Abstract
The genome of Halobacterium sp. strain NRC-1 contains a large gene cluster, gvpMLKJIHGFEDACNO, that is both necessary and sufficient for the production of buoyant gas-filled vesicles. Due to the resistance of gas vesicles to solubilization, only the major gas vesicle protein GvpA and a single minor protein, GvpC, were previously detected. Here, we used immunoblotting analysis to probe for the presence of gas vesicle proteins corresponding to five additional gvp gene products. Polyclonal antisera were raised in rabbits against LacZ-GvpF, -GvpJ, and -GvpM fusion proteins and against synthetic 15-amino-acid peptides from GvpG and -L. Immunoblotting analysis was performed on cell lysates of wild-type Halobacterium sp. strain NRC-1, gas vesicle-deficient mutants, and purified gas vesicles, after purification of LacZ fusion antibodies on protein A and β-galactosidase affinity columns. Our results show the presence of five new gas vesicle proteins (GvpF, GvpG, GvpJ, GvpL, and GvpM), bringing the total number of proteins identified in the organelles to seven. Two of the new gas vesicle proteins are similar to GvpA (GvpJ and GvpM), and two proteins contain predicted coiled-coil domains (GvpF and GvpL). GvpL exhibited a multiplet ladder on sodium dodecyl sulfate-polyacrylamide gels indicative of oligomerization and self-assembly. We discuss the possible functions of the newly discovered gas vesicle proteins in biogenesis of these unique prokaryotic flotation organelles.
Gas vesicles are gas-filled subcellullar buoyancy organelles that are commonly synthesized by halophilic archaea (haloarchaea) and cyanobacteria, as well as some other prokaryotes (6, 30). In haloarchaea, ultrastructural analysis of gas vesicles showed a predominant lemon-shaped form about 300 nm in length and 200 nm in width (9, 21). The proteinaceous gas vesicle membrane is extremely stable, rigid, gas permeable, and lipid free (16, 30). The function of gas vesicles in cell buoyancy is dependent on exclusion of water from the interior of the structure, which is thought to be a consequence of the hydrophobicity of the interior surface of the vesicle membrane (30). Although gas vesicles are easily isolated by flotation, biochemical characterization has been hampered by their extreme resistance to solubilization, and only two proteins, GvpA and GvpC, have previously been detected (10, 12, 26). Consequently, the biogenesis of gas vesicles has been speculated to occur through the self-assembly of one or at most two proteins (16, 26).
Genetic analysis of high-frequency spontaneous gas vesicle-deficient mutants in haloarchaea indicated that gas vesicle formation is complex (6, 25). In Halobacterium sp. strain NRC-1, a large gene cluster (gvpMLKJIHGFEDACNO) was found to be subject to IS element-mediated rearrangements in spontaneous gas vesicle-deficient mutants, implicating more than a dozen genes in vesicle formation (3, 13, 14). Insertion mutagenesis in Halobacterium species confirmed that most of the gvp genes are required for wild-type gas vesicle formation (4). A suitably engineered gvp gene cluster programmed the formation of gas vesicles in Escherichia coli, indicating that this gene cluster is not only necessary but also sufficient for vesicle formation (7, 8). Complete genome sequencing of Halobacterium sp. strain NRC-1 revealed additional complexity, with two similar but distinct copies of gvp gene clusters carried on the NRC-1 minichromosomes, pNRC100 and pNRC200 (20, 21). The pNRC200 cluster, however, was incomplete, lacking the gvpM gene, and was silent in strain NRC-1. For the pNRC100 gvp gene cluster, two divergent promoters were identified, with transcription of 10 genes, gvpD to -M, occurring in the leftward direction and the transcription of three or four genes, gvpA, -C, -N, and -O, occurring in the rightward direction (3, 6, 10, 13, 14).
