmis. 1993. Analysis of the mRNA structure of the Pseudomonas putida TOL meta-fission pathway operon. Biochim. Biophys. Acta 1216:227-236. [DOI] [PubMed] [Google Scholar]
- 12.Mosqueda, G., and J. L. Ramos. 2000. A set of genes encoding a second toluene efflux system in Pseudomonas putida DOT-T1E is linked to the tod genes for toluene metabolism. J. Bacteriol. 182:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 14.Nasser, W., S. Reverchon, and J. Robert-Baudouy. 1992. Purification and functional characterization of the KdgR protein, a major repressor of pectinolysis genes of Erwinia chrysanthemi. Mol. Microbiol. 6:257-265. [DOI] [PubMed] [Google Scholar]
- 15.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1:516-523. [DOI] [PubMed] [Google Scholar]
- 17.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44:2233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-positive bacteria and the mycobacteria. Antimicrob. Agents Chemother. 44:2595-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-González, A. Rojas, W. Terán, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 20.Ramos, J. L., E. Duque, P. Godoy, and A. Segura. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 180:3323-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos, J. L., E. Duque, M.-J. Huertas, A. Haïdour. 1995. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J. Bacteriol. 177:3911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas, A., A. Segura, M. E. Guazzaroni, W. Terán, A. Hurtado, M. T. Gallegos, and J. L. Ramos. 2003. In vivo and in vitro evidence show that TtgV is the specific regulator of the TtgGHI multidrug and solvent efflux pump of Pseudomonas putida. J. Bacteriol. 185:4755-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rojas, A., E. Duque, G. Mosqueda, G. Golden, A. Hurtado, J. L. Ramos, and A. Segura. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 183:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rojo, F. 1999. Repression of transcription initiation in bacteria. J. Bacteriol. 181:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saier, J., and I. Paulsen. 2001. Phylogeny of multidrug transporters. Cell Dev. Biol. 12:205-213. [DOI] [PubMed] [Google Scholar]
- 26.Spaink, H. P., R. J. H. Okker, C. A., Wijffelman, E. Pees, and B. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 27.Sunnaborg, A., D. Klumpp, T. Chung, and D. C. LaPorte. 1990. Regulation of the glyoxylate bypass operon: cloning and characterization of iclR. J. Bacteriol. 172:2642-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terán, W., A. Felipe, A. Segura, A. Rojas, J. L. Ramos, and M. T. Gallegos. 2003. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 47:3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu, E. W., J. R. Aires, and H. Nikaido. 2003. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 185:5657-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976-980. [DOI] [PubMed] [Google Scholar]
- 31.Zgurskaya, H. I., and H. Nikaido. 1999. AcrA is a highly asymmetric monomeric protein capable of spanning the periplasm. J. Mol. Biol. 285:409-420. [DOI] [PubMed] [Google Scholar]