Abstract
The FliG protein is a central component of the bacterial flagellar motor. It is one of the first proteins added during assembly of the flagellar basal body, and there are 26 copies per motor. FliG interacts directly with the Mot protein complex of the stator to generate torque, and it is a crucial player in switching the direction of flagellar rotation from clockwise (CW) to counterclockwise and vice versa. A primarily helical linker joins the N-terminal assembly domain of FliG, which is firmly attached to the FliF protein of the MS ring of the basal body, to the motility domain that interacts with MotA/MotB. We report here the results of a mutagenic analysis focused on what has been called the hinge region of the linker. Residue substitutions in this region generate a diversity of phenotypes, including motors that are strongly CW biased, infrequent switchers, rapid switchers, and transiently or permanently paused. Isolation of these mutants was facilitated by a “sensitizing” mutation (E232G) outside of the hinge region that was accidentally introduced during cloning of the chromosomal fliG gene into our vector plasmid. This mutation partially interferes with flagellar assembly and accentuates the defects associated with mutations that by themselves have little phenotypic consequence. The effects of these mutations are analyzed in the context of a conformational-coupling model for motor switching and with respect to the structure of the C-terminal 70% of FliG from Thermotoga maritima.
Escherichia coli swims by spinning its four to eight flagella (1, 38), which are distributed randomly around the surface of the cell (28). When the flagella turn counterclockwise (CCW), the left-handed helical flagellar filaments coalesce into a bundle that pushes the cell in a gently curved path known as a run. When one or more flagella rotate clockwise (CW), the bundle comes apart (29, 49) and the cells undergo sudden, random changes in direction that are called tumbles. Alternation of CCW and CW flagellar rotation produces a series of runs and tumbles that generates a three-dimensional random walk. Suppression of CW rotation by signals from chemoreceptors enables a cell to migrate up an attractant gradient or down a repellent gradient (41).
Most of the ∼40 flagellar proteins are involved in the assembly of the basal body, hook, and filament. Three proteins have been shown to be directly involved in torque generation: MotA, MotB, and FliG. Null mutations in motA and motB produce paralyzed flagella (3, 4). MotA and MotB are located in the cell membrane (36) surrounding the MS ring (2 2), and they form the transmembrane channel that delivers protons to the motor (3, 4, 5, 42, 51). The large C-terminal periplasmic domain of MotB has a putative peptidoglycan-binding domain that is believed to anchor the Mot proteins to the peptidoglycan cell wall (9, 11, 17, 51).
FliG is mounted on the cytoplasmic face of the MS ring (15), which is composed of the single polypeptide FliF (50). FliG also contacts the C ring, which contains FliM and FliN (16, 21, 34). FliG, FliM, and FliN form the switch-motor complex (52), which is required for torque generation, flagellar assembly, and switching the direction of flagellar rotation (19, 39, 53).
FliG from E. coli (FliGEc contains 330 residues and can be separated into two stable, overlapping domains (25). A fliG mutant with a stop codon at position 246 has paralyzed flagella, whereas a ΔfliG strain is not flagellated. Certain charged residues in the carboxyl-terminal region of FliG are essential for motility (26) and interact electrostatically with residues of opposite charge (55) in the cytoplasmic loop between transmembrane helices 2 and 3 of MotA (54). The picture of FliG that has emerged is that the amino-terminal region of FliG is required for flagellar assembly, whereas the carboxyl-terminal domain of FliG is required for torque generation.
The C-terminal 126 residues of E. coli FliG can be expressed as a stable, soluble cytoplasmic fragment (25). The crystal structure of the C-terminal motility domain of FliG from Thermotoga maritima (FliGTm) has been determined (27). A more recent publication describes the structure of a larger C-terminal region that includes the putative hinge between the domains (8).
The FliM protein plays an essential role in switching between CCW and CW rotation (39, 44, 47). FliM interfaces with the chemotactic circuit via the response regulator CheY (7, 33, 48). The default direction of the motor is CCW, and binding of CheY-phosphate (2) or a mutationally activated form of CheY (37) to FliM increases the probability of CW rotation. FliM also associates with FliG (31, 32, 33, 48). Conformational changes in FliM upon binding CheY-phosphate are presumably transmitted to FliG, but the mechanism by which these proteins communicate is uncertain.
The suppression of missense mutations in motB (17) and motA (6, 18) by mutations in fliG has been interpreted to mean that a misalignment of MotA and FliG caused by mot mutations can be compensated for by mutations in fliG. Three of these suppressors map to a part of fliG that encodes what is predicted to be a linker between the motility and assembly domains (17). Elements within this region, which encompasses residues 183 to 196, are predicted to function as a flexible hinge whose position determines how the motility domain of FliG presents itself to MotA. To assess how residue changes in the linker affect flagellar performance, we performed random PCR mutagenesis of codons 183 to 196 of a plasmid-borne fliG gene. Based on the phenotypes of our mutants, we conclude that a simple two-state (CW/CCW) model is inadequate to describe the full behavior of the flagellar motor.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strain DFB225 (26), which contains an in-frame deletion that removes the entire fliG coding region, was used as the host for fliG plasmids. The fliG gene was amplified from the chromosome of E. coli by using PCR. Unique HindIII and BamHI restriction sites were engineered into the 5′ and 3′ ends of fliG, respectively. The amplified gene was subsequently cloned into the low-copy-number plasmid pHSG576 (43) to form pSVW1, in which fliG is under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Plac promoter. The pSVW1 plasmid complements the fliG deletion in strain DFB225 to allow swarming in tryptone semisolid agar.
Localized random PCR mutagenesis.
The DNA sequence that encodes the putative hinge of fliG was mutagenized using the two-primer method described by Perlak (35) and Deng and Nickoloff (12). Plasmid pSVW1 was used as a substrate for mutagenesis, and a doped oligonucleotide was utilized to introduce missense mutations into the putative hinge. The level of doping was such that each of the three alternate bases was present as a 1% contaminant. In a subsequent effort to increase the likelihood of obtaining transversions to expand the range of amino acid substitutions we found, the mutagenesis protocol was altered by doping positions occupied by pyrimidines with purines only and positions occupied by purines with pyrimidines only, at a level of 3%. A counterselection primer was employed during mutagenesis to insert a mismatch within a unique and nonessential SpeI restriction site in pSVW1.
Isolation of mutants.