In addition to Halobacterium sp. strain NRC-1, gvp genes have been studied in related halophilic archaea, cyanobacteria, and gram-positive bacteria (2, 17, 25, 30). In some haloarchaeal strains, a functional second gvp cluster has been reported (28). In Haloferax volcanii, a halophile that does not naturally produce gas vesicles, transformation with Halobacterium genes indicated that eight genes, gvpM, -L, -K, -J, -G, -F, -A, and -O, comprise the minimal gene set necessary to program gas vesicle formation (22). Interestingly, many of the same genes were also discovered in bacteria containing gvp gene clusters. For example, the cyanobacterium Anabaena flos-aquae contains gvpA, -C, -N, -J, -K, and -L, and the gram-positive bacterium Bacillus megaterium contains gvpA, -N, -F, -G, -L, and -K (15, 17). The genome sequence of Streptomyces coelicolor also revealed the presence of a gene cluster with gvpO, -A, -F, -G, -J, -L, -S, and -K (2). These findings suggested that several gvp genes, including gvpA, -F, -G, -J, -L, and -M, are likely to be critical for gas vesicle formation in nearly all organisms producing these flotation organelles.
To address a possible role for the conserved gvp genes in gas vesicle structure, we raised antibodies and probed for the presence of five gene products in gas vesicles of the model halophile Halobacterium sp. strain NRC-1 by immunoblotting analysis. Our results clearly demonstrate that gas vesicles from Halobacterium sp. strain NRC-1 contain new gvp gene products not previously detected, including GvpF, GvpG, GvpJ, GvpL, and GvpM, which brings the total number of gas vesicle proteins to seven. Implications for the complexity of gas vesicle biogenesis are significant.
MATERIALS AND METHODS
Strains, culturing, and preparation of cell extracts and gas vesicles.
The Halobacterium strains used for this study are listed in Table 1 and include a sequenced wild-type strain, Halobacterium sp. strain NRC-1 (ATCC 700922/JCM11081); SD109, a completely gas vesicle-deficient mutant with a deletion of the entire gvp gene cluster (19); and derivatives containing the gvp gene cluster in plasmid pFL2 (4) with a kanamycin κ cassette inserted in specific gvp genes: SD109(pFL2gvpC::κ1), SD109(pFL2gvpF::κ5), SD109(pFL2gvpG::κ1), SD109(pFL2gvpJ::κ1), SD109(pFL2gvpL::κ6), and SD109(pFL2gvpM::κ1). Construction of these strains has been previously described (4).
TABLE 1.
Halobacterium species and strains used in this study
| Strain | Characteristics | Reference |
|---|---|---|
| NRC-1 | Wild-type, Vac+ | 21 |
| SD109 | Vac−, 67-kb pNRC100 ISH8-mediated deletion, ΔgvpMLKJIHGFEDACNO | 19 |
| SD109(pFL2gvpF::κ5) | Vac−, κ insertion 304 bp from 5′ end of gvpF | 4 |
| SD109(pFL2gvpG::κ1) | Partially Vac−, κ insertion 184 bp from 5′ end of gvpG | 4 |
| SD109(pFL2gvpJ::κ1) | Vac−, κ insertion 130 bp from 5′ end of gvpJ | 4 |
| SD109(pFL2gvpK::κ1) | Vac−, κ insertion 184 bp from 5′ end of gvpK | 4 |
| SD109(pFL2gvpL::κ6) | Vac−, κ insertion 538 bp from 5′ end of gvpL | 4 |
| SD109(pFL2gvpM::κ1) | Vac+, κ insertion 194 bp from 5′ end of gvpM | 4 |
For gas vesicle preparations, NRC-1 was cultured on CM+ (complete medium plus trace metals) agar plates with incubation at 42°C for 4 weeks and vesicles were prepared as previously described (5). Briefly, cells were collected from plates by washing with 15 ml of 1.0 mM MgSO4 containing 10-μg/ml DNase I and the cell suspension was incubated for 3 h at 37°C. After lysis, cell lysates were adjusted to 10% (wt/vol) NaCl, overlayed with 5% NaCl (wt/vol), and centrifuged at 60 × g overnight in a swinging bucket rotor. Next, intact floating gas vesicles were carefully collected and resuspended in 5% NaCl. The isolated gas vesicles were washed by centrifugally accelerated flotation three times.
