Abstract
Streptococcus pneumoniae secretes two different peptide pheromones used for intercellular communication. These peptides, which have completely unrelated primary structures, activate two separate signal transduction pathways, ComABCDE and BlpABCSRH, which regulate natural genetic transformation and bacteriocin production, respectively. Each signal transduction pathway contains a response regulator (ComE and BlpR, respectively) that activates transcription of target genes by binding to similar, but not identical, imperfect direct repeat motifs. In general the direct repeat binding sites are specific for one or the other of the two response regulators, ensuring that competence development and bacteriocin production are regulated separately. However, in the present study we show that the rate of transcription of an operon, encoding an ABC transporter of unknown function, can be stimulated by both peptide pheromones. We also show that this cross-induction is due to a hybrid direct repeat motif that can respond to both ComE and BlpR. To our knowledge this kind of convergent gene regulation by two separate two-component regulatory systems has not been described before in bacteria.
Quorum-sensing systems control several different important biological processes in bacteria, such as natural genetic transformation, virulence, bioluminescence, and sporulation. In gram-positive bacteria quorum-sensing is predominantly mediated by peptide pheromones instead of the N-acyl homoserine lactones used by the gram-negatives (8, 12, 13, 18). Two separate, but related, peptide-controlled quorum-sensing pathways have been discovered in Streptococcus pneumoniae (6). One of them, the ComABCDE pathway, regulates development of natural competence and has been relatively well characterized (2, 4, 9, 10, 21, 34). The other, more recently discovered pathway BlpABCSRH, regulates production of several class II bacteriocins and their immunity proteins (7, 25). Each pathway consists of a peptide pheromone, encoded by comC and blpC, their dedicated secretion apparatuses ComAB and BlpAB, and the two-component regulatory systems ComDE and BlpSRH. The peptide pheromones are both ribosomally synthesized as precursor peptides containing a double-glycine leader peptide at their N-terminal ends. Concomitant with export the double-glycine leaders of ComC and BlpC are removed by proteolytic domains in their respective ABC transporters ComA and BlpA. The mature pheromones, hereafter called the competence stimulating peptide (CSP) and bacteriocin inducing peptide (BIP), are both strain specific. Allelic variation of the comC and blpC genes exists within the species S. pneumoniae, giving rise to at least two different CSPs and four different BIPs (7, 11, 23, 36). The S. pneumoniae strain Rx used in the present study, which is closely related to strain R6 (33), produces CSP-1 consisting of 17 amino acids (NH2-EMRLSKFFRDFILQRKK-COOH) and BIP-1 consisting of 27 amino acids (NH2-GWWEELLHETILSKFKITKALELPIQL-COOH).
Activation of the competent state takes place when the external concentration of CSP-1 reaches a threshold level of about 10 ng ml−1 (9). At this concentration phosphorylation of ComE by the histidine kinase ComD triggers transcription of the comX gene, which encodes an alternative sigma factor (14, 15). The ComX sigma factor controls a regulon encompassing the late genes, whose gene products are involved in DNA processing, uptake, and recombination. Phosphorylated ComE binds to a direct repeat motif found in the promoter regions of the comAB, comCDE (34), and probably also the comX operon (6, 14), and activates transcription of these genes. The presence of this motif in the promoters of the comAB and comCDE operons generates the autoinducing behavior typical for quorum-sensing systems.
Induction of bacteriocin production by BIP does not involve an alternative sigma factor. In this system target genes, i.e., the bacteriocin structural and immunity genes, most likely are under direct control of the response regulator as has been demonstrated for closely related systems regulating bacteriocin production in other lactic acid bacteria (7, 27). The promoter regions of all operons in the blp cluster contain direct repeat motifs highly similar to those recognized by ComE, and it is therefore reasonable to assume that BlpR, which shares 33% amino acid sequence identity with ComE, binds to these motifs. Unexpectedly, the genes encoding the two-component regulatory system BlpRH are cotranscribed with a third gene blpS. This gene encodes a protein that shares significant sequence homology with the C-terminal DNA binding domain of BlpR, but lacks the N-terminal receiver domain. The role played by BlpS in regulation of bacteriocin production is not known (7).
In the present work we demonstrate that the two quorum-sensing systems controlled by CSP-1 and BIP-1 can activate transcription of the same target genes. These genes, which encode an ABC transporter of unknown function, are constitutively transcribed, but the rate of transcription is further increased by the addition of CSP-1 or BIP-1. Our data strongly indicate that the two signal transduction pathways converge at the promoter of the target genes, where the response regulators ComE and BlpR bind to the same direct repeat motif.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The following pneumococcal strains were used in the study: CP1200 (24); CP1500, a novobiocin-resistant CP1200 derivative (29); and CP1415, an erythromycin-resistant comA knockout mutant of CP1200 unable to secrete CSP-1 (4). A CP1415 derivative with no endogenous β-galactosidase activity (ebg mutant) was constructed previously by transforming strain CP1415 with genomic DNA isolated from the pneumococcal strain R262 (disrupted in ebg by a spectinomycin cassette) (1, 2, 30) to make the mutant EK100 (Eryr Spr comA ebg).
Liquid cultures of S. pneumoniae were routinely grown in casein hydrolysate (CAT) broth (19) at 37°C in the presence of chloramphenicol (CHL) (2.5 μg ml−1), erythromycin (2 μg ml−1), novobiocin (10 μg ml−1), streptomycin (100 μg ml−1), spectinomycin (200 μg ml−1), and kanamycin (KAN) (100 μg ml−1) when appropriate. The Escherichia coli strain TOP 10 (Invitrogen) was grown in Luria-Bertani (LB) broth. When selecting for plasmid-encoded drug resistance in E. coli the following antibiotics were used at the indicated concentrations: ampicillin (100 μg ml−1), KAN (50 μg ml−1), and CHL (34 μg ml−1).