DFB225 cells were transformed with the mutagenized fliG-encoding plasmids, and potential mutants were identified by screening for mutant motility phenotypes in tryptone swarm agar (10 g of Difco Bacto-Tryptone, 5 g of NaCl, and 3.25 g of Difco Bacto-Agar, all per liter, 1 mM IPTG, and 30 μg of chloramphenicol /ml). Mutants were retested in swarm agar to confirm their phenotypes and to measure swarm diameters. Plasmids for sequencing were isolated by using the Qiagen plasmid purification protocol. The entire fliG gene was sequenced for each candidate mutant by using the ABI fluorescent dideoxy-sequencing protocol.
Immunoblotting.
DFB225 cells carrying pSVW1, pSVW2, or one of their mutant derivatives were grown with vigorous swirling at 32°C to mid-logarithmic phase (an optical density at 590 nm [OD590 nm] of about 0.6) in tryptone broth containing 1 mM IPTG. Cells were then pelleted and washed twice with distilled water (dH2O) and finally resuspended in 50 μl of sodium dodecyl sulfate (SDS) buffer (2% [wt/vol] SDS, 5% [vol/vol] 2-mercaptoethanol, 8.5% [vol/vol] glycerol, 60 mM Tris [pH 6.8], 0.0004% [wt/vol] bromophenol blue). Crude extracts were prepared by three alternating cycles of 5 min at −80°C and 5 min of boiling. Extracts from equal numbers of cells (based on the OD590 nm values) were separated by SDS-10% polyacrylamide gel electrophoresis and transferred to nitrocellulose. Immunoblots were probed with anti-FliG antiserum (25), visualized with alkaline phosphatase-conjugated, goat anti-rabbit antibody (Bio-Rad), and developed with SigmaFast (Sigma).
Determination of motility parameters.
To analyze the behavior of free-swimming cells, cultures of DFB225 carrying pSVW1, pSVW2, or one of their mutant derivatives were grown as for immunoblotting. The cultures were then kept on ice. Before their behavior was observed, the cells were diluted 20-fold into chemotaxis buffer (10 mM potassium phosphate, 0.1 mM EDTA, 1 mM methionine) at room temperature. After the temperature was allowed to equilibrate for at least 10 min, the cells were observed by phase-contrast microscopy at a magnification of ×1,000. Their behavior was evaluated by direct visual examination by an experienced observer ignorant of the identity of the strains.
Cells for tethering assays were grown under the same conditions as the cultures used to examine swimming. When the cultures reached an OD590 nm of 0.6, they were pelleted and washed twice with tethering buffer (0.01 M potassium phosphate, 0.1 M NaCl, 0.01 mM EDTA, 0.02 mM l-methionine, 0.02 mM sodium lactate [pH 7.0]) and then resuspended in 20 ml of tethering buffer. Flagellar filaments were sheared during six 10-s runs in a 50-ml stainless steel cup of a Waring blender operated at low speed. The cells were then pelleted by centrifugation, washed twice with tethering buffer, and resuspended in 2.5 ml of tethering buffer. Twenty microliters of each resuspension was mixed with 20 μl of antifilament antibody at a 1:500 dilution, and the entire volume was placed within a thin ring of Apiezon L grease on a 12 mm-diameter round glass coverslip. Each coverslip was incubated in a humidity chamber at 30°C for a minimum of 20 min. Following incubation, the coverslip was affixed to a flow chamber in which fresh tethering buffer was replaced at regular intervals. The cells were observed under phase contrast at a magnification of ×1,000 and videotaped. Numerous fields were taped for each strain, and the first 20 freely rotating cells encountered during video playback were analyzed. Cells were not analyzed if their radius of gyration was too small, if their motion was erratic, or if they met resistance by running into neighboring cells or other obstacles. Rapidly spinning cells were observed at one-quarter speed. Angular velocity, rotational bias, and switching frequency were measured over intervals of 10, 30, and 60 s, respectively.
Electron microscopy.
Overnight cultures were grown with vigorous swirling at 32°C in tryptone broth containing 1 mM IPTG and 30 μg of chloramphenicol/ml. The next morning, the cultures were diluted 1:100 into the same medium and grown to an OD590 nm of 0.6 to 0.8. Cells were washed on nitrocellulose filters with 150 ml of dH2O and resuspended in dH2O to an OD590 nm of approximately 0.4. Cells were stained for 10 s with 1% ammonium molybdate.
RESULTS
Isolation and initial characterization of mutations affecting the FliG hinge.
The E. coli chromosomal fliG gene was amplified from the chromosome by PCR using Taq polymerase and cloned into a low-copy-number plasmid (pHSG576) under control of the lac promoter. The resulting plasmid, pSVW1 (pfliG*), complemented the motility and chemotactic defects of the ΔfliG strain DFB225 and was used as a template for doped-oligonucleotide mutagenesis of fliG codons 183 to 196. Strain DFB225 was transformed with mutagenized, pooled pSVW1 DNA, and mutants were isolated by screening for altered swarm phenotypes on tryptone semisolid agar. After confirmation that the mutations were plasmid borne, the mutant fliG genes were sequenced to identify mutations.
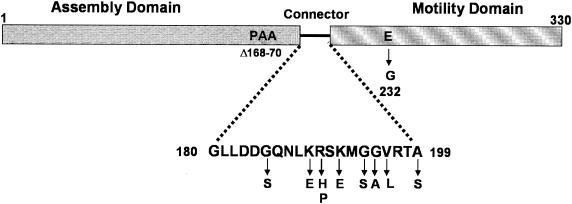
Mutants were independently isolated from approximately 3,000 screened transformant colonies. The plasmids contained six unique missense mutations that cause the following amino acid substitutions: K189E (five times), G195A (twice), and G185S, R190P, and V196L (once each). One plasmid had a G→T transversion that introduces an A199S substitution. This mutation is outside the mutagenized region but within the 7-base flanking region at the 3′ end of the primer. We also used site-directed mutagenesis to introduce three mutations earlier identified as suppressors of motB missense mutations (17) into pSVW1 and pSVW2. These mutations generate the substitutions R190H, K192E, and G194S. The positions of these substitutions within the FliG amino acid sequence are shown in Fig. 1.
FIG. 1.
Locations of residue substitutions in the connector between the assembly and motility domains of FliG. The residues within the expanded portion of the schematic of the protein primary sequence are within or immediately flank the region of FliG that corresponds to the long α helix that connects the assembly and motility domains (see the text). Residue substitutions in this region are indicated below the wild-type sequence. Residues Lys-192 to Gly-95 comprise the putative hinge. The positions of the Pro-168 through Ala-170 residues that are deleted in an extremely CW-biased mutant of S. enterica (46) and the position of the E232G substitution introduced during PCR cloning of fliG (see text) are also indicated.