For preparation of whole-cell extracts, NRC-1 and gas vesicle mutant strains were grown in liquid cultures in CM+ medium, with mutants under selective conditions (10-μg/ml mevinolin; Sigma Corp., St. Louis, Mo.), in an illuminated New Brunswick Scientific G25 incubator shaker at 42°C. Ten-milliliter cultures (optical density at 600 nm of 1.2) were harvested by centrifugation (8,000 rpm for 10 min at 4°C) in a Beckman Avanti centrifuge using a JA2550 rotor. Pellets were resuspended in 0.5 ml of sterile distilled water containing 1 mM phenylmethylsulfonyl fluoride (freshly prepared), 10-μg/ml DNase I was added, and the lysates were incubated at 37°C for 30 min and dialyzed against 4 liters of distilled water at 4°C. Protein concentrations were determined using a dye-binding assay (Bio-Rad, Hercules, Calif.).
Production and purification of antibodies against LacZ-Gvp fusion proteins.
Expression of LacZ-Gvp fusion protein in E. coli and production of rabbit polyclonal antisera used in this study were previously described (9, 10). For this work, antisera were further purified by affinity chromatography over a protein A-Sepharose 4B column and two serial passages over a β-galactosidase-agarose column. For chromatography on a protein A-Sepharose 4B affinity column (Zymed Laboratories, San Francisco, Calif.), 0.5 ml of gel was washed four times with 20 mM sodium phosphate (pH 7.0) by centrifugation at 1,000 rpm for 5 min in a Beckman Avanti centrifuge using a JA2550 rotor. Antisera were diluted 1:1 with phosphate-buffered saline (PBS) containing 150 mM NaCl, 2.5 mM KCl, 10 mM sodium phosphate monobasic, and 1.7 mM potassium phosphate dibasic (pH 7.4) and bound with the equilibrated gel for 1 h at 4°C with gentle shaking. Subsequently, the affinity gel was applied to a column and washed with 5 to 10 column volumes of 20 mM phosphate buffer until the A280 of the last wash was less than 0.02. The bound antibodies were eluted with 0.1 M acetic acid (pH 2.8), and 0.5-ml fractions were collected into tubes containing 50 μl of 2 M Tris-HCl (pH 8). Absorbance of purified fractions was recorded at 280 nm. For long-term storage, purified antisera were dialyzed overnight against PBS and sodium azide was added to 0.05% as a preservative.
For purification of antisera on β-galactosidase columns, β-galactosidase affinity gel was first prepared by coupling cyanogen bromide-activated 4% beaded agarose with purified β-galactosidase enzyme (Sigma, St. Louis, Mo.). For coupling 300 U of β-galactosidase, the enzyme was dissolved in 10 ml of coupling buffer containing 0.1 M NaHCO3 and 0.5 M NaCl (pH 8.5). The cyanogen bromide-activated agarose beads were washed and allowed to swell in 1 mM HCl at 4°C for 30 min. Subsequently, beads were washed four times with 1 mM HCl to preserve the activity of reactive groups, followed by washing with 10 column volumes of distilled water and 1 volume of coupling buffer. The washed agarose beads were transferred into coupling buffer containing β-galactosidase and allowed to bind with the activated resin overnight at 4°C. The unbounded β-galactosidase was removed by washing with uncoupling buffer, and unreacted groups were blocked by washing with 0.2 M glycine (pH 8.0) for 2 h at room temperature. Glycine was removed by extensive washing with coupling buffer followed by additional washes with a solution containing 0.1 M acetate buffer (pH 4.0) and 0.5 M NaCl. The β-galactosidase affinity gel was stored in 1 M NaCl at 4°C.
For affinity chromatography of LacZ-Gvp antibodies using the β-galactosidase-agarose gel, a 2-ml column was equilibrated with 10 column volumes of 10 mM Tris-HCl (pH 7.5) followed by 5 column volumes of 10 mM Tris-HCl (pH 8.8). One milliliter of protein A column-purified Gvp antibodies was loaded onto an equilibrated column and allowed to bind for 30 min with gentle shaking. Unbound Gvp antibody fractions (0.5 ml) were collected first, followed by elution of β-galactosidase antibodies with 100 mM glycine (pH 2.5), and eluted samples were neutralized by addition of 50 μl of 1 M Tris-HCl (pH 8.0).