DNA purification, PCR, and DNA sequencing.
Isolation of pneumococcal genomic DNA was carried out using Qiagen genomic tip-100 as described by the manufacturer with some minor modifications in the lysis step: 40 ml of an overnight culture was harvested at an optical density at 550 nm (OD550) of 0.5, and the pellet was resuspended in 3.5 ml of lysis buffer B1 (Qiagen) containing RNase A (1.4 mg ml−1; Sigma), lysozyme (2.3 mg ml−1; Sigma), protease (0.5 mg ml−1; Qiagen), and Mutanolysin (115 U ml−1; Sigma). After lysis at 37°C for 30 to 60 min the remaining steps of the DNA purification protocol were carried out as described in the standard procedure (Qiagen). The plasmid pEVP3 (5) and derivatives were isolated using the alkali lysis method (28). The Expand High Fidelity PCR system kit (Roche) was used in all PCR amplifications. DNA sequencing was prepared using the BigDye terminator cycle sequencing kit (Applied Biosystems) and analyzed on a Perkin-Elmer ABI Prism 377 sequencer.
Construction of recombinant plasmids and mutants.
To be able to disrupt the qsrAB and comM operons, and at the same time insert the lacZ reporter behind the promoters of these pneumococcal operons, internal fragments of the qsrB and comM genes were amplified by PCR and cloned into the pEVP3 plasmid. The pEVP3 plasmid, which contains a CHL marker, will replicate in E. coli but not in S. pneumoniae. Consequently, when pneumococci are transformed with this plasmid, only bacteria with the plasmid incorporated into the chromosome by homologous recombination will grow in the presence of CHL.
Initially, two different qsrAB reporter mutants were constructed; one with the pEVP3 plasmid inserted into the middle of the qsrB gene (qsrB::pEVP3), and another with the pEVP3 plasmid inserted into the promoter region of the qsrAB operon. DNA fragments from these regions (Fig. 1) were PCR amplified using CP1200 genomic DNA as template. The sequences of the primers used in the present work are derived from the complete genome sequence of the Norwegian serotype 4 strain available in GenBank (31). The primers eiv.1 (5′-TAATATGCATCAGCTTAATCGTTCAACTGCTTCCTTGTC-3′) and eiv.2 (5′-TAATGGATCCGCCAGACCACCTACAACATAGATCCC-3′) were used to amplify a 328-bp internal fragment of the qsrB gene, whereas 4144gal.1 (5′-ATTAAGGATCCGTGATAAATCCATTATACAGCAGCAAAC-3′) and 4144gal.2 (5′-ATTAAATGCATCTCTGATGG-ACGTAATTTATGGCTAG-3′) were used to amplify a 693-bp fragment immediately upstream of the box element in the promoter region of the qsrAB operon (Fig. 1). These fragments were ligated into the pCR 2.1 TOPO vector using the pCR II TOPO cloning kit (Invitrogen), and transformed into supercompetent E. coli TOP 10 cells (Invitrogen). Using restriction enzymes NsiI and BamHI (Fermentas MBI) the fragments were excised from the pCR II TOPO vector and ligated into the pEVP3 plasmid immediately upstream of the promoterless lacZ gene. Derivatives of pEVP3 were propagated in E. coli TOP 10 cells followed by transformation of S. pneumoniae strain EK100 with purified plasmid to target recombination at specific sites of the chromosome. The resulting qsrB::pEVP3 insertion-duplication mutant was termed EK4144, whereas the insertion-duplication mutant integrating immediately upstream of the box element was termed OE4144. The same procedure was followed when constructing a comM::lacZ reporter fusion mutant (EK4252) and a comM knockout mutant (EK4253). Primers 4252.1 (5′-TAATGGATCCGTTTAGTCAAACTCATCGACAAAGG-3′) and 4252.2 (5′-TAAATATGCATCAATGACCAATACAAGATCTCGG-3′) were used to amplify a 582-bp internal comM fragment that covers almost the entire length of the comM gene. This fragment was ligated into the pEVP3 vector in the correct orientation relative to the lacZ gene, and the resulting construct was then used to make the EK4252 mutant. The primers comM.50 (5′-CCATAGTGTCCTAGCCAGTAAGAAGTC-3′) and comM.51 (5′-CCAGCATAGCAAGAAAATCGCGCACG-3′) were used to amplify a smaller 229-bp internal comM fragment, which, in contrast to the 582-bp fragment, was suitable for constructing a comM knockout mutant (EK4253). These fragments were cloned as described above. All insertion-duplication mutants were checked by PCR using an internal pEVP3-specific primer pEVP3.1 (5′-CGTTTGTTGAACCATTATATCACATTATCC-3′) in combination with one of the primers 4144gal.1, eiv.1, 4252.1, or comM.50.
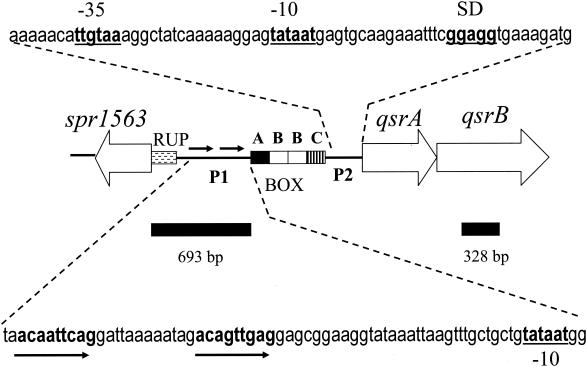
FIG. 1.