All mutant plasmid-borne fliG genes, as well as the parental pSVW1 plasmid, contained a second mutation that causes an E232G substitution (Fig. 1). It was presumably introduced during PCR cloning, since the mutation is not in the chromosomal fliG gene of strain RP437. We therefore used site-directed mutagenesis to introduce Glu-232 into all plasmid-borne mutant fliG genes. The true fliG+ plasmid was called pSVW2. DFB225/pSVW2 colonies swarmed appreciably better than DFB225/pSVW1 colonies, and swarming was also improved by this change in most mutants containing pSVW1-derived plasmids (compare swarm data in Tables 1 and 2). The E232G substitution appears to sensitize FliG to the effects of substitutions in the linker region. For example, the V196L and A199S mutants would not have been noticed had the mutations originally occurred in pSVW2.
TABLE 1.
Motility parameters of FliG single mutants
| FliG variant | Relative swarm diama | Swimming behaviorb | Flagella/cellc | Rotational velocityd | Rotational biase | Switching frequencyf | Location of mutated residueg |
|---|---|---|---|---|---|---|---|
| FliG+ | 100 | >90%, R/T | 3.2 ± 0.6 | 8.4 ± 0.6 | 70 ± 4.9 | 42 ± 4.4 | None |
| G185S | 47 | >90%, T | 1.1 ± 0.3 | 7.1 ± 0.7 | 7 ± 1.3 | 22 ± 4.1 | α-Helix E |
| K189E | 80 | >90%, R/T | 1.1 ± 0.4 | 8.0 ± 0.4 | 69 ± 5.4 | 40 ± 3.3 | α-Helix E |
| R190H | 82 | >90%, r/T | 1.1 ± 0.2 | 9.4 ± 0.7 | 51 ± 7.9 | 15 ± 3.5 | α-Helix E |
| R190P | 40 | >90%, R/t | 1.5 ± 0.3 | 2.7 ± 0.4 | 68 ± 9.5 | 10 ± 4.4 | α-Helix E |
| K192E | 109 | ∼50%, r/T | 0.6 ± 0.2 | 8.2 ± 0.6 | 54 ± 4.6 | 38 ± 2.9 | Hingeh |
| G194S | 113 | ∼50%, r/T | 1.1 ± 0.3 | 7.5 ± 0.8 | 84 ± 3.7 | 17 ± 2.7 | Hinge |
| G195A | 0 | ∼50%, T | 5.1 ± 0.6 | 9.6 ± 0.7 | 1 ± 0.5 | 3 ± 1.7 | Hinge |
| V196L | 91 | >90%, R/T | 3.1 ± 0.5 | 8.1 ± 0.4 | 85 ± 2.3 | 33 ± 2.7 | α-Helix F |
| A199S | 100 | >90%, r/t | 3.3 ± 0.5 | 5.7 ± 0.3 | 75 ± 3.3 | 77 ± 5.9 | α-Helix F |
The swarm diameters were measured as described in Materials and Methods, and the mean diameter for each mutant strain was calculated. These mean diameters are expressed as a percentage of the mean diameter determined for swarms made by the parental DFB225/pSVW2 (FliG+) strain.
The swimming behavior of cells in a mid-exponential-phase culture grown at 30°C in tryptone broth was viewed at room temperature under phase contrast at a magnification of ×400. The entries reflect the subjective evaluation of an experienced observer. Percentages of motile cells are given along with an assessment of the trajectories of the cells: R, run only; T, tumble only; R/T, wild-type run-tumble behavior; R/t, long runs, brief tumbles; r/T, short runs, extended tumbles; r/t, both runs and tumbles, but runs are short and tumbles are at least as brief as in wild-type cells.
The number of flagella clearly attached to cells in electron micrographs of a negatively stained preparation was counted for a random selection of 20 cells per mutant strain. The mean number ± the standard error about the mean for the 20 cells is given.
Rotation velocities were determined as described in Materials and Methods for 20 rotating cells of each strain. The mean value (in revolutions per second) ± the standard error about the mean for the 20 cells is shown.
The fraction of time spent in CCW rotation was recorded as described in Materials and Methods for the same 20 cells for which rotational velocities were determined. The mean value ± the standard error about the mean for the 20 cells is shown.
The number of reversals per minute was scored as described in Materials and Methods for the same 20 cells for which rotational velocities were determined. The mean value ± the standard error about the mean for the 20 cells is shown.
The positions of the residues are given based on the alignment of the FliGTm and FliGEc amino acid sequences and the positions of the corresponding residues in FliGTm within the published FliGTm crystal structure (8).
The hinge refers to the four residues in FliGEc that correspond to the four residues between helix E and helix F of FliGTm that are unresolved in the crystal structure.
TABLE 2.
Motility parameters of FliG double mutants
| FliG variant | Relative swarm diama | Swimming behaviorb | Flagella/cellc | Rotational velocityd | Rotational biase | Switching frequencyf | Location of mutated residueg |
|---|---|---|---|---|---|---|---|
| E232G | 52 | ∼50%, slow R/T | 0.3 ± 0.5 | 4.9 ± 0.4 | 92 ± 2.2 | 22 ± 6.2 | α-Helix H |
| G185S/E232G | 43 | >90%, r/T | 1.0 ± 0.3 | 2.8 ± 0.5 | 60 ± 8.0 | 74 ± 18 | See Table 1 |
| K189E/E232G | 13 | ∼15%, R/T | 0.3 ± 0.1 | 3.8 ± 0.6 | 97 ± 2.5 | 2 ± 1.1 | See Table 1 |
| R190H/E232G | 44 | ∼15%, slow r/T | 0.7 ± 0.1 | 1.5 ± 0.2 | 64 ± 7.0 | 23 ± 3.3 | See Table 1 |
| R190P/E232G | 13 | ∼25%, slow R | 0.6 ± 0.2 | 4.1 ± 0.5 | 45 ± 8.3 | 34 ± 7.0 | See Table 1 |
| K192E/E232G | 76 | ∼5%, slow r/T | 0 | 2.2 ± 0.2 | 80 ± 4.6 | 34 ± 8.5 | See Table 1 |
| G194S/E232G | 28 | ∼2%, slow r/T | 0 | 3.7 ± 0.7 | 72 ± 4.7 | 38 ± 6.2 | See Table 1 |
| G195A/E232G | 0 | Nonmotile | 0 | NRCi | NRC | NRC | See Table 1 |
| V196L/E232G | 5 | Jiggle onlyh | 0.6 ± 0.3 | 3.5 ± 0.5 | 46 ± 7.8 | 40 ± 5.9 | See Table 1 |
| A199S/E232G | 0 | <1%, slow r/T | 0.5 ± 0.1 | NRC | NRC | NRC | See Table 1 |
Mean diameters determined as for Table 1 and expressed as a percentage of the mean diameter of swarms made by the DFB225/pSVW2 strain.