Production of antibodies against synthetic peptides of gas vesicle proteins.
The antisera against GvpG and GvpL proteins were raised by synthesizing peptides from potentially immunogenic regions of the respective proteins (Sigma-Genosys, Woodlands, Tex.). Potentially immunogenic peptides were identified using the Genetics Computer Group program PlotStructure (www.accelrys.com). The antigenic peptide sequence for GvpG was 45-EVGERSDEEYQQRKQ-60, and that for GvpL was 180-KQSDQRLQELKRERR-195. The peptides were conjugated to keyhole lymphet hemocyanin via a disulfide linkage with an additional cysteine residue at the N termini of the GvpG and GvpL peptides. The conjugated peptides were combined with Freund's adjuvant and used for multiple immunizations of New Zealand White rabbits. Six immunizations were conducted biweekly, and the last bleed was collected 7 days following the final immunization. The antibody titers of GvpG and GvpL antisera to the synthetic peptides were 1:300,000 and 1:400,000, respectively.
Electrophoresis and immunoblotting analysis.
For immunoblot analysis of cell lysates or purified gas vesicles, cell lysates containing 150 μg of protein or 150 μg of gas vesicle protein were mixed with an equal volume of 2× sample loading buffer (60 mM Tris-HCl [pH 6.8], 20% glycerol, 0.2 M dithiothreitol [DTT], 2% sodium dodecyl sulfate [SDS], 0.02% bromophenol blue) and boiled for 4 min (11). After cooling to room temperature, the samples were briefly vortexed and electrophoresed on a 12% polyacrylamide-SDS gel for 90 min at 100 V at room temperature on a Novex (San Diego, Calif.) Xcell SureLock electrophoresis cell.
The electrophoretically resolved protein bands were electroblotted onto a 0.45-μm-pore Immobilon-NC nitrocellulose membrane (Millipore Corp., Boston, Mass.) for 1 h at 100 V using a Bio-Rad mini gel blotter. After electroblotting, membranes were washed twice for 5 min with PBS buffer, blocked for 1 h with 3% bovine serum albumin (Sigma Corp.), in PBS buffer, and washed five times for 5 min each with PBS containing 0.01% Tween 20. The washed membrane was incubated for 1.5 h at room temperature under gentle shaking with Gvp antibodies diluted 1:100 (GvpG, GvpJ, and GvpM) or 1:150 (GvpF and GvpL). The membrane was then washed five times for 5 min each with PBS buffer containing 0.01% Tween 20 and incubated with goat anti-rabbit secondary antibodies (Jackson Immuno Research, West Grove, Pa.) labeled with alkaline phosphatase diluted (1:2,000) in a solution containing 3% bovine serum albumin in PBS buffer. For detection of the protein bands, membrane was incubated in Sigma Fast (5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium) according to the manufacturer's specification.
RESULTS AND DISCUSSION
Immunoblotting analysis of GvpJ and GvpM.