Organization of the S. pneumoniae R6 qsrAB operon encoding an ABC transporter of unknown function. The operon contains a competence-inducible promoter (p1) with a direct-repeat motif recognized by ComE (double arrows), and a constitutive housekeeping promoter (p2). The two promoters are separated by a box element, consisting of the subunits A, B, B, and C (16). Partial DNA sequences of the promoters are shown, with −10 regions, the −35 region, and a putative ribosome-binding site (SD) underlined. The ComE recognition motif is indicated by arrows. Solid bars represent DNA fragments cloned into the pEVP3 vector to target insertion into the pneumococcal genome by homologous recombination. The upstream spr1563 locus encodes a degenerate transposase that is preceded by a repeat unit of pneumococcus (RUP) (20).
The comE gene of the EK4144 strain was inactivated by specific allelic replacement via double-crossover recombination. The pFW13 plasmid (derived from pFW11), which will replicate in E. coli but not in S. pneumoniae, contains two multiple cloning sites (MCS-1 and MCS-2) flanking a KAN resistance gene (22). The primers comE.56 (5′-TTAAGCTAGCATCTTTCGTTTCAGATATGGTAAGTACG-3′) and comE.57 (5′-TTAAGACGTCCATCCAATATTCTCTCTAGTCTCACTTGATG-3′) were used to amplify an ∼800-bp fragment corresponding to the C-terminal end of comD. This fragment, which contains an NheI restriction site at one terminus and an AatII site at the other, was ligated into the corresponding sites in MCS-1. Following the same procedure the primers comE.53 (5′-ATTACCATGGTCTCAAAAGTGATTGACAATTAGCAAG-3′) and comE.55 (5′-ATTACATATGGCTATGGTACAATTACTGATGGAACAGCC-3′) were used to amplify a PCR fragment of similar size corresponding to the region downstream of the comE gene. This fragment was ligated into the NcoI and NdeI sites of MCS-II. The resulting recombinant plasmid was propagated in E. coli TOP 10 cells and purified using standard methods. Then a PCR was carried out with the comE.56 and comE.55 primers and the purified plasmid as template. The resulting fragment was about 3,400 bp long and contained the two PCR fragments described above flanking the KAN resistance gene. This linear fragment was used to transform the EK4144 strain. Transformants were selected on agar plates containing KAN (100 μg ml−1), followed by PCR to verify that the Knr cassette had been integrated correctly in the genome of the transformants, i.e., by replacing the comE gene. The primers tArg2 (5′-CATAGCTCAGCTGGATAGAGCATTCGCCTTC-3′) and FW13.6 (5′-CATTTATTTACCTCCTTTTGGTTACCTCAC-3′), which are complementary to the Arg-tRNA gene upstream of the comCDE operon and the promoter region of the KAN resistance gene, respectively, were used for this purpose. By analyzing the resulting PCR products on an agarose gel we identified a mutant strain (EK 4145) containing the Knr cassette correctly integrated in the genome of stain EK4144.
The QuikChange site-directed mutagenesis kit (Stratagene) was used to make the mutants OE4145, OE4146, OE4147, OE4148, and OE4149. The pEVP3 plasmid harboring a 693-bp fragment corresponding to the region immediately upstream of the box element in the qsrAB promoter (Fig. 1) was used as template for PCR with complementary primers containing the desired nucleotide changes. All constructs were sequenced to verify that only the desired nucleotide changes had been introduced into the 693-bp insert (see Fig. 2). In one of the mutated plasmids it was discovered that a single base pair had been deleted by chance in the 12-bp spacer region of the direct repeat motif. This plasmid and the other mutated plasmids were then used to transform strain EK100. The resulting insertion-duplication mutants (OE4145, OE4146, OE4147, OE4148, and OE4149) were checked by PCR as described above.
FIG. 2.
Comparisons of ComE and BlpR recognition motifs in different promoters. These recognition motifs (degenerated direct repeats) are written in capital letters. Conserved bases in the direct repeat motifs are outlined in bold with the consensus sequences indicated below. (A) ComE binding sites in the promoters of the comCDE and comAB operons. (B) BlpR binding sites in the promoters of the blpT, blpABC, blpIJK, blpL, blpMNO, and blpXYZ operons (7). (C) The direct repeat motif found in the promoter region of the qsrAB operon. This direct repeat motif seems to be a hybrid between the consensus competence (A) and bacteriocin repeat (B) motifs. (C′) The specified mutations (underlined) were made in direct repeat motif upstream of the qsrAB operon in the strains OE4145 to OE4149. (D) The direct repeat motif in the promoter of the competence regulated membrane protein ComM.
Transformation of strain CP1415 and derivatives.
An overnight culture of the pneumococcal strain to be transformed was diluted to an OD550 of 0.05 in prewarmed (37°C) CAT broth. Before samples were withdrawn for transformation, reconstitution of growth was allowed by incubating 1-ml samples at 37°C for 30 min. Genomic DNA was added to a final concentration of 2 to 5 μg ml−1 together with 250 ng of CSP-1 ml−1. Cells were incubated for 2.5 h before plating on CAT-agar plates containing the appropriate antibiotics for selection. Transformation with plasmid DNA was carried out exactly as described above, using 10 to 20 μg of highly purified plasmid ml−1.
Assay of β-galactosidase activity.