Swimming behavior recorded as in Table 1.
The number of flagella per cell counted as for Table 1.
Rotation velocities determined as for Table 1.
Rotation biases recorded as in Table 1.
Rotation biases scored as in Table 1.
The residue corresponding to Gly-232 of the FliGEc sequence is in α-helix H in the crystal structure of FliGTm.
Cells of the V196L/E232G double mutant did not exhibit significant translational movement, nor did they actively tumble. However, they moved more chaotically than would be expected from Brownian motion alone, based on the behavior of ΔfliG strain DFB225 (data not shown).
NRC, no rotating cells in all microscopic fields observed.
Hinge mutations do not decrease total cellular FliG.
Immunoblots of crude extracts were used to determine the total amount of FliG present. All strains containing wild-type or mutant pSVW1 or pSVW2 contained the same amount of FliG (data not shown). Therefore, the mutant phenotypes observed are due to a decreased function rather than a decreased amount of the mutant proteins. We have no direct evidence that all of the mutant FliG proteins incorporate equally well into the motor. As discussed below, some of the mutations do affect flagellar assembly. However, because the portion of FliG that interacts with the MS ring is unaltered in all of the mutant proteins, we predict that defective interactions with FliM and/or FliN, rather than defective interactions with FliF, are responsible for the mutant phenotypes we observed.
Swarm phenotypes of single and double mutants.
DFB225/pSVW1 swarms were 52% the diameter of DFB225/pSVW2 swarms (Tables 1 and 2). The G195A substitution in either plasmid eliminated swarming. The remaining pSVW2 mutants formed swarms with ≥45% the diameter of those made by DFB225/pSVW2. The A199S/E232G double mutant did not swarm, and the K189E/E232G, R190P/E232G, and V196L/E232G mutant swarms had ≤13% the diameter of DFB225/pSVW2 swarms.
Behavior of free-swimming cells.
Subjective analyses of the swimming behavior of DFB225 cells carrying pSVW2 or one of its mutant derivatives or of DFB225 cells carrying pSVW1 or one of its mutant derivatives are given in Tables 1 and 2, respectively. The swimming behavior of each mutant was generally consistent with its swarming properties and the rotational behavior of tethered cells of the mutant.
Effects of fliG hinge mutations on flagellar assembly.
Mutations in FliG often affect flagellar assembly (25, 52). It therefore seemed possible that reduced motility might be a consequence of this effect. We therefore examined negatively stained preparations of whole cells to determine the average number of flagellar filaments on cells expressing various plasmid-encoded mutant FliG proteins. The mean number of filaments associated with cells of strain DFB225/pSVW2 was 3.2 (Table 1), slightly more than half the mean number (5.9) of flagella per cell of the wild-type strain, RP437. Except for the G195A, V196L, and A199S mutants, all of the single substitutions decreased the number of flagella to ≤1.5 per cell. The E232G substitution also had a severe effect, since DFB225/pSVW1 cells had an average of only 0.3 flagellum per cell (Table 2). Correspondingly low numbers of flagella, or none at all, were seen with the double mutants.
Analysis of tethered cells.
Tethered DFB225 cells containing pSVW1, pSVW2, or one of their derivatives were analyzed to assess torque generation, rotational bias, and switching frequency of the mutant motors.
(i) Rotational velocity.
The G185S, K189E, R190H, K192E, G194S, G195A, and V196L mutant cells rotated at speeds ±20% those of DFB225/pSVW2 cells (Table 1) or cells of the wild-type parental strain, RP437 (data not shown). The mean speeds of A199S and R190P mutant cells were 70 and 30% of the wild-type speed, respectively. The E232G substitution slowed rotation significantly (Table 2), and very few K192E/ E232G, G194S/E232G, or V196L/E232G cells rotated. No cells producing the G195A/E232G and A199S/E232G proteins spun. Surprisingly, R190P/E232G double mutant cells rotated faster than those of the R190P single mutant.
(ii) Directional bias of rotation.
DFB225/pSVW2 cells had a mean 70% CCW bias (Table 1), somewhat higher than the 55% CCW bias of tethered cells of strain RP437 (data not shown). The K189E, R190P, and A199S mutants exhibited similar biases. The G194S and V196L substitutions increased the CCW bias to 85%, whereas the G185S and G195A substitutions introduced a very strong CW bias. The E232G substitution increased the CCW bias in otherwise wild-type FliG or in FliG with the K189E and K192E substitutions (Table 2), and it caused a dramatic increase in CCW rotation in combination with G185S (from 7 to 60% CCW bias). In contrast, E232G decreased CCW rotation modestly in combination with the G194S substitution and more strongly in combination with the R190P and V196L substitutions. Cells expressing the G195A/E232G and A199S/E232G FliG proteins did not rotate.
(iii) Switching frequency.
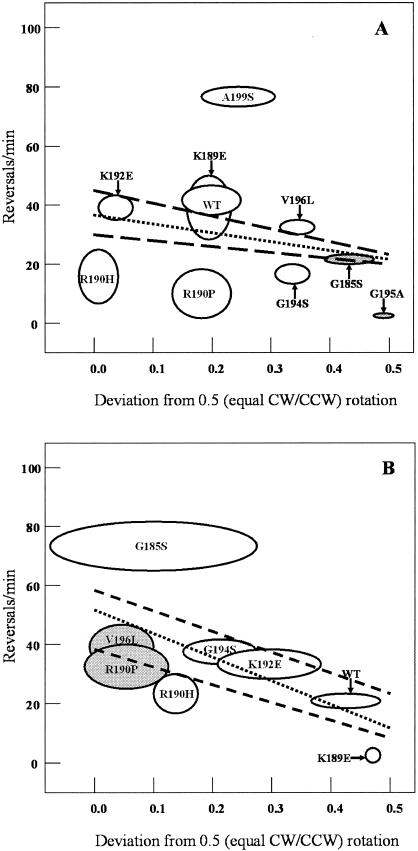
The number of reversals per minute of single-mutant cells is shown in Table 1. DFB225/pSVW2 cells reversed a mean of 42 times per minute. The K189E, K192E, and V196L mutants had similar frequencies (33 to 40/min). The A199S mutant exhibited an increased mean reversal frequency of 77/min. The R190H and R190P mutants reversed relatively infrequently (15 and 10/min, respectively), although they had nearly wild-type CCW biases of 51 and 68%, respectively. The G194S mutant, although modestly CCW biased (84%), also reversed less often than expected (17/min) (Fig. 2). Cells of the strongly CW-biased G185S mutant made fewer reversals (22/min), and the G195A mutant was essentially locked in CW rotation.