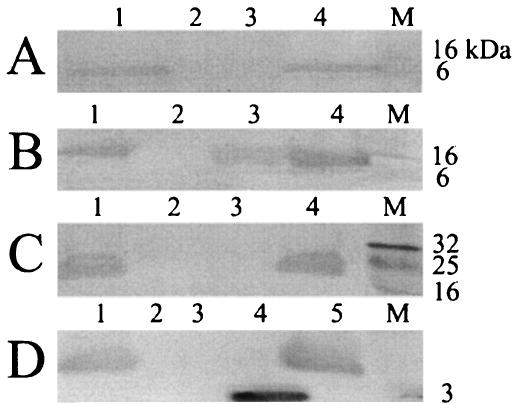
Our previous results established the likely existence of a small family of proteins (pfam741), including gas vesicle protein, GvpA, GvpJ, and GvpM (Fig. 1A) (14). However, only the GvpA gene product had been shown to be present in gas vesicles by immunoblotting (10). To determine whether the gvpJ and gvpM gene products are indeed gas vesicle proteins, we used extensively purified antisera for immunoblotting analysis. Figure 2A shows the results with GvpJ antiserum, and Fig. 2B shows the results with GvpM antiserum. These experiments detected the presence of two distinct proteins between 6- and 16-kDa molecular mass markers in the wild-type NRC-1 strain and in a gas vesicle (lanes 1 and 4, respectively), but not in the gas vesicle-deficient strain SD109 (lane 2). In Fig. 2A, for the GvpJ Western analysis, a gas vesicle protein of 9 to 12 kDa was detected, consistent with the predicted molecular mass of 11,983 Da based on the gvpJ gene sequence (14). The protein was absent in the gvpJ::κ1 mutant (lane 3), confirming its identity as GvpJ. In Fig. 2B, a gas vesicle protein of 9 to 13 kDa was detected, consistent with the predicted molecular mass of 9,248 Da for GvpM (14). In the gvpM::κ1 mutant, a similar-size protein was observed (lane 3). This result is consistent with the location of the κ insert near the 3′ end of the gvpM gene, which predicts the production of an essentially wild-type-size protein (4). These results show, in addition, that the antisera against GvpJ and GvpM are monospecific, as no cross-reactivity could be observed against the other proteins in this family. Thus, our results of immunoblotting with GvpJ and GvpM antisera show both GvpJ and GvpM to be gas vesicle proteins.
FIG. 1.
Alignment of gas vesicle gene families. (A) Partial sequence alignment of GvpJ, GvpA, and GvpM gas vesicle proteins from Halobacterium sp. strain NRC-1. (B) Partial sequence alignment of GvpF and GvpL proteins from Halobacterium sp. strain NRC-1. The numbering for GvpJ (A) and GvpF (B) and the consensus for the aligned sequences are shown above.
FIG. 2.
Identification of Gvp proteins by immunoblot analysis. (A) Whole-cell lysates of Halobacterium sp. strain NRC-1 (lane 1), SD109 (lane 2), and SD109 (pFL2gvpJ::κ1) (lane 3) and purified gas vesicles (lane 4) were electrophoresed, transferred to membrane, and probed with GvpJ antibodies. (B) As in panel A, except lane 3 contains whole-cell lysate of Halobacterium sp. strain SD109(pFL2gvpM::κ1) and the blot was probed with GvpM antibodies. (C) As in panel A, except lane 3 contains whole-cell lysate of Halobacterium sp. stran SD109(pFL2gvpF::κ5) and the blot was probed with GvpF antibodies. (D) As in panel A, except lane 3 contains whole-cell lysate of Halobacterium sp. strain SD109(pFL2gvpG::κ1), lane 4 contains synthetic GvpG peptide, and the blot was probed with GvpG antibodies. Prestained protein standards are displayed in lanes marked M, and molecular masses are indicated.
Immunoblotting analysis of GvpF.
The gvpF gene is conserved in gas vesicle-forming microbes and is essential for gas vesicle formation in Halobacterium sp. strain NRC-1 (Table 1) (4, 6). The GvpF protein has some sequence similarity to GvpL, and both contain coiled-coil domains (pfam 6386) (Fig. 1B). To probe for the GvpF gene product, purified GvpF antiserum was used for immunoblotting analysis (Fig. 2C). The results show the presence of a protein with an apparent molecular mass of 23 to 28 kDa in the wild-type NRC-1 strain and in gas vesicles (lanes 1 and 4, respectively), but not in the gas vesicle-deficient SD109 or gvpF::κ5 mutant strains (lanes 2 and 3). Since the predicted molecular mass of GvpF is 23,962 Da, the size of the observed band is consistent with GvpF. Our results show, therefore, that the GvpF protein is detectable by immunoblotting analysis and is a component of purified gas vesicles from Halobacterium sp. strain NRC-1.