Overnight cultures of the respective reporter strains were diluted 40 to 100 times in fresh prewarmed (37°C) CAT medium, and incubated until the culture reached and OD550 of 0.2 to 0.3. Then, 1-ml samples were collected, and peptide pheromones were added to a final concentration of 250 ng ml−1 (or 500 ng ml−1). Samples were incubated for 30 min at 37°C before lysis was initiated by the addition of 10 μl of 10% Triton X-100 to a final concentration of 0.1% (vol/vol). After 15 min at 37°C the pneumococcal cells were completely lysed. ONPG (o-nitrophenyl-β-d-galactopyranoside) hydrolysis was initiated by the addition of 250 μl of fresh 5× Z buffer (5 mM MgCl2, 250 mM β-mercaptoethanol, 50 mM KCl, 0.3 M Na2HPO4 · 7H2O, 0.2 M NaH2PO4 · H2O [pH 7] and 4 mg ml−1 ONPG) to the lysed cells followed by incubation at 30°C for 25 to 40 min. The β-galactosidase reaction was quenched with 300 μl of 1 M Na2CO3,, and hydrolysis of ONPG was recorded in a spectrophotometer at 420 nm. Enzyme activity was calculated according to the method of Miller (17).
Sequence analysis.
All databank searches were carried out using the program BLAST (3), and the alignment of the amino acid sequences of ComE and BlpR was performed with ClustalW (32).
RESULTS AND DISCUSSION
The qsrAB genes and natural transformation.
Using electrophoretic mobility shift assays (EMSAs) we have previously shown that a ComE binding site is present in the promoter region of a pneumococcal ABC transporter (Fig. 1) of unknown function (34). Later it was shown by Rimini et al. (26), using microarray-based identification of competence-induced genes, that transcription of the genes encoding this transporter is upregulated about sixfold, 5 min after competence induction. To determine if the ABC transporter, termed quorum-sensing regulated transporter (QsrAB) (accession no. AAL00365 and AAL00366), is essential for competence development in S. pneumoniae, we disrupted the gene encoding the membrane domain of the ABC transporter (QsrB) by site-directed insertion-duplication mutagenesis. The qsrB mutant (EK4144) was made in the ComA-deficient strain EK100 (30), which is not able to develop the competent state without exogenously added CSP-1. The EK100 strain also lacks endogenous β-galactosidase activity (egb mutant). Subsequent transformation assays with the qsrB knockout mutant, and genomic DNA from a novobiocin resistant (Novr) strain (S. pneumoniae CP1500), revealed that the loss of the QsrB gene product had no effect on the transformability of the mutant strain (results not shown). The vector (pEVP3) used above for site-directed insertion-duplication mutagenesis, carries a lacZ reporter gene suitable for expression in transcriptional fusions in S. pneumoniae (5). Since the qsrB mutant carrying this lacZ reporter gene lacks endogenous β-galactosidase activity, the reporter gene can be used to study transcriptional activation of the qsrAB operon when competence is induced in the pneumococcal culture by the addition of CSP-1. Surprisingly, we found that the QsrAB transporter is highly expressed in noncompetent cells, and that induction of competence by the addition of CSP-1 at 250 ng ml−1 further upregulated transcription from the qsrAB promoter by about 100% (Table 1). These findings show that the QsrAB transporter is involved in housekeeping functions in the bacterial cell, but is produced in higher amounts during the competent state. The competence-inducible promoter containing the ComE binding site is located upstream of the box elements (16) approximately 360 bp from the start codon of the qsrA gene (Fig. 1). In addition, there is a potential housekeeping promoter located between the box elements and the qsrA gene (Fig. 1), which most likely is used for constitutive expression of the ABC transporter.
TABLE 1.
β-Galactosidase assays measuring rate of transcription from qsrAB and comM promoters in the presence of CSP-1 and/or BIP-1
| Strain | Promoter | Pheromone | Activity (Miller units)a |
|---|---|---|---|
| EK4144 | qsrAB (p1 and p2)b | None | 80.4 ± 2.19 |
| CSP-1 | 176.7 ± 1.7 | ||
| BIP-1 | 120 ± 4.33 | ||
| CSP-1 + BIP-1 | 217 ± 2.14 | ||
| 2 × CSP-1 | 181.3 ± 2.5 | ||
| OE 4144 | qsrAB (p1)c | None | 0.58 ± 0.07 |
| CSP-1 | 13.3 ± 0.99 | ||
| BIP-1 | 4.89 ± 0.61 | ||
| CSP-1 + BIP-1 | 15.25 ± 1.18 | ||
| OE 4145 | qsrAB (p1)d | None | 0.41 ± 0.06 |
| CSP-1 | 10.12 ± 1.16 | ||
| BIP-1 | 10.60 ± 1.80 | ||
| CSP-1 + BIP-1 | 14.84 ± 1.87 | ||
| OE 4146 | qsrAB (p1)d | None | 0.46 ± 0.02 |
| CSP-1 | 11.60 ± 0.98 | ||
| BIP-1 | 1.01 ± 0.24 | ||
| CSP-1 + BIP-1 | 11.63 ± 1.31 | ||
| OE 4147 | qsrAB (p1)d | None | 0.82 ± 0.16 |
| CSP-1 | 7.55 ± 0.38 | ||
| BIP-1 | 3.24 ± 0.80 | ||
| CSP-1 + BIP-1 | 10.01 ± 1.52 | ||
| OE 4148 | qsrAB (p1)d | None | 0.79 ± 0.23 |
| CSP-1 | 8.11 ± 0.65 | ||
| BIP-1 | 3.19 ± 0.50 | ||
| CSP-1 + BIP-1 | 9.25 ± 0.43 | ||
| OE 4149 | qsrAB (p1)d | None | 0.20 ± 0.1 |
| CSP-1 | 0.30 ± 0.06 | ||
| BIP-1 | 0.34 ± 0.03 | ||
| CSP-1 + BIP-1 | 0.34 ± 0.03 | ||
| EK4252 | comM | None | 2.42 ± 0.12 |
| CSP-1 | 17.64 ± 0.12 | ||
| BIP-1 | 2.18 ± 0.80 | ||
| CSP-1 + BIP-1 | 19.48 ± 0.58 | ||
| 2 × CSP-1 | 15.95 ± 0.58 |
β-Galactosidase activities are given in Miller units, and are the means ± standard errors of triplicate samples.
