Abstract
The genome sequence of Halobacterium sp. strain NRC-1 encodes genes homologous to those responsible for conferring resistance to arsenic. These genes occur on both the large extrachromosomal replicon pNRC100 (arsADRC and arsR2M) and on the chromosome (arsB). We studied the role of these ars genes in arsenic resistance genetically by construction of gene knockouts. Deletion of the arsADRC gene cluster in a Halobacterium NRC-1 Δura3 strain resulted in increased sensitivity to arsenite and antimonite but not arsenate. In contrast, knockout of the chromosomal arsB gene did not show significantly increased sensitivity to arsenite or arsenate. We also found that knockout of the arsM gene produced sensitivity to arsenite, suggesting a second novel mechanism of arsenic resistance involving a putative arsenite(III)-methyltransferase. These results indicate that Halobacterium sp. strain NRC-1 contains an arsenite and antimonite extrusion system with significant differences from bacterial counterparts. Deletion analysis was facilitated by an improved method for gene knockouts/replacements in Halobacterium that relies on both selection and counterselection of ura3 using a uracil dropout medium and 5-fluoroorotic acid. The arsenite and antimonite resistance elements were shown to be regulated, with resistance to arsenic in the wild type inducible by exposure to a sublethal concentration of the metal. Northern hybridization and reverse transcription-PCR analyses showed that arsA, arsD, arsR, arsM, arsC, and arsB, but not arsR2, are inducible by arsenite and antimonite. We discuss novel aspects of arsenic resistance in this halophilic archaeon and technical improvements in our capability for gene knockouts in the genome.
The halophilic archaeon (haloarchaeon) Halobacterium sp. strain NRC-1 is an excellent model for postgenomic analysis of heavy metal resistance. Its genome is completely sequenced, and a large number of genetic tools are available for characterization of this extreme halophile (3, 9, 10). It is easily grown in the laboratory in hypersaline medium containing about a 10-fold concentration of seawater (2), and its natural environment is usually rich in heavy metals, many of which are toxic to cells. The genome sequence of Halobacterium sp. strain NRC-1 revealed multiple putative metal ion transporter genes, including arsenic, cadmium, copper, cobalt, zinc, and iron (9), indicating an excellent ability to handle metal ions in its environment. However, none of these hypothetical genes has been shown to be functional, and very few studies have been directed at the understanding of heavy metal resistance in haloarchaea.
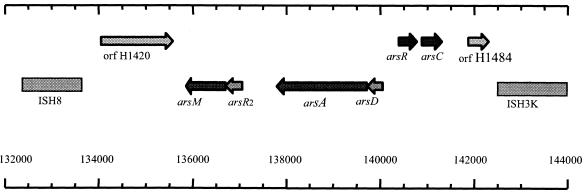
The Halobacterium sp. strain NRC-1 genome contains a 2-Mb chromosome and two megaplasmids/minichromosomes, pNRC100 and pNRC200 (191 and 365 kb, respectively) (6-9). The pNRC replicons share 145 kb of identity, including large inverted repeats that recombine to produce inversion isomers in the cell. The replicons also contain unique DNA, with a 45-kb segment within the large single-copy region of pNRC100 coding for an ars gene cluster including arsADRC (Fig. 1). Annotation of the Halobacterium sp. strain NRC-1 genome sequence also identified another putative ars gene, arsB, located on the chromosome. Recently, we identified two additional ars gene homologs near arsADRC on pNRC100, a second arsR gene (arsR2) and another gene, named arsM (Fig. 1). These genes were thought to be involved in arsenic resistance because of their homology to previously characterized genes (8, 9, 12).
FIG. 1.
The ars gene cluster of Halobacterium sp. strain NRC-1. A 14-kb region of pNRC100 (bp 132000 to144000) (8) from wild-type Halobacterium sp. strain NRC-1 is shown containing the genes putatively responsible for arsenic resistance in this organism. Genes and open reading frames are shown as arrows to scale, with rightward or leftward transcriptional direction indicated. The positions of two IS elements flanking the gene cluster are indicated by boxes.
The ars operons of gram-negative and gram-positive bacteria have been found on both plasmids and chromosomes, and the majority of these determinants are three-gene arsRBC operons (12). The arsR gene encodes the 13-kDa As(III)- and Sb(III)-responsive regulator, and arsB encodes a transporter responsible for proton-gradient-dependent extrusion of As(III) and Sb(III). Additional resistance to arsenate As(V) requires an arsenate reductase encoded by arsC. Some operons contain two additional genes, arsA and arsD, conferring increased resistance. ArsA is an ATPase that is activated by As(III) or Sb(III). ArsA binds to ArsB and converts the arsenite permease into a primary ATP-driven arsenite pump. ArsD is an additional transcriptional repressor unrelated in primary sequence but binding to the same regulatory region as ArsR (1). ArsD has been shown to regulate the maximal expression of the ATP-requiring pump, which itself can be toxic to the cell if overexpressed (12).
