Abstract
A temperate, type IV pilus-dependent, double-stranded DNA bacteriophage named DMS3 was isolated from a clinical strain of Pseudomonas aeruginosa. A clear-plaque variant of this bacteriophage was isolated. DMS3 is capable of mediating generalized transduction within and between P. aeruginosa strains PA14 and PAO1, thus providing a useful tool for the genetic analysis of P. aeruginosa.
Pseudomonas aeruginosa is an opportunistic human pathogen that infects individuals with weakened immune systems, such as hospitalized patients and those suffering from severe burns or other traumatic skin damage or from cystic fibrosis. P. aeruginosa is also a model organism for the study of bacterial biofilms (3). Two of the best-characterized strains of P. aeruginosa are PAO1 (7) and PA14 (11).
While bacteriophages are a useful tool for the genetic analysis of bacteria, we were not aware of any bacteriophage capable of mediating generalized transduction for strain PA14. Furthermore, bacteriophages with broader host ranges might also be capable of transductional gene exchange between strains (1, 8). We report the identification and characterization of a bacteriophage of P. aeruginosa that is capable of generalized transduction within and between Pseudomonas strains PAO1 and PA14.
Isolation and initial characterization of phage DMS3.
Based on work with Myxococcus spp., we rationalized that we could trigger the lytic phase of a temperate bacteriophage present in Pseudomonas isolates by prolonged starvation (C. Manoil, personal communication). To test this hypothesis, ∼70 clinical isolates identified as P. aeruginosa but otherwise uncharacterized were obtained from the Dartmouth Hitchcock Medical Center. The clinical isolates were grown in Luria-Bertani (LB) medium for 48 h at 37°C, the cells were subsequently pelleted by centrifugation at 17,000 × g for 5 min, and the supernatant was transferred to a clean tube. To kill the remaining bacteria, several drops of chloroform were added to the supernatant and the mixture was vortexed for 30 s. The resulting potential phage lysates were stored at 4°C.
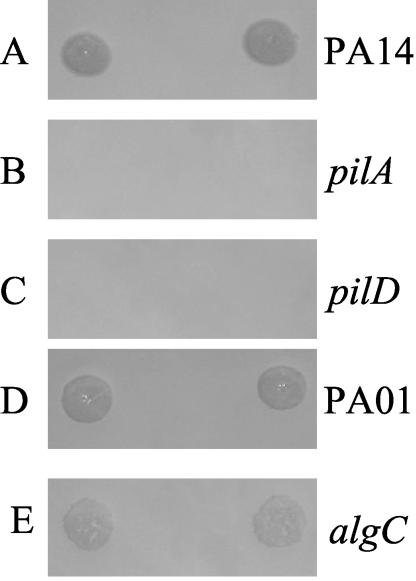
A phage lysate derived from one of the P. aeruginosa clinical isolates was found to be capable of infecting P. aeruginosa strains PAO1 and PA14 and four P. aeruginosa clinical isolates, as demonstrated by the zones of lysis resulting from phages spotted onto bacterial lawns. Bacterial lawns were produced by adding ∼5 × 108 CFU of P. aeruginosa from an overnight LB broth culture to 3 ml of LB top agar (0.8%) (12). The inoculated top agar was overlaid on an LB agar plate and allowed to solidify. Figure 1A shows the zones of lysis resulting from DMS3 spotted onto a lawn of PA14. Zones of lysis were also observed for DMS3 spotted onto lawns of PAO1 (Fig. 1D) and four clinical isolates (data not shown). The bacteriophage capable of infecting these bacterial strains was plaque purified and designated DMS3. DMS3 has a stable titer (as measured by PFU) over a period of several years when stored at 4°C in the LB medium used to prepare the lysate.
FIG. 1.
Plaque formation by DMS3. Five microliters of a lysate of phage DMS3 was spotted in duplicate onto lawns of P. aeruginosa wild-type and mutant strains. The zone of lysis (or clearing) on the plate indicates that DMS3 was able to infect the wild-type P. aeruginosa PA14 strain (A). In contrast, the phage was incapable of producing a zone of lysis on P. aeruginosa PA14 mutants defective in type IV pilus biogenesis (B and C). Both P. aeruginosa PAO1 (D) and the P. aeruginosa PAO1 algC mutant (E) supported formation of zones of lysis.