Activity is the sum of the activities of the competence-inducible promoter (p1) and the housekeeping promoter (p2).
Activity is the activity of only the competence-inducible promoter (p1).
Activity is that of the p1 promoter containing different mutations.
Searches in databases for the closest homologue of the QsrAB transporter with a known function, identified an ABC transporter from Bacillus firmus that extrudes Na+ ions (35). The ATP-binding domain of this transporter (NatA) shares 41% identity with QsrA, whereas the membrane domain NatB is 22% identical to QsrB. It is therefore possible that QsrAB is an Na+ exporter, but this must of course be verified experimentally.
Transcriptional activation of the qsrAB operon via two independent quorum-sensing pathways.
To check the specificity of the transcriptional activation of the qsrAB operon by CSP-1, we performed the β-galactosidase assay described above with CSP-2, CSP-3165, BIP-1, and BIP-2 (Table 2). CSP-1 and CSP-2, which share about 50% identity, are the two major competence pheromone types (pherotypes) found among pneumococcal strains (23, 36). When CSP-2 is used instead of CSP-1 to induce competence in S. pneumoniae strain Rx very few or no transformants are obtained, demonstrating that the ComD receptor of the Rx strain recognizes CSP-2 poorly. In accordance with these findings, cross-induction by CSP-2 in the β-galactosidase assay is negligible (Table 2). The competence pheromone CSP-3165, which is produced by Streptococcus gordonii strain NCTC 3165 (11), is completely inactive with respect to competence induction in S. pneumoniae. Our results show that CSP-3165 is inactive in the β-galactosidase assay as well, demonstrating that the pheromone-induced transcriptional activation of the qsrAB operon is pherotype specific. BIP-1 and BIP-2 are encoded by different alleles of blpC, and induce bacteriocin production in separate strains of S. pneumoniae. In parallel to CSP-1 and CSP-2, cross-induction experiments show that these peptide pheromones are strain specific (7, 25). As expected, the BIP peptides, which possess primary structures that are completely unrelated to CSP-1, showed no competence-inducing activity at all. It was therefore very surprising to find that the BIP-1 pheromone induces a significant increase in expression of the QsrAB transporter (Tables 1 and 2). The BIP-2 pheromone, on the other hand, is without activity (Table 2), demonstrating that the effect of BIP-1 is specific and most likely mediated by the BlpABCSRH pathway. Since BIP-1 is completely inactive with respect to competence induction it is unlikely that the effect of this peptide on QsrAB expression is mediated by the ComDE pathway. To rule out the possibility of cross-phosphorylation of ComE by BlpH, the comE gene of the qsrAB insertion-duplication mutant (EK4144) was disrupted by a Kn marker flanked with ∼800-bp regions of homologous DNA. Transcription of the qsrAB operon could still be upregulated by addition of BIP-1 in this mutant (EK4145), but it had become insensitive to CSP-1 (Table 3). The additive effect of CSP-1 and BIP-1 had also disappeared. This result demonstrates that the effect of BIP-1 is mediated by the BlpR, and not by ComE.
TABLE 2.
β-Galactosidase assays demonstrating specificity of the CSP-1 and BIP-1 pheromones in stimulating the rate of transcription of the qsrAB operon in the EK4144 straina
| Pheromone(s) | Pheromone sequence | Activity (Miller units)b |
|---|---|---|
| None | 50.5 | |
| CSP-1 | N-EMRLSKFFRDFILQRKK-C | 89.8 |
| 2 × CSP-1 | 87.9 | |
| CSP-2 | N-EMRISRIILDFLFLRKK-C | 52.1 |
| CSP-3165 | N-SQKGVYASQRSFVPSWFRKIFRN-C | 51.8 |
| BIP-1 | N-GWWEELLHETILSKFKITKALELPIQL-C | 70.7 |
| BIP-2 | N-GLWEDLLYNINRYAHYIT-C | 50.5 |
| CSP-1 + BIP-1 | 104.8 |
The experiment was repeated several times with similar results.
Sum of the activities of the competence-inducible promoter (p1) and the housekeeping promoter (p2).
TABLE 3.
β-Galactosidase assays measuring the rate of transcription from the qsrAB promoter in the presence of CSP-1 and/or BIP-1a
| Strain | Promoter | Pheromone | Activity (Miller units)a | |
|---|---|---|---|---|
| EK4144 | qsrAB (p1 and p2) | None | 138.1 ± 1.28 | |
| CSP-1 | 215.7 ± 3.02 | |||
| BIP-1 | 153.8 ± 4.71 | |||
| CSP-1 + BIP-1 | 229.7b | |||
| EK4145 | qsrAB (p1 and p2) | None | 126.9 ± 1.89 | |
| CSP-1 | 128.5 ± 0.47 | |||
| BIP-1 | 152.6 ± 0.22 | |||
| CSP-1 + BIP-1 | 150.7 ± 2.35 |
The strains employed (EK4144 and EK4145) contain a transcriptional fusion between the qsrB gene and lacZ (prtB::lacZ). In addition, the comE gene has been disrupted in the EK4145 strain (ComE−). β-Galactosidase activities are given in Miller units and are the means ± standard errors of triplicate samples. Results are based on the sum of the activities of the competence-inducible promoter (p1) and the housekeeping promoter (p2).