In Halobacterium sp. strain NRC-1, the arsADRC has an apparent divergent operon structure with arsDA transcribed leftward and arsRC transcribed rightward (Fig. 1). This is a highly unusual operon structure and surprisingly does not contain a transporter resembling ArsB. The location of ArsD suggested that it might regulate ArsA in Halobacterium (12). Interestingly, the single recognizable ArsB family homolog in Halobacterium is coded for at a separate chromosomal locus and is rather distantly related to ArsB sequences in bacteria. Two additional pNRC100 genes with possible involvement in arsenic resistance are located immediately downstream of the arsDA genes: a putative operon encoding a second member of the ArsR family (arsR2) and a putative methyltransferase (arsM) with homology to a recently identified arsenite(III)-methyltransferase from mammals (5).
To determine if the Halobacterium sp. strain NRC-1 ars genes contribute to arsenic resistance in this archaeon, we have employed a genetic approach using an improved method of gene knockout (10). We generated deletions of both pNRC100 ars loci, a putative arsenite(III)-methyltransferase gene and a putative chromosomally encoded arsB gene; determined their sensitivity to arsenite As(III), arsenate As(V), and antimonite Sb(III); and established the induction characteristics of the ars genes in the wild type. Significantly, our results indicate that multiple mechanisms of arsenic resistance are operating in this haloarchaeon.
MATERIALS AND METHODS
Strains, culturing, and deletion construction.
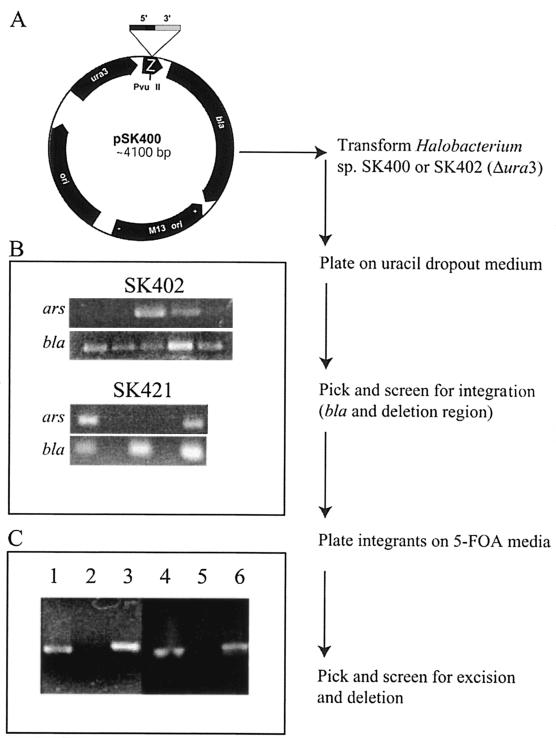
Escherichia coli strain DH5α was used as a host for plasmid constructions (13). Plasmid pSK400 (pseudonym pMPK408; a gift from M. P. Krebs) (10, 11), containing the 1.3-kb PshAI fragment with the ura3 gene of Halobacterium sp. strain NRC-1 cloned into the EcoRV site of Litmus 28, was used as a vector for construction of gene knockouts (Fig. 2). All oligonucleotide primers used for gene knockouts are listed in Table 1. Halobacterium sp. strain NRC-1 genomic DNA was used as a template for initial PCR amplification. Inserts in plasmids pSK402, pSK421, and pSK431 were checked by sequencing.
FIG. 2.
Gene knockout strategy and strain construction. (A) Map of plasmid pSK400 (10), a Litmus 18 derivative containing the Halobacterium sp. strain NRC-1 ura3 gene used as the knockout vector. The position of target gene fragments containing 5′ and 3′ flanking regions cloned into the PvuII site in the 3′ region of the lacZα gene fragment (Z) is shown above pSK400. (B) PCR assay of transformants grown on dropout SKURA medium (plasmid integrants) using appropriate ars primers (arsA reverse and arsC reverse for SK402 and 2599 reverse and 2603 forward for SK421) and bla forward and reverse primers. Positive isolates containing both PCR fragments are visible (two of five for SK402 and two of three for SK421). (C) PCR assays of isolates grown on 5-FOA plates that result from plasmid excision and deletion of ars genes (ΔarsADRC for strain SK402, lanes 1 to 3, and ΔarsB for strain SK421, lanes 4 to 6). Lanes 1 and 4 contain PCR products with a positive control (tbpC primers), lanes 2 and 5 contain products with ura3 gene primers, and lanes 3 and 6 contain products with ars primers (as in panel B).
TABLE 1.