FIG. 2.
Relationship between reversal frequency and directional bias for mutant flagellar motors. The absolute value of the mean deviation from 50% CCW rotation for a population of 20 cells of each mutant strain is shown on the abscissa. The mean number of reversals per minute for the same population of 20 cells of each mutant strain is plotted on the ordinate. (A) Strains containing plasmids derived from pSVW2 (FliG+). The ovals show the standard errors about the means for the values on the x and y axes. Deviations above 50% CCW rotation are indicated by open ovals; deviations below 50% are indicated by gray ovals. The FliG substitution in each mutant is indicated. WT is DFB225/pSVW2. The dotted line is a linear regression of the data points, and the dashed lines are the linear regression lines for the mean x and y values plus one standard error and minus one standard error. The data are from Table 1. (B) As in panel A, except that the strains contain plasmids derived from pSVW1 and the data are from Table 2.
The E232G substitution had little effect on reversal frequencies (Table 2) except in the K189E/E232G strain, which had a 97% CCW bias. R190H/E232G and R190P/E232G cells had reversal rates (22 and 34/min, respectively) that were higher than those of the corresponding single mutants. G185S/E232G cells exhibited an unusual behavior. Their mean reversal frequency (75/min) was high, and their mean CCW bias over the population of tethered cells examined was 60%, which is close to that of wild-type cells. However, some individual G185S/E232G cells were extremely CCW biased, others were very CW biased, and yet others spent nearly equal amounts of time in CW and CCW rotation. Some cells in each of these categories often stopped briefly without changing their direction of rotation. Although we scored these events as reversals, they may better be thought of as pauses in the sense used by Lapidus et al. (24) and Eisenbach et al. (14).
DISCUSSION
Based on genetic analyses of the E. coli and Salmonella enterica serovar Typhimurium fliG genes (17, 19, 25, 52) and a computer-generated prediction of secondary structure, Garza et al. (17) proposed that residues 183 to 196 of E. coli FliG constitute a flexible hinge between the N-terminal assembly and C-terminal motility domains of the protein. To study the role of this region in motor behavior, we mutagenized codons 183 through 196 of a plasmid-borne copy of fliG obtained by PCR cloning from the chromosome of wild-type E. coli strain RP437. The pSVW1 plasmid constructed in this way complemented strain DFB225 (ΔfliG) for swarming in tryptone semisolid agar. It was used as the template for random PCR mutagenesis. We subsequently found that pSVW1 contains a mutation that causes the E232G substitution in FliG. This adventitious change accentuated the effects of residue substitutions in the mutagenized region and facilitated detection of other mutations. The codon specifying the wild-type Glu-232 residue was introduced into pSVW1 to create plasmid pSVW2.
Identity of mutations affecting the FliG hinge.
The six random missense mutations identified cause the residue substitutions G185S, K189E, R190P, G195A, V196L, and A199S. Mutations causing the R190H, K192E, and G194S substitutions, which were previously found as suppressors of a motB missense mutation (17), were also introduced into pSVW1 and pSVW2.
Effects of mutations on torque generation.
Of the single substitutions, only R190P decreased the mean rotation speed by more than 50%—from 8.4 ± 0.6 revolutions per second (rps) to 2.7 ± 0.4 rps (Table 1). The A199S substitution caused a more modest reduction to 5.7 ± 0.3 rps. Since the rotational velocity of a tethered cell is directly proportional to the torque generated by the motor of the tethering flagellum (30), we conclude that most of these mutations by themselves have little influence on the force generated by the motor.
DFB225/pSVW1 cells rotated at 4.9 ± 0.4 rps (58% of the wild-type speed), and the mean rotational velocities of the double mutants were yet lower (Table 2). The most severe effect in mutants that produced rotating cells was seen with R190H/E232G (1.5 ± 0.2 rps). However, R190P/E232G cells spun faster than R190P cells (4.1 ± 0.5 versus 2.7 ± 0.4 rps). The reason that the E232G substitution decreases torque in most cases is unknown, although defective interaction with FliM is apt to be at least partially responsible (see below).
Effects of mutations on directional bias.
Some of the single mutations altered the CW/CCW ratio of rotation (Table 1). Tethered DFB225/pSVW2 cells had a 70% CCW bias. The flagella of two mutants spun primarily CW: G185S (7% CCW bias) and G195A (≤1% CCW bias). Two other mutants, G194S and V196L, had increased CCW biases (84 and 85%). The remaining single substitutions in the linker region had no significant effect on directional bias.
The mean CCW bias of DFB225/pSVW1 cells was 92% (Table 2). The E232G substitution had a mixed effect on directional bias in combination with second mutations. The CCW bias increased in four instances (G185S, K189E, R190H, and K192E) and decreased in three others (R190P, G194S, and V196L). No rotating cells were seen with A199S/E232G. Finally, G195A/E232G cells do not produce any flagella. Indeed, the E232G substitution uniformly and substantially decreased the number of flagellar filaments observed by electron microscopy (Table 2).
Effects of mutations on reversal frequency.
To visualize the effects of the mutations on switching rates in a way that corrects for rotational bias, we plotted reversal frequency as a function of absolute directional bias (deviation from 50% CCW rotation) for each mutant (Fig. 2) and calculated a linear regression.
The data fit the linear regression reasonably well for most strains except at the extremes of CW and CCW bias, where reversal frequencies rapidly collapse to zero. The advantage of plotting the data this way is that the outliers stand out. For example, in Fig. 2A the R190H, R190P, and G194S mutants clearly reverse less often than would be expected based on their directional bias, whereas the A199S mutant reverses more often than expected. Of the double mutants with wild-type directional biases, G185S/E232G reversed more often than expected and fell significantly outside the predicted distribution (Fig. 2B). As expected, the 97% CCW-biased K189E/E232G mutant seldom reversed.