Immunoblotting analysis of GvpG and GvpL.
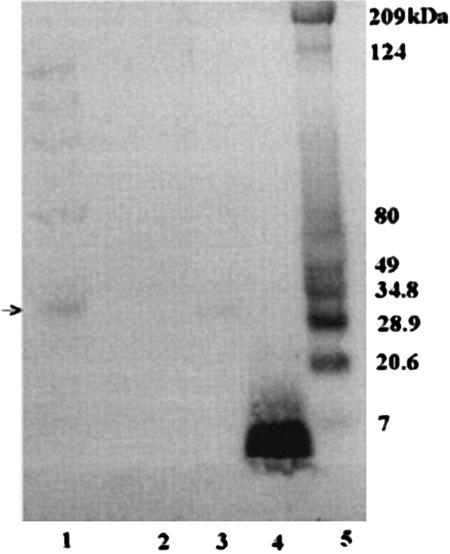
Like GvpF, both GvpG and GvpL are acidic proteins, and their predicted molecular masses are 10,014 and 31,994 Da, respectively (14). Insertional inactivation of the gvpG gene resulted in greatly reduced levels of gas vesicles, whereas disruption of gvpL completely blocked the formation of gas vesicles (4, 6). Immunoblotting analysis was conducted to probe for GvpG and GvpL using respective antisera raised against 15-amino-acid antigenic peptides of each protein. The results with the GvpG antiserum show that a protein band of 9 to 12 kDa was identified in cell lysates of Halobacterium sp. strain NRC-1 and purified gas vesicles (Fig. 2D, lanes 1 and lane 5, respectively) and a smaller band was detected in the lane containing the GvpG peptide (lane 4). However, the band in the cell lysate of the pFL2gvpG::κ1 mutant strain was absent due to the disruption of the gvpG gene (Fig. 2D, lane 3), confirming the identity of the protein band as GvpG. Immunoblotting analysis with GvpL antiserum detected the presence of a major 30- to 32-kDa protein band in purified gas vesicles and cell lysate of Halobacterium sp. strain NRC-1 (Fig. 3, lanes 1 and 3, respectively), but not in the lysate of pFL2gvpL::κ6 mutant (Fig. 3, lane 2). Interestingly, the purified gas vesicle lane contained, in addition to the major band, a ladder of bands with apparent molecular masses in multiples of about 30 kDa, consistent with formation of oligomers of GvpL in the SDS-polyacrylamide gel. We conclude therefore that both GvpG and GvpL are Halobacterium sp. strain NRC-1 gas vesicle proteins detectable by immunoblotting.
FIG. 3.
Identification of GvpL protein (arrow) by immunoblot analysis. Lanes: 1, purified gas vesicles; 2, SD109(pFL2gvpL::κ6); 3, Halobacterium sp. strain NRC-1 cell lysate; 4, synthetic GvpL peptide; and 5, prestained protein standards. Molecular masses are given on the right.
Possible functions of Gvp proteins.
Our finding of two proteins with sequence similarity to GvpA in gas vesicles is intriguing and suggests their direct involvement in the structure of the gas vesicle membrane. For example, this family of related but distinct gas vesicle proteins, GvpA, GvpJ, and GvpM, may be capable of packing in variable ratios and producing coils of various radii. Electron microscopic examination previously indicated that membrane subunits form a shallow coil (23). Varying the protein composition may be one way of generating diverse coil radii, with small coils near the tips of the lemon-shaped vesicles and larger coils in the middle of the structure. For example, one protein could possibly be dominant in the small coils (e.g., GvpJ and/or GvpM) and a different protein (GvpA) could be dominant in the large coils. Small changes in the binding geometry could potentially provide the versatility necessary for the observed structural variation. This hypothesis is consistent with reports that GvpA protein sequence influences the shape of gas vesicles (1, 28).