Oligonucleotides used for gene knockouts
| Gene and orientation | Sequence (5′ → 3′) |
|---|---|
| arsA reverse | CGGTCACCTCAGTGATCGTC |
| arsA inverse | CTGCGAATCCCCTGTCGAGACGA |
| arsC inverse | GCGCGTTGTAGACCTGTTCG |
| arsC reverse | AACAGACACCACCGGAACGC |
| 2599 reverse | CTACGCGAAGAACTGTGCCA |
| arsB 5′ inverse | CGCTGACATGGTGCCACGGT |
| arsB 3′ inverse | TGGGGCGTCTAAGGCGGATA |
| 2603 forward | ATGGAACAAGTGTGGGCAGA |
| 5175 reverse | CTATTTGAAATCCTGAAAGA |
| arsM 3′ inverse | GACTGACCGGCCGAATCG |
| arsM 5′ inverse | CATGGTTGCCTCGCAGGT |
| 5176 forward | ATGGTTCAGGACGCAACCGC |
| ura3 forward | ATGAGCTTCGTCGAGGAACT |
| ura3 reverse | CTACCGGTGGCGGTTCAGGC |
| bla forward | ATGAGTATTCAACATTTCCG |
| bla reverse | TTACCAATGCTTAATCAGTG |
| tbpC forward | ATGACGGTCGAGATTGCGAA |
| tbpC reverse | TTAAACTAATTCTTGGACTT |
For deletion of the arsADRC genes on pNRC100, the arsA reverse and inverse primers and the arsC inverse and reverse primers (Table 1) were used to PCR amplify 496- and 554-bp flanking regions, respectively (pNRC100 coordinates 137293 to 137788 and 141270 to 141823). The two PCR fragments were cloned sequentially into the PvuII site in the lacZ gene of pSK400 to form pSK401 and pSK402, respectively. Three nucleotides (CTG) were added at the 5′ end of the arsA inverse primer in order to reconstitute the PvuII site after cloning the first PCR fragment. In plasmid pSK402, a 3,482-bp region including all of arsA, arsD, and arsR, and all but 40 nucleotides of the 3′ coding sequence of arsC is deleted (pNRC100 deletion coordinates 137789 to 141269).
For deletion of the chromosomal arsB gene, a 2,927-bp PCR fragment containing the gene and 5′ and 3′ flanking regions was amplified using the 2599 reverse and 2603 forward primers (Table 1, chromosomal coordinates 1943386 to 1946312) and cloned into the PvuII site of pSK400 to form pSK420. PCR was carried out on the pSK420 template using the arsB 5′ and 3′ inverse primers (Table 1), and the linear product was circularized by ligation to form plasmid pSK421. This plasmid contained 658 bp of 5′ arsB flanking DNA and 353 bp of 3′ flanking DNA. It deleted all but the first three and the last three codons of arsB, resulting in an in-frame deletion (chromosomal coordinates 1944043 to 1945881).
Similarly, for deletion of the pNRC100 arsM (5177) gene, a 1,384-bp fragment product resulting from PCR amplification with the 5175 reverse and 5176 forward primers containing the gene plus flanking DNA (Table 1, pNRC100 coordinates 135676 to 137059) was first cloned into the PvuII site of pSK400 to form pSK430. Plasmid pSK431was generated by circularization of a PCR product using the arsM 5′ and 3′ inverse primers (Table 1), which resulted in a deletion spanning the region between the first and last codons of the arsM gene (pNRC100 coordinates 135868 to 136704) and including 272 bp of 5′ flanking DNA and 355 bp of 3′ flanking DNA.
Halobacterium sp. strain SK400 (Δura3), a uracil auxotroph, is a derivative of wild-type strain NRC-1 (ATCC 700922) and was used as the parent for construction of the arsADRC deletion (3, 10). SK400 is a pseudonym for MPK414 and was a gift from M. P. Krebs. SK400 was transformed with pSK402, and deletion strains were isolated first on uracil-deficient dropout medium (SKURA), containing (per liter) 250 g of NaCl, 20 g of MgSO47 · H2O, 3 g of Na citrate, 2 g of KCl, 10 g of nitrogen base (Sigma-Aldrich Corp., no. Y0626), 1.92 g of Dropout formula (Sigma-Aldrich Corp., no. Y1501), pH 7.0. First-crossover integrants (with plasmid integration) were confirmed by screening for presence of bla and/or ura3 genes by PCR (Table 1). Integrants were plated on rich CM+ media containing 0.25-mg/ml 5-fluoroorotic acid (5-FOA; Toronto Research Chemicals, Inc.; no. F595000) and (per liter) 250 g of NaCl, 20 g of MgSO4 · 7H2O, 3 g of Na citrate, 2 g of KCl, and 10 g of neutralized peptone (Oxoid L34), pH 7.2, plus trace metals (2). Halobacterium cultures were grown at 42°C with illumination, with plates containing 20-g/liter agar, and liquid cultures were shaken in an Innova 4230 rotary shaker (New Brunswick Scientific, Edison, N.J.) at 200 to 220 rpm. Second-crossover recombinants were screened to confirm loss of the ura3 gene and for the presence of either the deleted or wild-type knockout target gene by PCR (Table 1). The resulting strain was named SK402. Construction of arsB and arsM deletion strains was done with the appropriate plasmid (pSK421 and pSK431, respectively) using the above method. These strains were named SK421 (ΔarsB) and SK431 (ΔarsM).
Growth effects of arsenite, arsenate, and antimonite.
A single colony of each strain was inoculated into 5 ml of CM growth medium and incubated with agitation at 42°C for 1 week. On the seventh day, sodium arsenite was added to a 1 μM concentration to the culture and the culture was incubated with shaking for another 6 h. Addition of sodium arsenite was done since the substrate may have been altered by the 7 days of growth and induction might be necessary by that time. Cells were then transferred with a 1:500 dilution to each 6-ml tube of medium containing different concentrations of arsenite and antimonite and incubated for another 14 days. Aliquots of 100 μl were taken from the culture, and the optical density at 600 nm (OD600) of each was monitored for 1 or 2 days. The growth curves were performed with different concentrations of As(V), As(III), and Sb(III).