Effects on motor assembly, swimming, and swarming.
The G185S, K189E, R190H, R190P, K192E, and G194S substitutions all decreased the mean number of flagella from 3.2 ± 0.6 per cell for DFB225/pSVW2 to ≤1.5 per cell. This decreased flagellation is not clearly reflected in the swarm diameters or swimming behavior of these mutants (Table 1). For example, the K189E, R190H, K192E, and G194S mutants form essentially wild-type swarms, and the first two produce >90% motile cells. We conclude that swarm formation is quite forgiving of moderate defects in flagellar assembly, perhaps because the best swimmers out of a genetically uniform population determine the rate at which the swarm rings advance.
The double mutants all had an average of ≤1.0 flagellum per cell (Table 2), and with three of them (K192E/E232G, G194S/E232G, and G195A/E232G) no flagella were found on 20 randomly chosen cells. This result is consistent with the very small number of motile cells in cultures of these three mutants (Table 2). In contrast, V196L/E232G and A199S/E232G cells were almost all nonmotile but had as many or more flagella than DFB225/pSVW1 cells. Thus, most of the flagellar motors of V196L/E232G and A199S/E232G cells must be nonfunctional.
A conformational-coupling model for directional switching.
A theoretical treatment of directional control in the flagellar motor has been published recently (13). Based on the arguments advanced in that publication and the performance of the flagella of our hinge mutants, we have concluded that, at least under some conditions, motor switching behavior cannot be entirely accounted for by a simple two-state model. We have simplified our discussion by considering only conformational changes in the 26 FliG subunits (15, 20, 40) in the rotor. These events can be viewed as the output of the switching event. These changes in FliG conformation are strongly influenced by the fraction of the 34 FliM subunits per motor (45) that are bound to the tumble (CW rotation) regulator CheY-phosphate (10, 37).
We propose that the motor can exist in multiple states. At one extreme, all of the subunits are oriented to allow CW rotation. At the other extreme, all of the subunits are oriented to allow CCW rotation. Between these extremes are 25 hypothetical states in which 1 to 25 subunits can be in the CW orientation, with the remainder being in the CCW orientation. (This minimal estimate assumes that only the number of CW- and CCW-oriented subunits, rather than the distribution of CW- and CCW-oriented subunits around the ring, is relevant to overall motor behavior.)
Reversals normally occur within a few milliseconds without a significant pause (1, 2, 23). This rapid switching is explained by Duke et al. (13) as the consequence of a high degree of allosteric coupling among the individual switch elements, a phenomenon they describe as conformational spread in a protein ring. The basic idea is that the conformation of one subunit strongly influences the conformation of one of its neighbors in an asymmetric fashion such that changes propagate in one direction or the other around the ring. Thus, even though each FliG subunit can occupy only two possible conformations, there are 27 possible states of the motor: all subunits in the CW orientation, all subunits in the CCW orientation, or 25 different intermediate ratios of CW and CCW subunits. The last condition would correspond to a transition state that is normally very brief but potentially becomes significant in mutant motors in which allosteric coupling among FliM or FliG subunits is impaired (see below).
Evaluation of the behavior of mutant motors with respect to the conformational-coupling model.
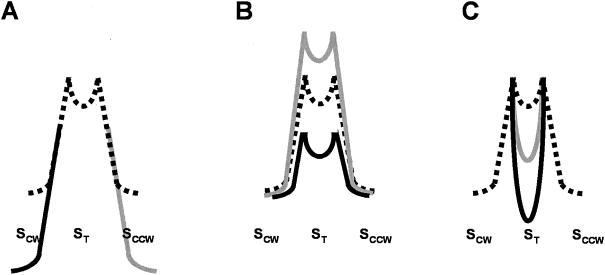
The diverse phenotypes associated with different residue replacements in the hinge region of FliG can be evaluated in the context of the model shown in Fig. 3. Mutations that increase the stability (i.e., decrease the free energy) of either the CW or the CCW conformation of FliG are expected to generate motors that are CW or CCW biased relative to the wild-type motor. Note that this result could arise from making one extreme state more stable than in the wild type, one extreme state less stable, or a combination of these two effects.
FIG. 3.
Schematic diagrams showing the relative free-energy levels of the CW (SCW), CCW (SCCW), and transition (ST) states of motor rotation. Free energy increases upward; therefore, more stable states are lower. (A) The energy diagram for a wild-type motor with a mean 50% CCW bias is depicted with a black dotted line. The SCW and SCCW states are at the same energy level, and the ST state is shown as a minor dip in the activation-energy barrier between the SCW and SCCW states. The more-stable SCW and SCCW states for strongly CW-biased and CCW-biased motors, respectively, are indicated by black and dark gray solid lines. (B) The energy diagram for a wild-type motor is as in panel A. Motors with higher and lower reversal frequencies are shown with black and dark gray solid lines, respectively. (C) The energy diagram for a wild-type motor is as in panel A. The energy diagram for a frequently pausing motor is shown in dark gray. Note that the ST state occupies a lower free-energy level than in the wild-type motor. The motor becomes trapped for longer periods in the ST state. For a paralyzed motor, the ST state is at a lower free-energy level than either the SCW or SCCW state (solid black line) and the motor is permanently stuck.
Mutations that increase the total activation energy barrier between the two extreme states should decrease the reversal frequency, as is seen with the R190H, R190P, and G194S mutants (Fig. 2A). Mutations that decrease the activation energy should increase the reversal frequency, as is seen with the A199S mutant (Fig. 2A). One might expect that one could differentiate between a mutant motor in which one directional state becomes more stable than in the wild type as opposed to a mutant motor in which one directional state becomes less stable based on reversal frequencies. If the former is true, reversals of cells whose flagella are spinning in the dominant direction should be infrequent and reversals of cells whose flagella are spinning in the other direction should occur at the normal frequency. In the latter case, reversals of cells whose flagella are spinning in the dominant direction should occur at the normal frequency and reversals of cells whose flagella are spinning in the other direction should occur more often than normal. We did not distinguish between CW-to-CCW and CCW-to-CW reversals in our tally of switches per minute. However, since tethered cells of the most strongly biased mutants reversed only rarely (Tables 1 and 2), we favor the interpretation that a directionally biased mutant motor is more stable than a wild-type motor in the conformation that corresponds to the predominant direction of rotation.