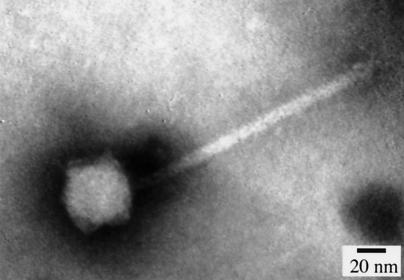
Transmission electron microscopy (TEM) was performed to determine phage morphology. TEM grids (copper; size, 300 mesh) were coated with 0.4% Formvar and then floated on 10-μl spots of phage lysates followed by negative staining for 4 min using 20 μl of 2% phosphotungstate (pH 4.0). After being stained, the grids were rinsed with water and air dried. The grids were viewed using a transmission electron microscope (JEM 100CX EM; JEOL Ltd.) at ×500,000 magnification. TEM showed that DMS3 has a tail approximately 110 nm in length and an icosahedral head approximately 50 nm in diameter (Fig. 2).
FIG. 2.
Electron micrograph of phage DMS3. A transmission electron micrograph of DMS3 negatively stained with phosphotungstate is shown.
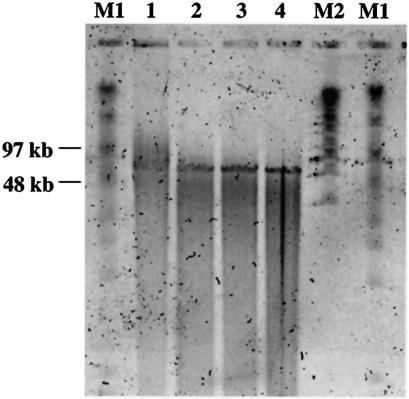
The phage chromosome was isolated as reported previously (12) to determine the size and nature of the nucleic acid. After the experiment was begun with 0.5 ml of a phage lysate (∼1010 PFU/ml), sodium dodecyl sulfate was added to achieve a concentration of 0.5% and proteinase K was added to achieve a concentration of 0.05 mg/ml. The solution was heated for 30 min at 65°C followed by four successive extractions with a 1:1 phenol-chloroform solution. The nucleic acid was then ethanol precipitated. Phage nucleic acid was identified as DNA on the basis of its sensitivity to digestion by restriction endonucleases. The nucleic acid isolated from the phage could be cleaved by restriction enzymes DpnII and AluI (New England Biolabs, Beverly, Mass.), which exclusively digest double-stranded DNA (dsDNA), suggesting that the nucleic acid of DMS3 is dsDNA. To determine the size of the phage genome, pulsed-field gel electrophoresis (PFGE) was performed as reported previously (4). Isolated phage DNA was treated with S1 nuclease (0 to 10 U) for 45 min at 37°C to linearize any circular DNA. Reactions were stopped with addition of cold EDTA (50 mM). Samples were electrophoresed in 1% PGFE-certified agarose (Bio-Rad) in 0.5× Tris-borate-EDTA. Electrophoresis was performed in a CHEF-DR II apparatus (Bio-Rad) at 14°C with a linearly ramped switching time of 1 to 12 s. A constant voltage of 5 V/cm (150 V) was used for 20 h. After electrophoresis, the gel was stained with SYBR Gold (Molecular Probes) and visualized on a STORM 860 apparatus (Amersham). The genome size of the phage is estimated on the basis of PFGE results to be between 50 and 65 kb (Fig. 3).
FIG. 3.
PFGE of phage DMS3 DNA treated with S1 nuclease. Isolated phage DNA was treated with S1 nuclease (lane 1, 0 U; lane 2, 0.5 U; lane 3, 1 U; lane 4, 10 U), which linearizes circular DNA. Lanes labeled M1 and M2 show size markers.
As previously reported (9), DMS3 is susceptible to host-controlled modification, as evidenced by efficiency of plating. As shown in Table 1, the titer of a DMS3 lysate was lower on a strain other than the one on which it was originally grown, suggesting that the phage DNA might be modified to escape restriction. Host-controlled modification of the phage was observed when it was grown on P. aeruginosa strain PAO1 or PA14. Although the basis of this host-controlled modification is unclear, for practical purposes these results suggest that phage grown on PA14 should be used to prepare lysates on strain PA14 and that phage grown on PA01 should be used to prepare lysates on strain PA01.
TABLE 1.
DMS3 is susceptible to host-controlled modification
| Strain on which the phage lysate was prepared | Lawn on which phage titration was performed | Titer (PFU/ml) |
|---|---|---|
| PA14 | PA14 | 5 × 1011 |
| PAO1 | 3 × 106 | |
| PAO1 | PAO1 | 1.1 × 108 |
| PA14 | 1.6 × 107 |
Type IV pili are the receptors for phage DMS3.