Northern blot analysis.
A single colony of Halobacterium sp. strain NRC-1 was inoculated into 5 ml of CM growth medium and incubated with agitation at 42°C for 1 week. One hundred microliters of the culture was transferred separately to 100 ml of growth medium and incubated for another 7 days. (The first batch was the control; to the second batch, 10 μM antimony potassium tartrate was added; and to the third batch, 25 μM sodium arsenite was added.) On the seventh day (OD600 of ∼1.0), 1 μM antimony potassium tartrate and 5 μM sodium arsenite were added to corresponding cultures, which were incubated for another 6 h. Total RNA was isolated according to the protocol of Dyall-Smith (4) with modification. Briefly, the cells were harvested by centrifugation at 5,000 rpm for 15 min at 4°C and resuspended in 1.25 ml of lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, 1 mM trisodium citrate, 1.5% sodium dodecyl sulfate [SDS], 35 μl of diethyl pyrocarbonate [DEPC]) followed by incubation at 37°C for 10 min and later on ice for 10 min. After centrifugation at 4°C for 15 min, 2.5 volumes of 100% ethanol was added to the supernatants and the mixture was incubated at −80°C for 30 min. The pellets were then resuspended with DEPC-treated water, and the RNAs were precipitated with 2.5 volumes of 4 M LiCl. The RNA concentration was determined by measuring A260 and separated on a 1.2% agarose-formaldehyde gel, transferred to nylon membranes, and hybridized following the manufacturer's instructions (MSI, Inc., Westborough, Mass.). Equal loading of total RNA (10 μg) was confirmed by ethylene blue staining of rRNAs in the transferred membranes (8). DNA probes for arsA, arsC, arsD, arsR, arsB, arsR2 (ORF5176), and arsM (5177) were amplified by PCR using gene-specific primers (Table 2), cut from agarose gels, and purified with the Qiaquick gel extraction kit (Qiagen, Valencia, Calif.). Each probe was randomly labeled with [α-32P]dCTP using the RadPrime DNA labeling system (Invitrogen, Carlsbad, Calif.) and used for hybridization with the RNA gel blot at 68°C in hybridization buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× Denhardt's solution, and 0.5% SDS. Membranes were washed once with 2× SSC-0.1% SDS at room temperature and twice with 0.2× SSC-0.1% SDS for 20 min at 68°C and exposed in Kodak BioMax MS film from overnight to 3 days at −80°C.
TABLE 2.
Oligonucleotides used for making probes
| Gene and orientation | Sequence (5′ → 3′) |
|---|---|
| arsA forward | ATGACTGCTACACAAACGCC |
| arsA reverse | TCACGACACCGTCACCTCCT |
| arsD forward | ATGACTCAACTCACCCTGTA |
| arsD reverse | TTACGCTTCCTGTGGGTCTG |
| arsR forward | ATGTCATCGACCGAGCGGTT |
| arsR reverse | TCATCTGCGGGATCCGTTCA |
| arsC forward | ATGTCTGCTGATTCGACGAC |
| arsC reverse | TTAGTCGTCGGGATTGAACT |
| arsB forward | ATGTCAGCGGTGTCACCACC |
| arsB reverse | TTAGACGCCCCAGAAGGCCG |
| arsM forward | ATGGTTCAGGACGCAACC |
| arsM reverse | CTATCCGATTCTATTTGAAA |
| arsR2 forward | ATGGTTCAGGACGCAACCGC |
| arsR2 reverse | TTACTCATGGTTGCCTCGCA |
| Universal 16S 338F | CTCCTACGGGAGGCAGCAG |
| Universal 16S 784R | GGACTACCAGGGTATCTAATCC |
| Archaeal 16S 27F | TCCGGTTGATCCTGCCGGAG |
| Archaeal 16S 685R | TTACGGGATTTCACTCCTAC |
RT-PCR analysis.
Total RNA was isolated as described above. One microgram of each RNA sample was digested with 2 U of DNase I (Novagen, Darmstadt, Germany) at 37°C for 30 min. After checking the complete digestion of DNA using an agarose gel, the RNA was precipitated with ethanol and resuspended in DEPC-treated water. Reverse transcription-PCR (RT-PCR) was performed using the forward and reverse arsC primers (Table 2), which yielded an ∼430-bp fragment. The universal 16S rRNA gene primers 338F and 784R (yielding an ∼440-bp fragment) and the archaeal 16S rRNA gene primers 27F and 685R (yielding an ∼650-bp fragment) were used in RT-PCR as positive controls. RT was performed using Superscript II reverse transcriptase (GIBCO BRL Cambridge, Mass.) at 42°C for 1 h with a mixture of primers (arcC reverse, 784R, and 685R) (Table 2). PCR was performed using the AmpliTaq Gold polymerase (Perkin Elmer, Foster City, Calif.) for 35 cycles at an annealing temperature of 52°C. RT-PCR products were analyzed by electrophoresis in a 2% agarose gel and ethidium bromide staining.
