Abstract
Border cell migration in the Drosophila ovary has emerged as a genetically tractable model for studying collective cell movement. Over many years border cell migration was exclusively studied in fixed samples due to the inability to culture stage 9 egg chambers in vitro. Although culturing late stage egg chambers was long feasible, stage 9 egg chambers survived only briefly outside the female body. We identified culture conditions that support stage 9 egg chamber development and sustain complete migration of border cells ex vivo. This protocol enables one to compare the dynamics of egg chamber development in wild type and mutant egg chambers using time-lapse microscopy and taking advantage of a multiposition microscope with a motorized imaging stage. In addition, this protocol has been successfully used in combination with fluorescence resonance energy transfer biosensors, photo-activatable proteins, and pharmacological agents and can be used with widefield or confocal microscopes in either an upright or inverted configuration.
Keywords: Border cell migration, Drosophila stage 9 egg chambers, Organ culture, Collective cell migration, Time-lapse live imaging
1. Introduction
Collective cell migration refers to the concerted movement of groups of cells. Unlike single moving cells such as fibroblasts or fish keratinocytes, collectively migrating cells maintain some level of adhesion among themselves during movement (1, 2). Though this kind of cellular movement is characteristic of several physiological processes during embryonic development (3), wound healing and tumor metastasis (1), it has been studied less extensively than the movements of single cells. Recently, a number of model systems have emerged for the study of collective movement using the powerful combination of genetic manipulations and live imaging (4, 5). One of these, border cell migration in the Drosophila ovary, is the focus of this chapter.
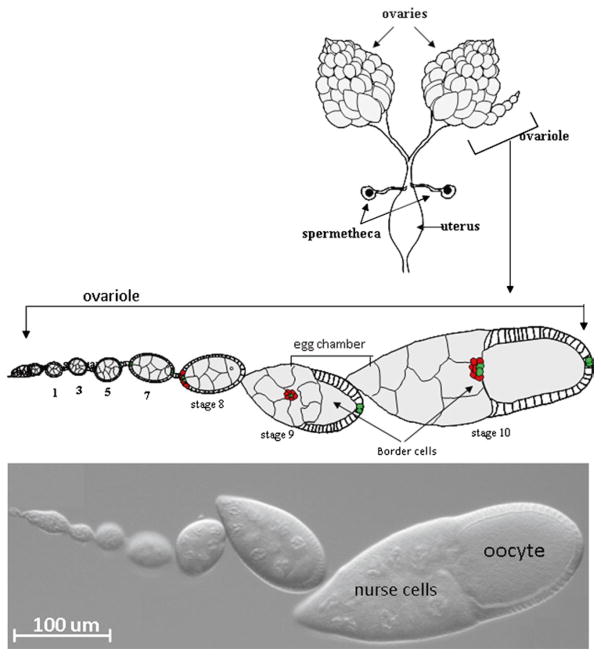
Drosophila females bear a pair of ovaries within the abdomen (Fig. 1). Each ovary consists of 15–20 strings of egg chambers of increasing stages of maturity, called the ovarioles. At the tip of each ovariole resides the germarium, which contains germline and somatic stem cells and their immediate progeny. Egg chambers assemble in the germarium when somatic follicle cells surround a cyst of 16 interconnected germline cells, one of which develops into the oocyte while the other 15 differentiate as support cells called nurse cells (6). Egg chambers bud off of the germarium and then grow and progress through 14 developmental stages (7). Whereas germline cells do not divide further, follicle cells continue to undergo mitotic divisions until stage 6 when they switch to endoreplication without cytokinesis (8). During early oogenesis at each end of each egg chamber a pair of specialized follicle cells differentiates into the polar cells (9). The polar cells secrete a cytokine, Unpaired, which activates JAK-STAT signaling in nearby follicle cells (10). In late stage 8 and early stage 9, anterior follicle cells (4–6 in number) that perceive the highest level of JAK-STAT signal round up (11, 12). These cells are the border cells.
Fig. 1.
Anatomy of the Drosophila ovary. Top – Schematic drawing of a pair of ovaries dissected from female fruit fly. A schematic drawing of an enlarged single ovariole containing egg chambers of the indicate stages of development. The bottom panel shows a DIC image of an ovariole with similar stages of egg chamber development.