A number of surface factors have been implicated as bacteriophage receptors. The ability of DMS3 to form plaques on a strain carrying a mutation in algC (Fig. 1E), which is required for the synthesis of the lipopolysaccharide O-antigen and alginate, indicates that neither of these molecules serves as a receptor for this bacteriophage (2, 5). While DMS3 could form a zone of lysis on wild-type P. aeruginosa PA14 (Fig. 1A), no such plaques formed on a pilA or pilD mutant (Fig. 1B and C). Both the PilA and PilD proteins are required for the synthesis of type IV pili. Therefore, DMS3 utilizes the type IV pilus as its receptor.
DMS3 mediates generalized transduction within and between P. aeruginosa strains.
Phage lysates were prepared, as reported previously (8), by combining 100 μl of phage (∼1 × 109 PFU) with 100 μl of the donor bacterial strain (∼5 × 108 CFU from an overnight, LB-grown culture) and calcium chloride (10 mM). After a 15-min incubation at room temperature, the mixtures were combined with 3 ml of LB top agar (0.8%) and overlaid onto fresh LB plates. After ∼14 to 16 h of incubation at 37°C, 4 ml of LB liquid medium was added to the LB top agar containing cells and phage and the top agar was scraped from the surface of the agar plate and transferred (along with the added LB liquid medium) to a 50-ml conical tube. An additional 3 ml of LB liquid medium was added to this mixture, which was then shaken slowly at room temperature for 4 h to allow release of the phage from the top agar. The lysates were centrifuged for 5 min at 17,000 × g to remove bacteria and agar, the supernatants were transferred to a new tube, and the remaining bacterial cells were lysed by the addition of several drops of chloroform. The chloroform-containing lysates were vortexed for 30 s and stored at 4°C. The resulting phage titers were typically ∼1010 to 1011 PFU/ml.
Transductions were performed as reported previously (8), with tetracycline or gentamicin resistance genes as the selectable markers carried by donor strains. Typically, 100 μl of an overnight LB-grown culture (∼5 × 108 CFU) was mixed with 100 μl of a phage lysate with a titer of 1010 to 1011 PFU/ml and calcium chloride was added to achieve a 10 mM concentration. After incubation for 15 min at 37°C, transduction mixtures were plated on LB agar medium supplemented with tetracycline (150 μg/ml) or gentamicin (100 μg/ml) as appropriate; transductants typically appeared after 48 h of incubation at 37°C. DMS3 mediated transduction within P. aeruginosa strains PAO1 and PA14 at an efficiency of ∼5 × 10−9 transductants/PFU. Representative transduction frequencies for a variety of markers are shown in Table 2. The addition of EGTA (10 mM) or citrate (10 mM) or altering the multiplicity of infection did not improve transduction frequencies. We were unable to demonstrate that phage DMS3 was capable of mediating generalized transduction of markers with strain PAK. At least 15 different genetic markers from strains PA14 or PAO1 mapping across the chromosome, including alleles mapping to the flgK, pilU (a hyper-piliated strain), sadB (a biofilm mutant), det-3 (a biofilm mutant), lasR, and lasI genes, were transduced using phage DMS3. Several of these transduction events were subsequently confirmed by PCR mapping and sequencing (10) or by phenotypic testing (e.g., flagellum-mediated swimming motility or biofilm development). These data demonstrate that DMS3 is capable of generalized transduction.
TABLE 2.
Representative transduction frequencies
| Donor strain | Recipient strain | Marker transduced | Frequencyb |
|---|---|---|---|
| PA14 | PA14 | flgK | 1.0 × 10−10 |
| PA14 | PA14 | pilU | 1.4 × 10−8 |
| PA14 | PA14 | sadBa | 1.0 × 10−8 |
| PA14 | PA14 | sad-107a | 2.0 × 10−9 |
| PA14 | PA14 | det-3a | 2.0 × 10−9 |
| PA01 | PA01 | lasI | 4.5 × 10−9 |
| PA01 | PA01 | lasR | 5.0 × 10−9 |
| PA01 | PA01 | sad-6a | 1.7 × 10−10 |
| PA01 | PA14 | sad-6a | 2.2 × 10−9 |
| PA14 | PAO1 | flgK | 5.0 × 10−10 |
Uncharacterized mutation involved in biofilm formation.
Frequency was calculated on the basis of transductions that yielded transductants. Some transductions yielded no transductions, especially for experiments using different donor and recipient strains.