RESULTS
Construction of ars deletion strains.
For deletion of the arsADRC gene cluster and the arsB and arsM genes on pNRC100, flanking regions were cloned into pSK400 (pMPK408), a knockout vector containing the Halobacterium sp. strain NRC-1 ura3 gene (Fig. 2A). In each case, the deletions were designed to remove the coding regions of the targeted genes essentially in their entirety. For deletion of the arsADRC gene cluster, Halobacterium sp. strain SK400 (NRC-1 Δura3) was transformed with the knockout plasmid pSK402, using the EDTA-polyethylene glycol method (3). We obtained 1,600 CFU per μg of pSK402 on uracil-dropout plates, while the mock transformation contained no colonies. The transformation efficiency was 5.5 × 105 per μg using control pNG168 DNA (3). PCRs designed to amplify both the bla gene (or ura3 gene) and the Δars derivative from the plasmid were used to assay DNA from 20 selected colonies. Twenty of 20 isolates contained the bla gene, and 9 of 20 contained the correct deleted ars gene (Fig. 2B). Two isolates containing both genes were streaked onto rich medium containing 5-FOA for counterselection against the ura3 gene and isolation of the deleted strain. Several hundred colonies were obtained, of which 10 were picked for further analysis. PCR amplification showed that 20% of the isolates had the smaller ars fragment corresponding to the deletion and had also lost the ura3 gene. The strain with deletion of the arsADRC genes is referred to as SK402 (Fig. 2C).
Similar methods were used for deletion of the chromosomal arsB gene and the pNRC100 arsM gene, with strain SK400 used as the parent for construction of the mutants. In both of these cases, a large fraction of isolates from the first crossover showed integration of the plasmid and about 40% of those also had the deleted ars gene. Selection for the second crossover on 5-FOA plates resulted in approximately 20% deleted strains. These strains were named SK421 (ΔarsB) and SK431 (ΔarsM) and displayed shorter PCR amplification fragments than wild-type NRC-1 (Fig. 2) (data not shown). Deletion of genes was also confirmed by Southern blot analysis using DNA from deletion strains against arsA, arsB, and arsM probes, respectively. In each case, the DNA of the deletion strain showed no hybridization with the corresponding probes but hybridization with the DNA from the wild type (data not shown).
Deletion of arsADRC leads to increased sensitivity to arsenite and antimony but not arsenate.
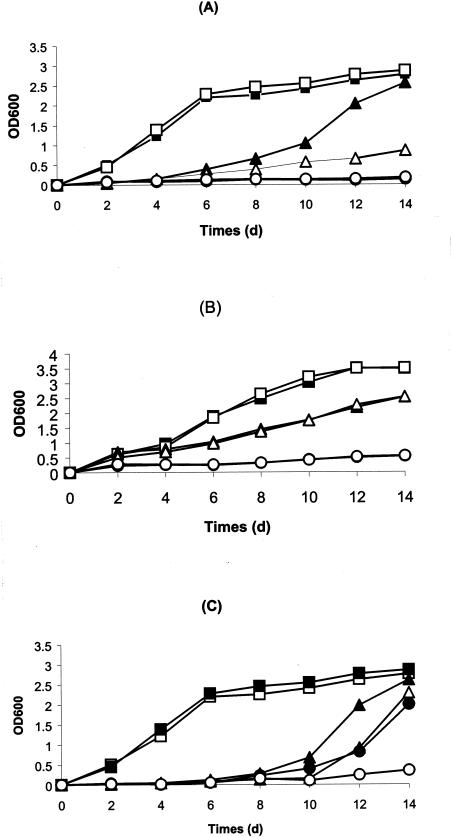
The arsADRC gene cluster on the minichromosome pNRC100 is similar to bacterial genes conferring resistance to arsenite, arsenate, and antimonite. This suggested that these genes might have a similar function in Halobacterium sp. strain NRC-1. In order to evaluate the function of this cluster in metal resistance, we constructed strain SK402 with a deletion of the entire arsADRC gene cluster. SK402 was found to be more sensitive to arsenite and antimonite but not to arsenate (Fig. 3).
FIG. 3.
Arsenite, arsenate, and antimonite resistance of Halobacterium sp. strains NRC-1 (solid symbols) and SK402 (open symbols). Growth with different concentrations of sodium arsenite (A), sodium arsenate (B) and antimony potassium tartrate (C) is plotted. Original cultures of each single colony from NRC-1 and SK402 were induced with 1 μM arsenite and diluted 1:500 into fresh broth with the indicated concentrations of As(III), As(V), or Sb(III). Cell growth was monitored at OD600 for 14 days with incubation at 42°C and shaking at 200 rpm. ▪ and □, no addition; ▴ and ▵, 0.1 mM arsenite (A), 10 mM arsenate (B), or 0.05 mM antinomite (C); • and ○, 0.15 mM arsenite (A), 20 mM arsenate (B), or 0.1 mM antinomite (C).