One or two of the cells extend protrusions in between the nurse cells (Movie 1). Some of these protrusions retract right away but sooner or later a protrusion attaches stably to the nurse cells and the border cell cluster detaches from the other follicle cells and from the basement membrane that surrounds the egg chamber (13, 14). The border cells migrate directly down the center of the egg chamber toward the oocyte, in response to secreted signals. One such signal is the PDGF- and VEGF-related factor 1 (PVF1), which binds to a receptor tyrosine kinase, PVR, expressed on the border cells (15, 16). PVR functions redundantly with epidermal growth factor receptor (EGFR) (15). Three ligands for the EGFR are expressed in the oocyte (16, 17), Spitz, Keren, and Gurken, all of which are TGFα homologs. Spitz and Keren mRNAs are distributed throughout the oocyte at stage 9 and these two ligands can redirect border cells when either one is misexpressed (16). Thus, these ligands seem to promote migration of the border cells to the oocyte. When the border cells get very near to the oocyte, they turn and move toward the dorsal side (17) (Movie 1). Grk mRNA and protein are restricted to the dorsal/anterior corner of the oocyte and promote the dorsal turn (17). It is unlikely that border cells sense Grk until they get near the oocyte because there is no dorsal bias to the migration before that point (16). Moreover, when Grk is expressed ectopically it is not sufficient to redirect border cells during the posterior migration (16).
The border cells cover a distance of approximately 150–200-μm in 4–6-h (13). Their migration speed is variable but may be a little faster in the beginning and a little slower near the end (14, 18). In the migrating cluster, individual border cells can change relative position within the group, while the polar cells remain in the center (13, 14). Until 2007, border cell migration was studied exclusively in fixed tissue due to the lack of suitable culture conditions for stage 9 egg chambers. Recently, we identified the culture conditions and subsequently optimized the imaging conditions for capturing the complete migration while minimizing phototoxicity (13) (Movie 1). This protocol has enabled more detailed phenotypic analysis and use of pharmacological agents, fluorescence resonance energy transfer (FRET) probes, and photo-activatable proteins (12, 19). In addition, this protocol can be used for studying other aspects of oogenesis including epithelial morphogenesis of follicle cells (20), RNA localization in the oocyte (21), and actin dynamics in nurse cells (22). Key features of the protocol are optimization of pH and addition of insulin, which may generally enhance cultures of Drosophila tissues including imaginal discs. Longer-term cultures (>6-h) may require perfusion systems to allow media exchange.
2. Materials
2.1. Culture Reagents and Equipment
Drosophila Schneider’s medium (Invitrogen, cat. no. 11720-034) supplemented with 15% fetal bovine serum (FBS).
Insulin, a 10-mg/ml (Sigma, cat. no. I5550) stock solution was prepared in acidified water (see Note 1) and used to supplement the culture media to a final concentration of 0.2-mg/ml.
(Optional) FM4-64 (Molecular Probes, cat. no. T-3166) lipophilic dye dissolved in water or DMSO (see Note 2) used for labeling plasma membranes of all cells.
A fluorescent reporter that marks the border cells, e.g., slbo-GAL4; UAS-mCD8GFP (23, 24) or slbo-GAL4 UAS-MoesinGFP (13).
Streptomycin/penicillin (Invitrogen, cat. no. 15140-122) 10,000-U/ml of penicillin G–sodium, 10,000-mg/ml streptomycin sulfate in 0.85% saline.
Greiner Lumox culture dish hydrophilic (50-mm) (Sigma, cat. no. Z376744).
Fisher brand no. 1 coverslips (Fisher, cat. no. 12-542-B).
Two pairs of very fine surgical forceps (FST-5 biologie) and a dissecting microscope.
Depression glass slides (Fisher Scientific, cat. no. 12 565B).
Halocarbon oil 27 (Sigma, H 8773).
Poly-D-lysine (Sigma, P1024), 10-mg/ml in PBS.
Dry baker’s Yeast dissolved in MilliQ water to form a thick paste.
Heat-filter KG1 (Chroma technologies).
BG38 IR suppression filter (Chroma technologies).
Neutral-density filters (ND 0.3) (Chroma technologies).
Widefield microscope. We use a Zeiss Axio Imager upright epifluorescence microscope and Plan-Apochromat 20×/N.A. 0.8 air objective. Illumination source is X-Cite 120 metal halide lamp. Filter sets: BP470/40, FT495, BP525/50 for Alexa 488/GFP; BP550/25, FT570, BP605/70 for Alexa 568/DsRed.
Confocal Microscope. We use a Zeiss 510-Meta inverted confocal microscope and Plan-Apochromat 63×/N.A. 1.4 oil objective with argon (488-nm) and HeNe (542-nm) lasers. Filter sets are FITC and TRITC.