Another attractive hypothesis is that one (or likely several) of the newly identified gas vesicle proteins nucleates the formation of gas vesicles in a complex destined to become the tips of the vesicles. An early investigation reported the formation of small biconical vesicles after collapse of the larger vesicles, suggesting that gas vesicle biogenesis is initiated at the tips of the cones, with growth occurring by the addition of gas vesicle protein subunits to the center of the structure (29). If so, GvpF, GvpG, and GvpL qualify as possible candidates for nucleation proteins. Both GvpF and GvpL, in particular, contain predicted coiled-coil domains (pfam 6386) known to be involved in self-oligomerization (18), a property that is ideal for initiating the formation of a new structure. In immunoblots, GvpL was found to display a ladder of protein bands, consistent with self-oligomerization (Fig. 3). However, the possibility that the ladder of protein bands results from binding of other proteins cannot be ruled out at this time. Although a similar property was not observed for GvpF, the presence of the coiled-coil domain suggests that it may complex with itself or other proteins under appropriate conditions.
Conclusions and future prospects.
Our results show that in addition to GvpA and GvpC, five new proteins, GvpF, GvpG, GvpJ, GvpL, and GvpM, complex in the process of gas vesicle biogenesis. Two hypothesized roles for the new proteins are in initiation of gas vesicle synthesis and in determination of the curvature of the vesicle membrane. Some of these Gvp proteins may also be required for assembly. Several additional gvp gene products beyond the seven identified thus far are also likely to be involved in gas vesicle formation. With the availability of immunological probes, genome sequence, expression vectors, and gene knockout and replacement methodology (5, 24, 27, 31), we now have in place many tools necessary for better understanding the process of gas vesicle biogenesis in the model archaeon, Halobacterium sp. strain NRC-1.
Acknowledgments
This work was supported by grant MCB-0296019 from the National Science Foundation.
We thank John T. Halladay for raising GvpF, GvpJ, and GvpM antisera.
REFERENCES
- 1.Beard, S., P. Hayes, F. Pfeifer, and A. Walsby. 2002. The sequence of the major gas vesicle protein, GvpA, influences the width and strength of halobacterial gas vesicles. FEMS Microbiol. Lett. 213:149-157. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.DasSarma, S., T. Damerval, J. G. Jones, and N. Tandeau de Marsac. 1987. A plasmid-encoded gas vesicle protein gene in a halophilic archaebacterium. Mol. Microbiol. 1:365-370. [DOI] [PubMed] [Google Scholar]
- 4.DasSarma, S., P. Arora, F. Lin, E. Molinari, and L. R-S. Yin. 1994. Wild-type gas vesicle formation requires at least ten genes in the gvp gene cluster of Halobacterium halobium plasmid pNRC100. J. Bacteriol. 176:7646-7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DasSarma, S., F. T. Robb, A. R. Place, K. R. Sowers, H. J. Schreier, and E. M. Fleischmann. 1995. Archaea: a laboratory manual—halophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.DasSarma, S., and P. Arora. 1997. Genetic analysis of gas vesicle gene cluster in haloarchaea. FEMS Microbiol. Lett. 153:1-10. [Google Scholar]
- 7.DasSarma, S., F. Morshed, E. Stuart, and S. Black. October. 1998. Recombinant gas vesicles and uses thereof. U.S. patent 5,824,309.
- 8.DasSarma, S., J. Halladay, and W. Ng. December. 1999. Recombinant vector and process for cell flotation. U.S. patent 6,008,051.
- 9.Halladay, J. T. 1992. Analysis of gas vesicle deficient mutants of Halobacterium halobium, identification of a gas vesicle gene cluster, and development of techniques to further investigate gas vesicle synthesis and assembly. Ph.D. thesis, University of Massachusetts, Amherst.