Deletion of the chromosomal arsB gene does affect resistance to antimonite but not to arsenite and arsenate.
The arsADRC operon contains all of the genes usually present in five-gene operons with the exception of arsB. A chromosomally encoded gene was annotated arsB as it putatively complemented the arsenic resistance system, although it is only distantly related to bacterial arsB genes, showing 38% amino acid identity and 56% amino acid similarity to the putative sodium/sulfate transporter (NP_898258.1) and 9% amino acid identity and 18% amino acid similarity to the arsB of E.coli K-12 (AAC76527). To examine the physiological role, if any, that this putative arsB gene plays in conferring resistance to arsenite and antimony, we constructed an arsB deletion strain, SK421. A slight increase in sensitivity to antimonite was observed, but strain SK421 did not show any additional arsenate or arsenite sensitivity compared to strain NRC-1 or SK402 (data not shown).
Deletion of the pNRC100 arsM gene results in increased sensitivity to arsenite.
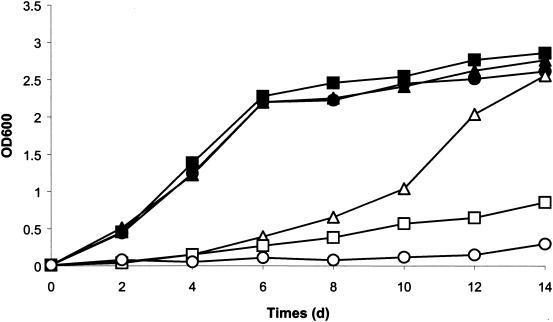
The arsM gene in pNRC100 encodes a putative arsenite(III)-methyltransferase. To determine if arsM is involved in conferring resistance to arsenite, arsenate, and antimony, we generated an arsM deletion creating strain SK431. This strain was more sensitive to arsenite but not to arsenate and antimonite when compared to strains NRC-1 and SK402 (Fig. 4). This difference was most pronounced if cells had been induced with sublethal concentrations of arsenite.
FIG. 4.
Arsenic resistance of Halobacterium strains NRC-1, SK402, and SK431. Original cultures of each single colony from NRC-1, SK402, and SK431 were induced with 1 μM arsenite and diluted 1:500 into fresh broth with 0.1 mM arsenite. Cell growth was monitored at OD600 for 14 days with incubation at 42°C and shaking at 200 rpm. For Halobacterium sp. strain NRC-1, ▴ represents no addition and ▵ represents 0.1 mM As(III). For Halobacterium sp. strain SK402, ▪ represents no addition and □ represents 0.1 mM As(III). For Halobacterium sp. strain SK431, • represents no addition and ○ represents 0.1 mM As(III).
Induction of ars genes by antimonite and arsenite.
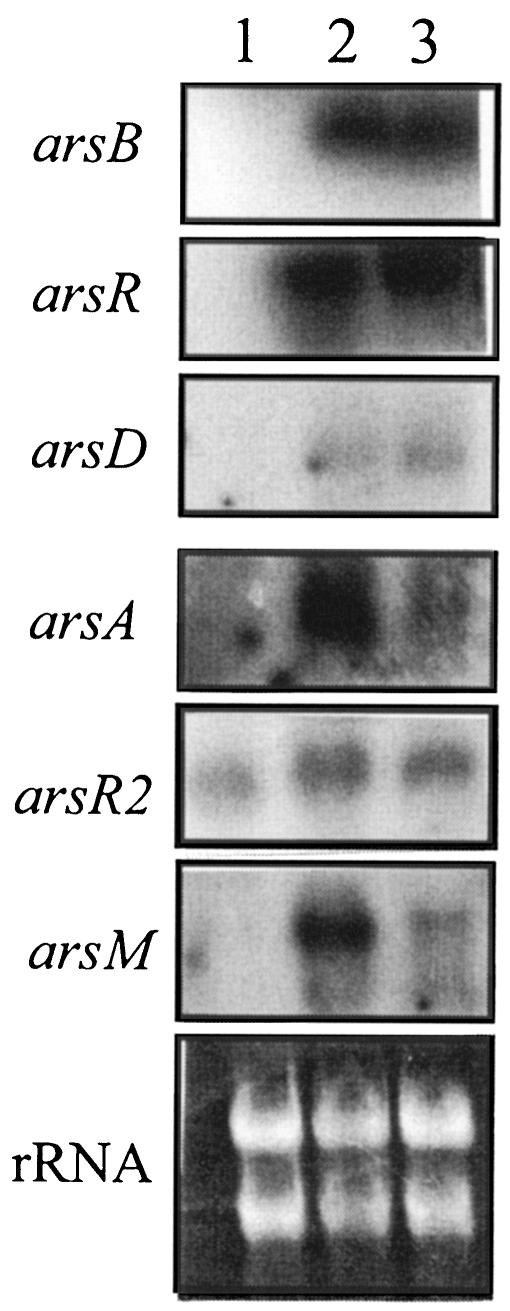
To understand regulation and expression patterns of individual genes in the pNRC100 ars gene cluster, cells were grown under differential conditions and transcripts were analyzed by Northern analysis. Using probes specific for individual ars genes, arsenite-dependent induction of transcription could be detected for arsA, arsD, and arsR (Fig. 5). These genes were also induced by the presence of antimonite. Interestingly, expression of chromosomal arsB appeared to be influenced by the presence of arsenite and antimonite, even though it only conferred slight antinomy resistance. In addition, we detected arsenite- and antimonite-dependent transcription of the arsM gene encoding a putative As(III)-methyltransferase (Fig. 4). Expression of arsR2 upstream of arsM appeared to be constitutive. The molecular size of each transcript corresponds approximately to the size of each gene (Fig. 5). The arsC gene was only weakly induced by arsenite, antimonite, and arsenate. This was not detected in a Northern analysis but only by using RT-PCR (Fig. 6).