Two pairs of forceps (Dumostar forceps #5).
2.2. Preparation of Culture Media
Drosophila S2 media was supplemented with 15% FBS and 0.6× streptomycin/penicillin. It was sterilized by passing through 0.2-μm filter.
The final pH of the media was adjusted to 6.95–7.00. Verify the pH of the media before every use (see Note 3).
Supplement with insulin to a final concentration of 0.2-mg/ml. Henceforth, the media with all the supplements would be referred as complete media.
(Optional) FM4-64 dye can be used at a final concentration of 9-μM to stain plasma membranes of all cells.
3. Method
3.1. Preparing Poly-D-Lysine Coated Coverslip
Five to ten Fisher brand No. 1 coverslips (22 × 22-mm), washed with milliQ water and dried. Layer the coverslip evenly with approximately 200-μl of 2-mg/ml of poly-D-lysine in PBS and dry it completely in a 37°C oven. Wash the coverslip couple of times in flowing tap water and subsequently rinse with MilliQ water and then dry.
3.2. Fattening Females Flies
Five to ten females (2–4 days old) of the desired genotype along with three to four males are transferred into a new fly vial with fresh fly food, a small amount of yeast paste and incubated at 25°C for 14–18-h (see Note 4).
3.3. Dissection of Ovaries to Egg Chambers
Immobilize 2–3 females flies either by exposure to CO2 or anesthetic ether.
Fill the cavity of the depression slide with complete culture media and place the slide under the dissecting microscope.
Transfer one immobile fly to the cavity slide.
Hold the thorax or upper abdomen of the immobile fly using a pair of forceps, submerge it in the medium, while with another pair of forceps grasp and peel off a bit of abdominal cuticle at the middle of the abdomen (Fig. 2a).
This reveals a large pair of ovaries, which are white and opaque. Grasp the base of the ovaries (the common oviduct) and pull them away from the rest of the fly and into the medium (Fig. 2b). Remove the carcass using the forceps and discard it on a Kimwipe. Keep the ovaries covered in medium at all times.
Repeat this process for all female flies, accumulating the ovaries in the complete medium in the depression slide itself. Be careful to handle the ovaries very gently (see Note 5).
With one pair of forceps, hold the posterior part of a single ovary. Use another pair of forceps to gently grasp the anterior tip of the ovary (Fig. 2c). Older egg chambers reside at the posterior while the germarium and early stage egg chambers occupy the anterior tip.
Gently pull the ovarioles out of the muscular sheath. As you pull, you will observe strings of egg chambers pop out of the muscular sheath and sometimes you will also get individual egg chambers (Fig. 2c, d). Repeat this action until you see five to ten stage 9 egg chambers. Never touch the stage 9 egg chambers directly with the forceps! You can use early or very late stage egg chambers to manipulate the position of stage 9 chambers (see Note 5).
Use a plastic transfer pipette to transfer small number of egg chambers to four separate locations on a poly-D-lysine-coated glass coverslip. The distance between the locations should be large enough so that during multiposition imaging only the egg chamber being imaged is in the field of view (see Note 6).
As the egg chambers are settling down, use the forceps/transfer pipette to gently arrange them so that they are evenly spread on the coverslip and not too close together (see Note 7). Manipulate the stage 9 egg chambers by holding/touching late stage or early stage egg chambers. Allow a minute for the egg chambers to adhere to the coverslip.
In the meantime, break a 22 × 22-mm coverslip into two halves. Align them approximately 1-cm apart on a lumox dish with help of paint brush. Transfer approximately 60–80-μl of the complete media underneath and between the broken coverslip. Remove excess media from underneath the broken coverslip if they start to float.
With the help of the forceps very slowly flip over the coverslip so that the egg chambers face downward. Then gently lower this coverslip to form a bridge between the two coverslip pieces such that the egg chambers face the lumox dish membrane (see Note 8).
Gently remove excess media from around the coverslips, taking care not to compress the egg chambers.
Seal the edges of the coverslips with a thin layer of halocarbon oil 27 to minimize evaporation of the media during imaging (see Note 9).
For the upright microscope, the glass spacers and coverslips are mounted on the top of lumox dish; for the inverted microscope, the sample is mounted on the bottom of the dish.
Fig. 2.