Mean of two samples.
BIP-1 does not stimulate transcription from all promoters containing ComE-like binding sites.
The BIP-1 peptide does not activate competence development in diluted cultures of S. pneumoniae strain CP1200, i.e., at cell densities below the level needed for autoinduction to take place (results not shown). This result implies that the direct repeat located in the comCDE promoter region must be ComE specific. If this binding site was recognized by BlpR, the addition of the BIP-1 peptide would immediately trigger the ComABCDE autoinduction mechanism, leading to rapid secretion of CSP-1 followed by induction of the competent state. Although it has not been proven experimentally, it is also likely that the direct repeat upstream of the comAB operon is ComE specific. To learn more about the properties of ComE-like binding sites, we used BLAST to search for additional direct repeat motifs in the genome of S. pneumoniae. A previously unrecognized motif was identified in the promoter region of a gene encoding an integral membrane protein. This protein has no close homologues in the sequence databases with a known function. However, Southern analysis demonstrated that it is present also in several strains of Streptococcus mitis and Streptococcus oralis (results not shown). Since it is a membrane protein and appeared to be competence induced, it was called ComM (accession no. AAL00565). The comM gene was disrupted by insertion-duplication mutagenesis (EK4253), using the same technique as described above. Disruption of the gene did not result in a detectable loss of transformability, indicating that the comM gene product is not involved in DNA uptake, processing or recombination. As the EK4252 mutant has the lacZ reporter gene inserted behind the comM promoter, it was possible to monitor the rate of transcription of this gene in the presence of the peptide pheromones. The results of the β-galactosidase assays showed that only CSP-1 induces transcription of the comM gene, demonstrating that the direct repeat located upstream of this gene is completely ComE specific (Table 1).
A hybrid direct repeat motif.
As discussed above, experimental evidence shows that BIP-1 activates expression of several bacteriocin operons by a quorum-sensing mechanism involving the BlpSRH two-component regulatory system (7). Interestingly, the putative DNA binding site of the response regulator BlpR is highly similar to the direct repeat sequence motif recognized by ComE (Fig. 2). The target site of ComE consists of two 9-bp imperfect direct repeats separated by a stretch of 12 nucleotides (34). The consensus sequence of the 9-bp direct repeat is 5′-(AGT)CA(GCT)TT(GT)(AG)G-3′, where the underlined positions are conserved. This consensus is based on the direct repeats found in the promoter regions of the comAB and comCDE operons (Fig. 2). EMSAs carried out with DNA fragments containing these direct repeat motifs demonstrated binding of ComE to both of them (34). A search for conserved sequence elements in the promoter regions of the operons (blpT, blpAB, blpIJK, blpL, blpMNO, and blpXYZ) regulated by BIP, revealed a closely related motif consisting of two 9-bp direct repeats separated by a stretch of 12 bp (7). In this case the consensus sequence of the 9-bp direct repeat is 5′-(CGA)C(ACT)ATTCAA-3′, where all positions are highly conserved except the first and third base pair from the 5′ end (Fig. 2). Several recent studies have shown that the 9-bp direct repeats are directly involved in binding of the response regulator. Substitutions of the most conserved nucleotide positions in the left repeat of the ComE motif with four adenylate residues resulted in a loss of ComE binding to this repeat while binding to the right repeat was maintained (34). Furthermore, studies carried out on a related two-component regulatory system controlling bacteriocin production in Lactobacillus plantarum, which included point substitutions in the direct repeats and DNase I footprinting analyses, demonstrated that the response regulator binds directly to the 9-bp repeat motif as a homodimer and that the conserved positions in the direct repeats are essential for binding (27). A comparison of the sequences recognized by ComE and BlpR show that they do not differ significantly in positions 1, 2, 3, 5, 6, and 8 from the 5′ end. In contrast, positions 4, 7, and 9 almost always differ between the two motifs, indicating that these positions are involved in discriminating between the two related response regulators. In other words, these positions most likely prevent effective binding of BlpR to the ComE-specific motif and vice versa.
The most straightforward interpretation of the results showing that transcription from the qsrAB promoter is constitutive, but can be further increased via two different signal transduction pathways, is to assume that the region upstream of the qsrAB genes contains three separate promoters. One of these promoters could be a constitutive housekeeping promoter, whereas the other two could be specific for ComE and BlpR, respectively. As expected, a potential housekeeping promoter (TTGTAA-17 bp-TATAAT) is located at the appropriate distance from the ribosome binding site upstream the qsrA gene (Fig. 1). However, searches for direct repeat motifs in the qsrAB promoter region revealed that only one such motif is present. This motif is located immediately upstream of the box element, approximately 300 bp from the housekeeping promoter (Fig. 1). Curiously, this direct repeat motif seems to be a hybrid between the motifs recognized by ComE and BlpR (Fig. 2). The right motif constitutes a typical ComE consensus motif, whereas the left repeat has more in common with the motif recognized by BlpR. This hybrid structure suggests a mechanism for the unusual transcriptional regulation observed for the qsrAB operon. It appears that ComE and BlpR both recognize and activate transcription from the same direct repeat motif. Converging quorum-sensing systems have previously been observed in Pseudomonas aeruginosa (37). However, these systems respond to acyl homoserine lactones, and are structurally and functionally unrelated to the two-component regulatory systems described here. Judging from the sequences of the left and right repeats of the direct repeat motif, ComE will bind with high affinity to the right repeat but poorly to the left. Presumably, it will be the other way around for BlpR, which will bind more strongly to the left than to the right repeat. Since the response regulators bind in a cooperative manner, binding of ComE and BlpR monomers to their respective high-affinity sites will presumably recruit and stabilize binding of the other monomer to the low-affinity site. By adding a mixture of CSP-1 and BIP-1 (250 ng ml−1 each) to a growing culture of S. pneumoniae we obtained a further increase in the transcriptional rate compared to the rate obtained with each peptide pheromone alone (Tables 1 and 2). This effect requires a combination of the two pheromones, as only a marginal increase in transcriptional rate was obtained when the CSP-1 concentration was doubled from 250 to 500 ng ml−1 (Tables 1 and 2). This result is best explained by assuming that ComE and BlpR can bind to the direct repeat motif upstream of qsrA as a ComE/BlpR heterodimer and that the heterodimer is bound with higher affinity than the ComE/ComE and BlpR/BlpR homodimers.