FIG. 5.
Northern blot analysis of ars genes in Halobacterium sp. strain NRC-1. Panels contain hybridization results for arsB, arsR, arsD, arsA, arsR2, and arsM are labeled. RNA was isolated from Halobacterium sp. strain NRC-1 grown in either standard CM medium (lane 1), medium supplemented with 25 μM As(III) (lane 2), or medium supplemented with 10 μM Sb(III) (lane 3). The arsC transcript was not detected by Northern blot (data not shown). The approximate molecular sizes of the transcripts were as follows (nucleotides): 1,800 for arsB, 400 for arsR, 300 for arsD, 1,950 for arsA, 400 for arsR2, and 800 for arsM. The panel labeled rRNA shows a stained gel as a control for the amount of RNA loaded.
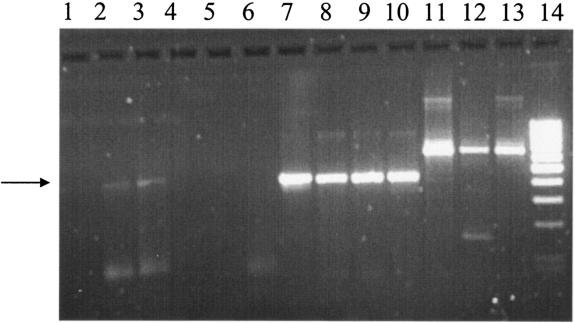
FIG. 6.
RT-PCR analysis of arsC gene expression in Halobacterium sp. strain NRC-1. A 2% agarose gel is shown after electrophoresis and staining with ethidium bromide. Lanes 1 to 7 contain PCR products using arsC primers. Lanes 1, 2, and 3 contain RT-PCR products from RNAs (after DNase I treatment) purified from Halobacterium sp. strain NRC-1 cultured in CM medium, 25 μM As(III), and 10 μM Sb(III), respectively. Arrows indicate the positions of the major RT-PCR products in lanes 2 and 3. Lanes 4, 5, and 6 contain PCR products using the same RNAs as in lanes 1, 2, and 3, respectively, after DNase I treatment but without RT. Lane 7 contains a positive control PCR fragment using genomic DNA from Halobacterium sp. strain NRC-1 as a template. Lanes 8 to 10 contain RT-PCR products using the same RNAs as in lanes 1, 2, and 3, respectively, but with universal 16S rRNA gene primers. Lanes 11 to 13 contain RT-PCR products using the same RNAs as in lanes 1, 2, and 3, respectively, with archaeal 16S rRNA gene primers. Lane 14 contains a 100-bp ladder as a size marker.
DISCUSSION
The halophilic archaeon Halobacterium sp. strain NRC-1 is an excellent model for postgenomic analysis. In the present study, we have conducted genetic analysis of arsenic resistance in Halobacterium using an improved method for gene knockouts. We examined genes located in a pNRC100 gene cluster, as well as an additional gene in the chromosome, which are homologous to previously characterized ars genes in bacteria and eukaryotes (5). Our results show that the pNRC100 arsADRC gene cluster is indeed important for conferring resistance to arsenite and antimonite. Interestingly, we also found that a gene encoding a mammalian arsenite(III)-methyltransferase homolog, named arsM, confers additional resistance to arsenite in Halobacterium sp. strain NRC-1, indicating a second novel mode of resistance in this archaeon.
Although our studies establish for the first time the existence of genetic determinants for heavy metal resistance in Halobacterium sp. strain NRC-1, the complete system for arsenic and antimonite resistance is not completely evident. A deletion of arsADRC should have no influence on resistance to arsenate. The arsC gene encoding a putative arsenate reductase is only weakly expressed, and therefore a deletion of arsADRC should have no or very little influence on resistance to arsenate. This is what we observed. The putative repressors, arsR and arsD, should also have no influence on arsenic resistance levels. The arsR and arsD gene products could control expression of genes outside of this gene cluster. However, deletion of a repressor should not lead to a more sensitive phenotype. Only arsA encoding a putative As(III)/Sb(III)-dependent ATPase might influence resistance to arsenite and antimonite. However, in bacterial systems, ArsA needs a transporter such as ArsB to extrude As(III) and Sb(III). Our results indicated that the closest homolog of bacterial ArsB transporters in Halobacterium sp. strain NRC-1 apparently has no role in arsenite resistance and only slightly increases antimonite resistance. Therefore, in addition to ArsB, this haloarchaeon may contain a novel transporter that is possibly unrelated to ArsB but with a similar function. This might be the reason for not observing an arsenite-sensitive phenotype in the arsB deletion.