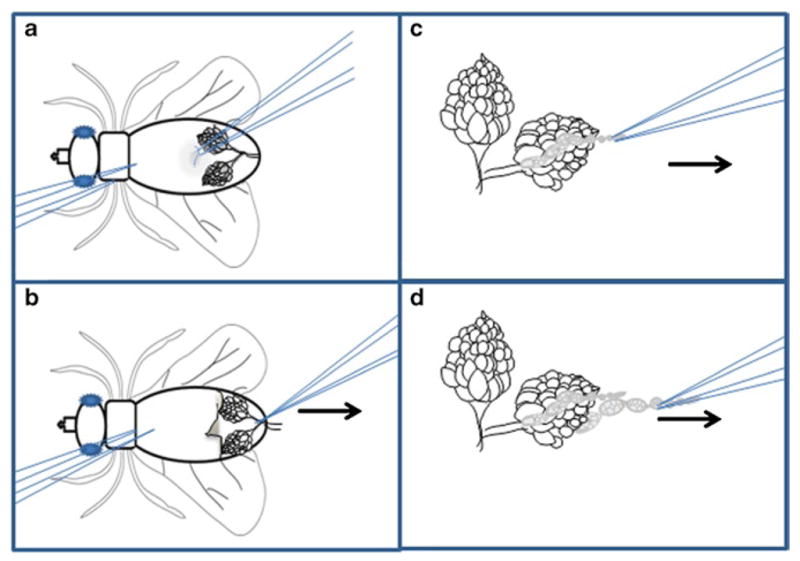
Ovary dissection technique. Schematic representation of egg chamber dissection. (a) The female fly should be immobilized on its back using the left forceps. The right forceps are used to pinch the soft cuticle of the ventral side of the abdomen. (b) Pull the cuticle toward the right (arrow ), revealing the ovaries. Grasp the common oviduct with the right forceps and pull to the right (arrow ) to free the ovaries from the carcass. (c) Immobilize the ovary pair by pinching the oviduct using the left forceps. Use the right forceps to grasp the tip of an ovariole and slowly pull to the right (arrow ). (d) Repeat until multiple ovarioles have been freed from the sheath.
3.4. Time-Lapse Imaging of the Egg Chambers
Transfer the lumox dish to a microscope stage equipped with a petri-dish holder.
Set up the center of the lumox dish as a reference point. Move to one of the locations and identify an egg chamber of the desired stage and mark its coordinates. Repeat this step for all the other locations avoiding egg chambers that have abnormal morphology. In addition avoid choosing egg chambers that are located near a germarium. The inherent pulsating movement of germarium might move the egg chamber during imaging.
Lower the light intensity using 25% Neutral Density filter. Include KG1 and BG38 filters to suppress heating of the sample (see Note 10). When imaging more than one egg chamber at a time, mark the position of the center of each border cell cluster and identify the correct exposure for each channel individually for each egg chamber. Try not to exceed 150-ms for each channel. With this exposure setting one can collect 16-z sections, 1.25-μm apart at 20× magnification for each channel. Egg chambers generally tolerate a time interval of 2.25-min between successive frames when imaging four egg chambers with 16-z sections for each of two channels.
Start the time-lapse image capture. Use modest speed of the mobile stage. We have successfully used Axiocam MRm camera mounted on a Zeiss Axioimager (upright, widefield) microscope with 20× magnification for 5–6-h. Higher magnification like 40× can be used for shorter total imaging times.
Between the acquisitions, it is worthwhile to refocus on the border cells as they move. In addition, look for signs of normal development such as oocyte growth, outer follicle cell rearrangement, dynamic changes in gene expression (e.g., expression of the slow border cells enhancer in centripetal cells), and of course border cell movement.
The lumox dish can be reused. After the experiment, carefully remove the coverslips and wash away the halocarbon oil by rinsing several times with ethyl alcohol.
After imaging, one can process the images in different ways depending upon the available software. Movie 1 shows a maximal intensity projection of the 16-z slices over the entire time interval of acquisition.
Footnotes
Insulin powder dissolves in slightly acidic condition. For preparing acidic water, dilute 1-μl of concentrated hydrochloric acid to 1-ml with milliQ water.
If using DMSO for dissolving FM4-64, the final dilution of DMSO in complete medium should be 1:1,000. Higher concentrations of DMSO, impedes border cell migration.
The pH of the medium is critical for the experiment. Low pH impedes border cell migration and high pH leads to early degeneration of the egg chamber.
Age of the female fly is very important. Newly hatched flies do not fatten very well. While the ovaries of older fly have large number of mature egg chambers.