- 10.Halladay, J. T., J. G. Jones, F. Lin, A. B. MacDonald, and S. DasSarma. 1993. The rightward gas vesicle operon in Halobacterium plasmid pNRC100: identification of the gvpA and gvpC gene products by use of antibody probes and genetic analysis of the region downstream of gvpC. J. Bacteriol. 175:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 12.Hayes, P. K., B. Buchholz, and A. E. Walsby. 1992. Gas vesicles are strengthened by the outer-surface protein, GvpC. Arch. Microbiol. 157:229-234. [DOI] [PubMed] [Google Scholar]
- 13.Jones, J. G., N. R. Hackett, J. T. Halladay, D. J. Scothorn, C.-F. Yang, W.-L. Ng, and S. DasSarma. 1989. Analysis of insertion mutants reveals two new genes in the pNRC100 gas vesicle gene cluster of Halobacterium halobium. Nucleic Acids Res. 17:7785-7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, J. G., D. C. Young, and S. DasSarma. 1991. Structure and organization of the gas vesicle gene cluster on the Halobacterium halobium plasmid NCR100. Gene 102:1017-1022. [DOI] [PubMed] [Google Scholar]
- 15.Kingsman, R., and P. K. Hayes. 1997. Genes encoding proteins homologous to halobacterial Gvps N, J, K, F and L are located downstream of gvpC in the cyanobacterium Anabaena flos-aquae. DNA Seq. 7:97-106. [DOI] [PubMed] [Google Scholar]
- 16.Krantz, M. J., and C. E. Ballou. 1973. Analysis of Halobacterium halobium gas vesicles. J. Bacteriol. 114:1058-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, N., and M. C. Cannon. 1998. Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli. J. Bacteriol. 180:2450-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupas, A. 1997. Predicting coiled-coil regions in proteins. Curr. Opin. Struct. Biol. 7:388-393. [DOI] [PubMed] [Google Scholar]
- 19.Ng, W.-L., P. Arora, and S. DasSarma. 1994. Large deletions in class III gas vesicle-deficient mutants of Halobacterium halobium. Syst. Appl. Microbiol. 16:560-568. [Google Scholar]
- 20.Ng, W. V., S. A. Ciufo, T. M. Smith, R. E. Bumgarner, D. Baskin, J. Faust, B. Hall, C. Loretz, J. Seto, J. Slagel, L. Hood, and S. DasSarma. 1998. Snapshot of a large dynamic replicon in a halophilic archaeon: megaplasmid or minichromosome? Genome Res. 8:1131-1141. [DOI] [PubMed] [Google Scholar]
- 21.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Offner, S., A. Hofacker, G. Wanner, and F. Pfeifer. 2000. Eight of fourteen gvp genes are sufficient for formation of gas vesicles in halophilic archaea. J. Bacteriol. 182:4328-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Offner, S., U. Ziese, G. Wanner, D. Typke, and F. Pfeifer. 1998. Structural characteristics of halobacterial gas vesicles. Microbiology 144:1331-1342. [DOI] [PubMed] [Google Scholar]
- 24.Peck, R. F., S. DasSarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 25.Pfeifer, F., K. Kruger, R. Roder, A. Mayr, S. Ziesche, and S. Offner. 1997. Gas vesicle formation in halophilic archaea. Arch. Microbiol. 167:259-268. [DOI] [PubMed] [Google Scholar]
- 26.Simon, R. D. 1980. Acrylamide gel electrophoresis of hydrophobic proteins: gas vacuole protein. Electrophoresis 11:172-176. [Google Scholar]
- 27.Stuart, E. S., M. Sremac, F. Morshed, and S. DasSarma. 2001. Antigen presentation using novel particulate organelles from halophilic archaea. J. Biotechnol. 88:119-128. [DOI] [PubMed] [Google Scholar]
- 28.Surek, B., B. Pillay, U. Rdest, K. Beyreuther, and W. Goebel. 1988. Evidence for two different gas vesicle proteins and genes in Halobacterium halobium. J. Bacteriol. 170:1746-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waaland, R. J., and D. Branton. 1969. Gas vacuole development in a blue-green alga. Science 163:1339-1341. [DOI] [PubMed] [Google Scholar]
- 30.Walsby, A. E. 1994. Gas vesicles. Microbiol. Rev. 58:94-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, G., S. P. Kennedy, S. Fasiludeen, C. Rensing, and S. DasSarma. 2004. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J. Bacteriol. 186:3187-3194. [DOI] [PMC free article] [PubMed]