One of the most interesting results that we obtained is the presence of an inducible second system conferring arsenite resistance in Halobacterium sp. strain NRC-1. This system is dependent on the arsM gene. As part of the current investigation, we found that this gene is organized as an apparent operon with an upstream arsR homolog (arsR2). The location of the arsR2 and arsM genes is less than 1 kb from the arsADRC cluster (Fig. 1). The ArsM protein sequence is similar to some methyltransferases in bacteria and mammals, with 60% amino acid similarity to the S-adenosylmethionine-dependent methyltransferase in Magnetospirillum magnetotacticum (ZP_00054798). The finding that deletion of arsM can increase sensitivity to arsenite indicates a novel mechanism of arsenic detoxification in this haloarchaeon. One possibility is that the As(III)-methyltransferase methylates intracellular arsenite, thereby creating a concentration gradient to the outside. Since methylated arsenic species are negatively charged or uncharged, As(III)-(CH3)x could move down a concentration gradient to the outside of the cell. This would be a resistance mechanism since it would lower the intracellular arsenic concentration. In another possible scenario, arsenite would be methylated to volatile trimethyl-arsine. This mode of action is reminiscent of mercury resistance where Hg2+ is reduced to elemental mercury that is also volatile. Trimethyl-arsine would leave the cell by diffusion and be dispersed into the atmosphere.
This investigation has demonstrated the power of genetics for postgenomic analysis of Halobacterium sp. strain NRC-1. We have improved our gene knockout and replacement method by utilizing both positive and negative selection of the ura3 gene, eliminating the need for mevinolin (lovastatin) selection previously used. This is advantageous, since mevinolin is relatively expensive and sometimes difficult to obtain, and it precludes the use of double selections necessary for more complex genetic manipulations. Our results also show for the first time that genes on pNRC100 are amenable to knockout and that the multiple copies of this extrachromosomal replicon, compared to the chromosome, are not a technical limitation to gene replacement. Moreover, we also show that multiple genes in a large gene cluster with a size of 4 kb may be deleted together in a single step. With these technical advances, we have established the basic requirement for arsenic resistance in an archaeon. Future studies using ars gene mutants constructed singly and in combination will be necessary to fully characterize the arsenic resistance system of Halobacterium sp. strain NRC-1.
Acknowledgments
This work was supported by NSF grants DEB-0196502 and MCB-0296017 to S.D. and grant EEC9908280 from NSF to C.R.
We thank Barry Rosen for suggestions and many fruitful discussions, Eileen Finnerty-Rae for technical assistance with Southern blot analysis, and Mark P. Krebs for gifts of Halobacterium sp. strain MPK414 and pMPK408.
REFERENCES
- 1.Chen, Y., and B. P. Rosen. 1997. Metalloregulatory properties of the ArsD repressor. J. Biol. Chem. 272:14257-14262. [DOI] [PubMed] [Google Scholar]
- 2.DasSarma, S., and P. Arora. 2002. Halophiles, p. 458-466. In Encyclopedia of life sciences, vol. 8. Nature Publishing Group, London, United Kingdom.
- 3.DasSarma, S., and E. M. Fleischmann. 1995. Halophiles. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 4.Dyall-Smith, M. 2001. The Halohandbook: protocols for halobacterial genetics, ed. 4.5. University of Melbourne, Melbourne, Australia.
- 5.Lin, S., Q. Shi, F. B. Nix, M. Styblo, M. A. Beck, K. M. Herbin-Davis, L. L. Hall, J. B. Simeonsson, and D. J. Thomas. 2002. A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. J. Biol. Chem. 277:10795-10803. [DOI] [PubMed] [Google Scholar]
- 6.Ng, W.-L., S. Kothakota, and S. DasSarma. 1991. Structure of the large gas vesicle plasmid in Halobacterium halobium: inversion isomers, inverted repeats, and insertion sequences. J. Bacteriol. 173:1958-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng, W.-L., and S. DasSarma. 1993. Minimal replication origin of the 200-kilobase Halobacterium plasmid pNRC100. J. Bacteriol. 175:4584-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng, W. V., S. A. Ciufo, T. M. Smith, R. E. Bumgarner, D. Baskin, J. Faust, B. Hall, C. Loretz, J. Seto, J. Slagel, L. Hood, and S. DasSarma. 1998. Snapshot of a large dynamic replicon in a halophilic archaeon: megaplasmid or minichromosome? Genome Res. 8:1131-1141. [DOI] [PubMed] [Google Scholar]
- 9.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peck, R. F., S. DasSarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 11.Peck, R. F., C. Echacarri-Erasun, E. A. Johnson, W. V. Ng, S. P. Kennedy, L. Hood, S. DasSarma, and M. P. Krebs. 2001. brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium. J. Biol. Chem. 276:5739-5744. [DOI] [PubMed] [Google Scholar]
- 12.Rosen, B. P. 1999. Families of arsenic transporters. Trends Microbiol. 7:207-212. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.