Two of the double-mutant motors (G185S/E232G and A199S/E232G) exhibited behaviors that we attribute to a disruption of normal conformational coupling among the FliG subunits. This result is in keeping with the generally deleterious effect on motor function that might be expected from random mutations.
A population of G185S/E232G cells showed no pronounced directional bias, but individual cells spun predominantly CW, predominantly CCW, or about equally in both directions (Table 2). Furthermore, many cells engage in frequent pauses, which we scored as reversals. Such longer pauses can be observed, even with wild-type cells, under some conditions (14, 24). We suggest that in the G185S/E232G motor these pauses are accentuated because the transition state, in which individual FliG subunits within a motor are in different conformations, sometimes persists long enough to be detected by eye.
The motors of the A199S/E232G double mutants may suffer a drastic loss of allosteric coupling among the FliG protomers (13). This effect might be particularly noticeable in tethered cells because generation of the high torque required to rotate a cell body may require greater synchrony among rotor subunits. The slower rotational speeds of tethered cells (Tables 1 and 2) of some of the mutants whose flagella are not paralyzed might reflect a more modest decrease in conformational coupling among FliG protomers, although there are clearly other equally plausible explanations. If the former explanation is correct, one might expect larger variations in rotational speed in these mutants. Since we did not specifically score variations in rotational velocity in our analysis of tethered cells, relatively subtle changes could easily have been missed.
The structural basis of mutant-motor phenotypes.
The crystal structure of the C-terminal 70% (residues 104 to 335) of FliG from T. maritima (FliGTm) has recently been determined (8, 27). FliGTm shows significant sequence identity to E. coli FliG (FliGEc), with residues 104 to 335 of FliGTm corresponding to residues 103 to 331 of FliGEc (see Fig. 1). Residues 237 through 331 of FliGEc (residues 239 to 335 of FliGTm) form a stable domain that has been shown to interact directly with the large cytoplasmic loop of MotA (54, 55). The N-terminal 100+ residues of FliG comprise a domain that is attached to the MS ring, but whose structure is unknown. The intervening region contains two well-defined subdomains that interact with FliM (32).
In the FliGTm crystal structure these two domains are connected by an extended α-helix (residues 172 to 193 in FliGTm, corresponding to residues 171 to 191 in FliGEc). This helix is flanked on its N-terminal end by a turn (residues 166 to 171 in FliGTm and 165 to 171 in FliGEc) that contains a Pro residue in both proteins. Residues 194 to 197 in FliGTm (residues 192 to 195 in FliGEc) comprise a turn. The conserved tandem Gly residues (Gly-196 and Gly-197 in FliGTm, Gly-194 and Gly-195 in FliGEc) may provide a flexion point for interdomain movement during switching events. The R190H and R190P substitutions, which are N terminal to the tandem Gly residues, and the A199S substitution, which is C terminal to them, decrease and increase the frequency of motor reversal, respectively. The significance of this correlation remains to be demonstrated.
Based on the FliGTm structure, Glu-232 of FliGEc is directly adjacent to a hydrophobic patch that is proposed to constitute a site of interaction with FliM (8). Also, a mutation in E. coli fliG that generates the E232V substitution decreases interaction of FliG with FliM (32). We therefore suspect that the defects in motor assembly and the changes in motor function associated with the E232G substitution are, at least in part, attributable to impaired interaction with FliM and might be reversed by overproduction of FliM and/or FliG (25). The possibility that the FliM interaction is weaker when FliG contains the E232G substitution could account for a longer duration of the transition state in some double-mutant motors.
A number of CW-biased fliG mutations had previously been shown to change residues in the interdomain region of FliG from S. enterica serovar Typhimurium (19) and E. coli (25). The most potent of these is an in-frame deletion of the codons for Pro-169, Ala-170, and Ala-171 in S. enterica serovar Typhimurium. This mutation confers such an extreme bias that the motor spins CW even in a strain lacking CheY (46). Binding of CheY-phosphate to FliM is likely to induce a conformational change in FliG similar to the one introduced by this deletion. One possibility is that FliM pushes into or recedes from the hydrophobic patch near residue Glu-232.
The structure of FliGEc has not been determined nor, to our knowledge, modeled from the crystallographic data for FliGTm. However, the ability to couple specific perturbations of motor function with single-residue changes within the hinge region of FliG, summarized in Tables 1 and 2, should help elucidate the structural basis for control of the direction of flagellar rotation.
Acknowledgments
We thank David Blair for supplying the ΔfliG strain and FliG antibody and Christos Savva for help with electron microscopy. David Blair, Howard Berg, and Greg Reinhart provided useful suggestions about various aspects of our interpretation of the results. We are also grateful to the anonymous reviewers of earlier submissions of the manuscript for their candid comments and valuable suggestions, which we have tried to incorporate as they intended. The late-night heroic effort of Arjan Bormans to get the TIFF files of the figures up to snuff for electronic submission is especially appreciated.
This work was supported by grant DAAG55-97-1-0380 from the Army Research Office.
REFERENCES
- 1.Berg, H. C. 1976. Does the flagellar rotary motor step?, p. 47-56. In R. Goldman, T. D. Pollard, and J. Rosenbaum (ed.), Cell motility, book A. Motility, muscle, and non-muscle cells. Cold Spring Harbor Laboratory, Cold Spring Harbor, N. Y.