As the CSP-1 and BIP-1 inducible promoter is located approximately 300 bp upstream of the housekeeping promoter, mRNAs transcribed from the inducible promoter will include a box element consisting of the subunits A, B, B, and C at its 5′ end (Fig. 1). To separate the activity of the competence-inducible promoter from that of the housekeeping promoter, insertion-duplication mutagenesis was used to insert the lacZ reporter gene between the direct repeat motif and the box element (Fig. 1). No constitutive transcription of the qsrAB genes was observed in this mutant (OE4144), demonstrating that the housekeeping promoter is located between the box element and the qsrA gene as predicted. However, transcriptional activation by both CSP-1 and BIP-1 was still evident, even though the level of transcriptional activation for some reason was reduced compared to that observed for the qsrB::lacZ reporter construct in strain EK4144 (Table 1). This reduction might be caused by the removal of the box element from the 5′ end of the mRNA. The function of the box elements, of which there are 127 in the serotype 4 isolate (31), is a mystery. It is possible that the presence of a box element in a particular mRNA will increase its half-life or increase the efficiency by which it is translated. However, further investigations are needed to clarify this matter.
To obtain further evidence that BlpR is binding to the same direct repeat as ComE, a series of mutants (OE4145 to OE4149) were constructed (Fig. 2). The OE4145 mutant contains two identical left repeats, whereas the OE4146 mutant contains two identical right repeats. The introduced changes in the direct repeat motifs of these mutants had little effect on the transcriptional activation elicited by CSP-1, indicating that ComE binds well to both repeats. In contrast, transcriptional activation by BIP-1 was increased by about 100% in the OE4145 mutant and almost abolished in the OE4146 mutant. These data demonstrate that BlpR binds strongly to the left repeat, but very poorly to the right repeat, suggesting that binding of BlpR to the right repeat in the wild-type promoter is stabilized by protein-protein interaction. As the left and right repeats only differ at nucleotide positions 4 and 7 from the 5′ end (Fig. 2), these adenylate and cytidylate residues must be very important for binding of BlpR. This is in excellent agreement with the BlpR recognition motifs found in the promoters of the blpT, blpAB, blpIJK, blpL, blpMNO, and blpXYZ operons (Fig. 2). In these promoters the adenylate and cytidylate residues at positions 4 and 7 are highly conserved, while they are absent in the promoters recognized only by ComE (Fig. 2).
Interestingly, adding both CSP-1 and BIP-1 to the cells has a synergistic effect on the level of transcription (Table 1). This synergistic effect has been lost in the OE4146 mutant, presumably because BlpR binds very poorly to the direct repeat motif upstream the lacZ gene in this mutant. It follows from this that the synergistic effect must be due to binding of BlpR to the left and ComE to the right repeat, indicating that the BlpR/ComE-promoter complex is stabilized by protein-protein interaction between BlpR and ComE.
A comparison of the direct repeat motifs in the comAB, comCDE, and comM promoters shows that the guanylate residue at position 9 from the 5′ end is highly conserved (Fig. 2). In contrast, adenylate residues are found in the corresponding position in the direct repeat motifs located upstream of the blpT, blpAB, blpIJK, blpL, blpMNO, and blpXYZ operons. We therefore speculated that the guanylate residue could be important for ComE binding. To investigate this possibility we replaced the conserved guanylate in the left repeat with an adenylate residue in mutant OE4147. In addition, we made a mutant (OE4148) where the conserved guanylate residues in both repeats were replaced by adenylate. By chance, a third mutant (OE4149) was obtained that contains a single-nucleotide deletion in the 12-bp spacer region separating the direct repeats (Fig. 2). Our results show that CSP-1-induced transcription of the reporter gene was significantly reduced in the OE4147 mutant compared to OE4144, whereas BIP-1-induced transcription was only slightly affected. This finding supports the idea that the conserved guanylate residue is involved in ComE binding. We therefore expected that the CSP-1-activated transcription should be further reduced in the OE4148 double mutant, but this turned out not to be the case. Interestingly, the addition of CSP-1 and/or BIP-1 did not activate transcription of the lacZ reporter gene at all in the OE4149 mutant. This striking result demonstrates that when the 12-bp spacing is decreased by just a single nucleotide, the promoter becomes nonfunctional and responds to neither ComE nor BlpR. This finding is in agreement with the results reported by Risøen et al. (27). They used EMSAs to study the properties of a closely related direct repeat motif located upstream of the plnA gene in L. plantarum. Their data show that formation of a dimeric complex was abolished when the 12-bp spacing was decreased by a single nucleotide. In sum, the fact that CSP-1- and BIP-1-activated transcription is equally affected by the single base pair deletion clearly shows that ComE and BlpR binds to the same direct repeat motif.
Concluding remarks.