Gentle handling of the ovaries is critical as even inconspicuous damage to egg chambers inhibits border cell migration.
Optimum distance between the egg chambers of interest is desired so as to reduce phototoxicity.
Evenly spread out egg chambers with empty space between is preferable to accommodate the increase in size of the egg chambers during imaging.
While lowering the coverslip with attached egg chambers, one needs to be very careful that ends of coverslip should align on the bridge to avoid any damage to the egg chambers.
Do not use a Nail Polish.
Addition of heat-filter KG, infra red suppression filter (BG38), and 25% neutral density suppresses phototoxicity of the sample during long-term imaging.
This volume includes electronic supplemental material that will appear on the website http://extras.springer.com/wells.
References
- 1.Ilina O, Friedl P. Mechanisms of collective cell migration at a glance. J Cell Sci. 2009;122:3203–8. doi: 10.1242/jcs.036525. [DOI] [PubMed] [Google Scholar]
- 2.Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Weijer CJ. Collective cell migration in development. J Cell Sci. 2009;122:3215–23. doi: 10.1242/jcs.036517. [DOI] [PubMed] [Google Scholar]
- 4.López-Schier H. Fly fishing for collective cell migration. Curr Opin Genet Dev. 2010;20:428–32. doi: 10.1016/j.gde.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Rørth P. Collective cell migration. Annu Rev Cell Dev Biol. 2009;25:407–29. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- 6.Spradling AC. Developmental Genetics of Oogenesis. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- 7.King RC. Ovarian Development in Drosophila melanogaster. Academic Press; New York: 1970. [Google Scholar]
- 8.Edgar BA, Orr-Weaver TL. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 9.Ruohola H, Bremer KA, Baker D, Swedlow JR, Jan LY, Jan YN. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell. 1991;66:433–449. doi: 10.1016/0092-8674(81)90008-8. [DOI] [PubMed] [Google Scholar]
- 10.Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107:831–41. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- 11.Montell DJ, Rørth P, Spradling AC. Slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- 12.Starz-Gaiano M, Melani M, Wang X, Meinhardt H, Montell DJ. Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev Cell. 2008;14:726–38. doi: 10.1016/j.devcel.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Prasad M, Montell DJ. Cellular and Molecular Mechanisms of Border Cell Migration Analyzed Using Time-lapse Live-cell Imaging. Dev Cell. 2007;12:997–1005. doi: 10.1016/j.devcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 14.Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rorth P. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–65. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- 15.Duchek P, Somogyi K, Jekely G, Beccari S, Rorth P. Guidance of cell migration by the drosophila PDGF/VEGF receptor. Cell. 2001;107:17–26. doi: 10.1016/s0092-8674(01)00502-5. [DOI] [PubMed] [Google Scholar]
- 16.McDonald JA, Pinheiro EM, Kadlec L, Schupbach T, Montell DJ. multiple EGFR ligands participate in guiding migrating border cells. Dev Biol. 2006;296:94–103. doi: 10.1016/j.ydbio.2006.04.438. [DOI] [PubMed] [Google Scholar]
- 17.Duchek P, Rorth P. Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science. 2001;291:131–33. doi: 10.1126/science.291.5501.131. [DOI] [PubMed] [Google Scholar]
- 18.Prasad M, Jang AC, Starz-Gaiano M, Melani M, Montell DJ. A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat Protoc. 2007;2:2467–73. doi: 10.1038/nprot.2007.363. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol. 2010;12:591–7. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horne-Badovinac S, Bilder D. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev Dyn. 2005;232:559–72. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- 21.Grunert S, St Johnston D. RNA localization and the development of asymmetry during Drosophila oogenesis. Curr Opin Genet Dev. 1996;6:395–402. doi: 10.1016/s0959-437x(96)80059-1. [DOI] [PubMed] [Google Scholar]
- 22.Hudson AM, Cooley L. Understanding the function of actin-binding proteins through genetic analysis of Drosophila oogenesis. Annu Rev Genet. 2002;36:455–88. doi: 10.1146/annurev.genet.36.052802.114101. [DOI] [PubMed] [Google Scholar]
- 23.Rørth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milán M, Benes V, Ansorge W, Cohen SM. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–57. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Bo J, Bridges T, Dugan KD, Pan TC, Chodosh LA, Montell DJ. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev Cell. 2006;10:483–95. doi: 10.1016/j.devcel.2006.02.003. [DOI] [PubMed] [Google Scholar]