- 2.Berg, H. C., and R. A. Anderson. 1973. Bacteria swim by rotating their flagellar filaments. Nature 245:380-382. [DOI] [PubMed] [Google Scholar]
- 3.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439-449. [DOI] [PubMed] [Google Scholar]
- 4.Blair, D. F., and H. C. Berg. 1991. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J. Mol. Biol. 221:1433-1442. [DOI] [PubMed] [Google Scholar]
- 5.Blair, D. F., D. Y. Kim, and H. C. Berg. 1991. Mutant MotB proteins in Escherichia coli. J. Bacteriol. 173:4049-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, T. F., S. Poulson, J. B. Gully, J. C. Empey, S. Van Way, A. Putnam, and D. Blair. 1999. Function of proline residues of MotA in torque generation by the flagellar motor of Escherichia coli. J. Bacteriol. 181:3542-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bren, A., and M. Eisenbach. 1998. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J. Mol. Biol. 278:507-514. [DOI] [PubMed] [Google Scholar]
- 8.Brown, P. N., C. P. Hill, and D. F. Blair. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun, S. Y., and J. S. Parkinson. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276-278. [DOI] [PubMed] [Google Scholar]
- 10.Cluzel, P., M. Surette, and S. Leibler. 2000. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science 287:1652-1655. [DOI] [PubMed] [Google Scholar]
- 11.DeMot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 12.Deng, W. P., and J. A. Nickoloff. 1992. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 200:81-88. [DOI] [PubMed] [Google Scholar]
- 13.Duke, T. A., N. L. Novere, and D. Bray. 2001. Conformational spread in a ring of proteins: a stochastic approach to allostery. J. Mol. Biol. 308:541-553. [DOI] [PubMed] [Google Scholar]
- 14.Eisenbach, M., A. Wolf, M. Welch, S. R. Caplan, I. R. Lapidu, R. M. Macnab, I. H. Alon, and O. Asher. 1990. Pausing, switching and speed fluctuation of the bacterial flagellar motor and their relation to motility and chemotaxis. J. Mol. Biol. 211:551-563. [DOI] [PubMed] [Google Scholar]
- 15.Francis, N. R., V. M. Irikura, S. Yamaguchi, D. J. DeRosier, and R. M. Macnab. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. USA 89:6304-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis, N. R., G. E Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 17.Garza, A. G., R. Biran, J. A. Wohlschlegel, and M. D. Manson. 1996. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J. Mol. Biol. 258:270-285. [DOI] [PubMed] [Google Scholar]
- 18.Garza, A. G., P. A. Bronstein, P. A. Valdez, L. W. Harris-Haller, and M. D. Manson. 1996. Extragenic suppression of motA missense mutations of Escherichia coli. J. Bacteriol. 178:6116-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irikura, V. M., M. Kihara, S. Yamaguchi, H. Sockett, and R. M. Macnab. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, C. J., R. M. Macnab, H. Okino, and S. Aizawa. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212:377-387. [DOI] [PubMed] [Google Scholar]
- 21.Khan, I. H., T. S. Reese, and S. Khan. 1992. The cytoplasmic component of the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 89:5956-5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan, S., M. Dapice, and T. S. Reese. 1988. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202:575-584. [DOI] [PubMed] [Google Scholar]
- 23.Kudo, S., Y. Magariyama, and S. Aizawa. 1990. Abrupt changes in flagellar rotation observed by laser dark-field microscopy. Nature 346:677-680. [DOI] [PubMed] [Google Scholar]
- 24.Lapidus, I. R., M. Welch, and M. Eisenbach. 1988. Pausing of flagellar rotation is a component of bacterial motility and chemotaxis. J. Bacteriol. 170:3627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd, S. A., and D. F. Blair. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 26:733-744. [DOI] [PubMed] [Google Scholar]
- 26.Lloyd, S. A., H. Tang, X. Wang, S. Billings, and D. F. Blair. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd, S. A., F. G. Whitby, D. F. Blair, and C. P. Hill. 1999. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400:472-475. [DOI] [PubMed] [Google Scholar]
- 28.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhart, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D. C.
- 29.Macnab, R. M., and N. K. Ornston. 1977. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J. Mol. Biol. 112:1-30. [DOI] [PubMed] [Google Scholar]
- 30.Manson, M. D., P. M. Tedesco, and H. C. Berg. 1980. Energetics of flagellar rotation in bacteria. J. Mol. Biol. 138:541-561. [DOI] [PubMed] [Google Scholar]
- 31.Marykwas, D. L., S. A. Schmidt, and H. C. Berg. 1996. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J. Mol. Biol. 256:564-576. [DOI] [PubMed] [Google Scholar]
- 32.Marykwas, D. L., and H. C. Berg. 1996. A mutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J. Bacteriol. 178:1289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathews, M. A., H. L. Tang, and D. F. Blair. 1998. Domain analysis of the FliM protein of Escherichia coli. J. Bacteriol. 180:5580-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oosawa, K., T. Ueno, and S. Aizawa. 1994. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J. Bacteriol. 176:3683-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perlak, F. J. 1990. Single step large scale site-directed in vitro mutagenesis using multiple oligonucleotides. Nucleic Acids Res. 18:7457-7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridgway, H. F., M. Silverman, and M. I. Simon. 1977. Localization of proteins controlling motility and chemotaxis in Escherichia coli. J. Bacteriol. 132:657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scharf, B. E., K. A. Fahrner, L. Turner, and H. C. Berg. 1998. Control of direction of flagellar rotation in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 95:201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silverman, M., and M. Simon. 1974. Flagellar rotation and the mechanism of bacterial motility. Nature 249:73-74. [DOI] [PubMed] [Google Scholar]
- 39.Sockett, H., S. Yamaguchi, M. Kihara, V. M. Irikura, and R. M. Macnab. 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol. 174:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sosinsky, G. E., N. R. Francis, D. J. DeRosier, J. S. Wall, M. N. Simon, and J. Hainfeld. 1992. Mass determination and estimation of subunit stoichiometry of the bacterial hook-basal body flagellar complex of Salmonella typhimurium by scanning transmission electron microscopy. Proc. Natl. Acad. Sci. USA 89:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhart, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 42.Stolz, B., and H. C. Berg. 1991. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 173:7033-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 44.Tang, H., and D. F. Blair. 1995. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J. Bacteriol. 177:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, D. R., D. G. Morgan, and D. J. DeRosier. 1999. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. USA 96:10134-10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Togashi, F., S. Yamaguchi, M. Kihara, S. I. Aizawa, and R. M. Macnab. 1997. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 179:2994-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toker, A. S., M. Kihara, and R. M. Macnab. 1996. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J. Bacteriol. 178:7069-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toker, A. S., and R. M. Macnab. 1997. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J. Mol. Biol. 273:623-634. [DOI] [PubMed] [Google Scholar]
- 49.Turner, L., W. S. Ryu, and H. C. Berg. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182:2793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ueno, T., K. Oosawa, and S. Aizawa. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein. J. Mol. Biol. 227:672-677. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, M. L., and R. M. Macnab. 1990. Co-overproduction and localization of the Escherichia coli motility proteins MotA and MotB. J. Bacteriol. 172:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaguchi, S., H. Fujita, A Ishihara, S. Aizawa, and R. M. Macnab. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamaguchi, S., S. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. Macnab. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, J., and D. F. Blair. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273:428-439. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, J., S. A. Lloyd, and D. F. Blair. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 95:6436-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]