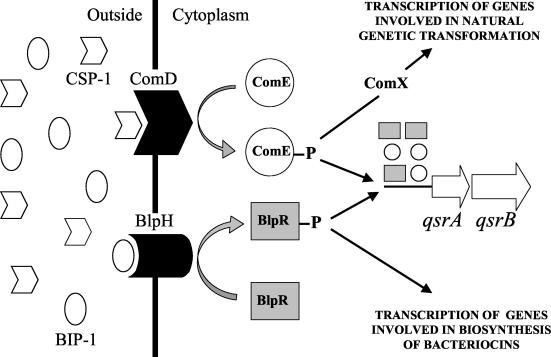
In the present study we have shown that transcription of an operon encoding the ABC transporter QsrAB can be upregulated via two independent quorum-sensing systems. Our results strongly indicate that these systems, ComABCDE and BlpABCSRH, converge at a common binding site for the ComE and BlpR response regulators (Fig. 3). If QsrAB is a sodium pump, as homology searches suggest, its biological function might be to protect S. pneumoniae against osmotic stress. Both secretion of membrane-active bacteriocins and translocation of DNA across the cell wall and cytoplasmic membrane are likely to elicit osmotic stress, and such stress could therefore be the common denominator that explains why both CSP-1 and BIP-1 stimulate expression of QsrAB. Whatever the biological function of this particular system, hybrid motifs that are recognized by response regulators from two different two-component regulatory systems, have not been described before in bacteria.
FIG. 3.
Model depicting the mechanism of the convergent transcriptional activation of the qsrAB operon by the ComCDE and BlpCSRH two-component regulatory systems. ComD and BlpH, histidine kinase receptors; ComE and BlpR, response regulators; ComX, alternative sigma factor; QsrAB, ABC transporter of unknown function.
Acknowledgments
This work was supported by a grant from The Research Council of Norway.
REFERENCES
- 1.Alloing, G., C. Granadel, D. A. Morrison, and J. P. Claverys. 1996. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol. Microbiol. 21:471-478. [DOI] [PubMed] [Google Scholar]
- 2.Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75-83. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandler, M. S., and D. A. Morrison. 1987. Competence for genetic transformation in Streptococcus pneumoniae: molecular cloning of com, a competence control locus. J. Bacteriol. 169:2005-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 6.Claverys, J. P., and L. S. Håvarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:d1798-d1814. [DOI] [PubMed] [Google Scholar]
- 7.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 9.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 11.Håvarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Håvarstein, L. S. 2003. Intercellular communication in Gram-positive bacteria depends on peptide pheromones and their histidine kinase receptors, p. 341-363. In M. Inouye and R. Dutta (ed.), Histidine kinases in signal transduction. Academic Press, San Diego, Calif.
- 13.Kleerebezem, M., L. E. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 14.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo, P., and D. A. Morrison. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin, B., O. Humbert, M. Camara, E. Guenzi, J. Walker, T. Mitchell, P. Andrew, M. Prudhomme, G. Alloing, R. Hakenbeck, D. A. Morrison, G. J. Boulnois, and J. P. Claverys. 1992. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 20:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 18.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 19.Morrison, D. A., S. A. Lacks, W. R Guild, and J. M. Hageman. 1983. Isolation and characterization of three new classes of transformation-deficient mutants of Streptococcus pneumoniae that are defective in DNA transport and genetic recombination. J. Bacteriol. 156:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oggioni, M. R., and J. P. Claverys. 1999. Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology 145:2647-2653. [DOI] [PubMed] [Google Scholar]
- 21.Pestova, E. V., L. S. Håvarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 22.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lütticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 23.Pozzi, G., L. Masala, F. Iannelli, R. Manganelli, L. S. Håvarstein, L. Piccoli, D. Simon, and D. A. Morrison. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravin, A. W. 1959. Reciprocal capsular transformations of pneumococci. J. Bacteriol. 77:296-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reichmann, P., and R. Hakenbeck. 2000. Allelic variation in a peptide-inducible two-component system of Streptococcus pneumoniae. FEMS Microbiol. Lett. 190:231-236. [DOI] [PubMed] [Google Scholar]
- 26.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36:1279-1292. [DOI] [PubMed] [Google Scholar]
- 27.Risøen, P. A., O. Johnsborg, D. B. Diep, L. Haemoen, G. Venema, and I. F. Nes. 2001. Regulation of bacteriocin production in Lactobacillus plantarum depends on a conserved promoter arrangement with consensus binding sequence. Mol. Gen. Genomics 265:198-206. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 29.Shoemaker, N. B., and W. R. Guild. 1974. Destruction of low efficiency markers is a slow process occurring at a heteroduplex stage of transformation. Mol. Gen. Genet. 128:283-290. [DOI] [PubMed] [Google Scholar]
- 30.Steinmoen, H., E. Knutsen, and L. S. Håvarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 99:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiraby, G., M. S. Fox, and H. Bernheimer. 1975. Marker discrimination in deoxyribonucleic acid-mediated transformation of various pneumococcus strains. J. Bacteriol. 121:608-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ween, O., P. Gaustad, and L. S. Håvarstein. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33:817-827. [DOI] [PubMed] [Google Scholar]
- 35.Wei, Y., A. A. Guffanti, and T. A. Krulwich. 1999. Sequence analysis and functional studies of a chromosomal region of alkaliphilic Bacillus firmus OF4 encoding an ABC-type transporter with similarity of sequence and Na+ exclusion to the Bacillus subtilis NatAB transporter. Extremophiles 3:113-120. [DOI] [PubMed] [Google Scholar]
- 36.Whatmore, A. M., V. A. Barcus, and C. G. Dowson. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whiteley, M., and E. P. Greenberg. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]