We also observed cross-strain transductions between strains PAO1 and PA14. The frequency of cross-strain transductions between strains PAO1 and PA14 was ∼2 × 10−9 transductants/PFU. These frequencies were determined using three markers: lasI, flgK, and sad-6 (a mutation in an uncharacterized gene required for motility; see Table 2). Cross-strain transductions were performed using fresh lysates prepared from single-phage plaques propagated on the desired donor strain. Fresh lysates (with titers of ∼1011 PFU/ml) used the day they are prepared are essential for successful cross-strain transductions. The frequency of transduction events mediated by DMS3 appears to be at the borderline at which transductions can be detected; thus, it is helpful to perform multiple, replicate transductions.
Phage immunity tests of lysogens were performed as reported previously (9); the results of these tests suggested that DMS3 is a temperate (class 1) bacteriophage capable of lysogeny (6). This presents a problem in that transductants may be lysogens, which would prevent their use in subsequent transductions. Therefore, all transductants were assessed for phage-mediated immunity by the cross-streaking assay described previously (12), and only those transductants susceptible to phage infection were used for subsequent studies.
Isolation of a clear-plaque variant of DMS3.
A clear-plaque variant of DMS3 is useful in that it would form a more readily discernible plaque and allow for more-robust visualization of phage sensitivity in cross-streak assays. Phage DMS3 was mutagenized with UV (∼2.5 J/m2), resulting in a ∼100-fold reduction of PFU. The mutagenized phage was plated on P. aeruginosa PAO1, and a single clear-plaque isolate was identified among ∼6,500 plaques screened with a dissecting microscope. This clear-plaque isolate is still capable of mediating generalized transduction.
Summary.
We have identified and characterized a generalized transducing phage that is capable of mediating transduction within and between strains of P. aeruginosa. Our data indicate that this temperate phage contains a dsDNA chromosome of ∼50 to 65 kb, infects via type IV pili, and has an icosahedral phage-head morphology. A clear-plaque variant of this bacteriophage was isolated. DMS3 is a useful genetic tool for genetic studies of P. aeruginosa strains PA01 and PA14.
Acknowledgments
We are grateful to J. Schwartzman for providing us with the P. aeruginosa clinical isolates and to C. Manoil for suggesting the approach of obtaining phage from bacterial isolates. We also thank Chuck Daghlian and Louisa Howard at the Rippell Electron Microscopy Facility at Dartmouth College for their assistance with electron microscopy.
This research was supported by funds from the M. Bovaird Center for Studies in Molecular Biology and Biotechnology (W.A.R.), by Dartmouth College and the Arnold and Mabel Beckman Foundation Beckman Scholars Program (J.M.B.), and by the National Science Foundation (CAREER 9984521) and The Pew Charitable Trusts (G.A.O.). G.A.O. is a Pew Scholar in the Biomedical Sciences.
REFERENCES
- 1.Blahova, J., K. Kralikova, V. Krcmery, Sr., A. Mikovicova, and N. Bartonikova. 1998. Two high-frequency-transduction phage isolates from lysogenic strains of Pseudomonas aeruginosa transducing antibiotic resistance. Acta Virol. 42:175-179. [PubMed] [Google Scholar]
- 2.Coyne, M. J., Jr., K. S. Russell, C. L. Coyle, and J. B. Goldberg. 1994. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176:3500-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster, P. L., and W. A. Rosche. 1999. Increased episomal replication accounts for the high rate of adaptive mutation in recD mutants of Escherichia coli. Genetics 152:15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg, J. B., M. J. Coyne, Jr., A. N. Neely, and I. A. Holder. 1995. Avirulence of a Pseudomonas aeruginosa algC mutant in a burned-mouse model of infection. Infect. Immun. 63:4166-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holloway, B. W., J. B. Egan, and M. Monk. 1960. Lysogeny in Pseudomonas aeruginosa. Aust. J. Exp. Biol. Med. Sci. 38:321-330. [DOI] [PubMed] [Google Scholar]
- 7.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen, E. C., H. S. Schrader, B. Rieland, T. L. Thompson, K. W. Lee, K. W. Nickerson, and T. A. Kokjohn. 1998. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 64:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilbane, J. J., and R. V. Miller. 1988. Molecular characterization of Pseudomonas aeruginosa bacteriophages: identification and characterization of the novel virus B86. Virology 164:193-200. [DOI] [PubMed] [Google Scholar]
- 10.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to the study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 11.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.