Abstract
Induction of biosynthesis of the photosystem in anoxygenic photosynthetic bacteria occurs when the oxygen concentration drops. Control of this induction takes place primarily at the transcriptional level, with photosynthesis genes expressed preferentially under anaerobic conditions. Here, we report analysis of the transcriptional control of two photosynthesis promoters, pucBA and crtI, by the PpsR factor in Rubrivivax gelatinosus. This was accomplished by analyzing the photosystem production in the wild type and in the PPSRK (ppsR::Km) mutant grown under anaerobic and semiaerobic conditions and by assessing the β-galactosidase activity of lacZ transcriptionally fused to promoters possessing the putative PpsR-binding consensus sequences. It was found that under semiaerobic conditions, inactivation of the ppsR gene resulted in overproduction of carotenoid and bacteriochlorophyll pigments, while the production of LH2 was drastically reduced. The β-galactosidase activity showed that, in contrast to what has been found previously for Rhodobacter species, PpsR acts in R. gelatinosus as an aerobic repressor of the crtI gene while it acts as an activator for the expression of pucBA. Inspection of the putative PpsR-binding consensus sequences revealed significant differences that may explain the different levels of expression of the two genes studied.
Anoxygenic photosynthetic bacteria are remarkable for their metabolic versatility. Indeed, they can grow under different regimes and develop the required machinery for growth under aerobic respiration, anaerobic respiration, and photosynthesis (anaerobiosis and light). This faculty requires tight control to ensure production of the desired complexes and rapid adaptation to changes in environmental conditions. Under anaerobiosis and light, these bacteria use their photosystem to convert light to chemical energy (6). The photosystem is composed of three types of pigment-protein complexes whose function is to capture light energy and convert it into electron and proton flows. The two light-harvesting (LH) antennae (LH1 and LH2) capture light and transfer energy to the third complex, the reaction center (RC), in which charge separation occurs. The cytochrome bc1 complex is then required for oxidation of the quinone pool and generation of the proton gradient (11).
Rubrivivax gelatinosus is a facultative phototrophic nonsulfur bacterium belonging to the β subclass of purple bacteria. As is the case for well-studied Rhodobacter species belonging to the α subclass, it can grow under aerobic conditions in the dark, as well as under photosynthetic conditions. The genes coding for the structural subunits of LH1, RC, LH2, and cytochrome bc1 complexes have been cloned and sequenced (17-19). These subunits are encoded by the puf, puc, and pet operons, respectively. The nucleotide sequence of the photosynthesis gene cluster has been determined for two strains of R. gelatinosus, strain IL-144 (13) and strain 1 (accession numbers AY234384 and AH012710). Most of the genes encoding carotenoids and bacteriochlorophylls are present on the same photosynthesis gene cluster, but their localization and organization are quite different from those described for Rhodobacter (13, 21). As in most species of anoxygenic photosynthetic bacteria, the transition of R. gelatinosus from aerobiosis to an anaerobiosis regime results in a significant induction of the biosynthesis of photosystem components.
The expression of photosynthesis genes has mainly been studied in Rhodobacter sphaeroides and Rhodobacter capsulatus. The control of gene expression in response to oxygen tension occurs principally at the transcriptional level, with photosynthesis genes expressed preferentially under anaerobic conditions. This regulation requires the recruitment of several regulatory proteins, including the two-component Prr/Reg regulatory system, PpsR/CrtJ, AerR, AppA, the fnrL gene product, IHF, and TspO (for a review, see references 5 and 32).
It was shown that PpsR from R. sphaeroides (CrtJ in R. capsulatus) is a major negative factor that represses the expression of many photosystem genes (puc, bch, and crt) under aerobiosis. This protein contains a helix-turn-helix domain found in various DNA binding proteins, and deletion of this domain results in a nonfunctional PpsR protein (22). PpsR blocks transcription by binding to the two TGT-N12-ACA consensus sequences positioned either upstream of or overlapping the promoters of the puc, bch, and crt genes (7, 8, 25). The length of the intergenic sequence between the two consensus sequences seems to be important for CrtJ/PpsR function. In fact, repression by CrtJ involves cooperative interactions between two CrtJ molecules bound to adjacent consensus sequences. Mutations in the spacing between the two consensus sequences abolish repression by CrtJ (26).
In this paper, we report the analysis of the transcriptional control of two photosynthesis gene promoters, pucBA (LH2 subunits) and crtI (carotenoid biosynthesis), by the PpsR transcription factor in R. gelatinosus. Our results show that PpsR exerts not only negative but also positive control over these photosynthesis genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
Escherichia coli strains were grown at 37°C in Luria-Bertani medium (27). R. gelatinosus strain 1 (30) and R. sphaeroides 2.4.1 were grown at 30°C in malate medium (1) either photosynthetically (light, 3,000 lx; anaerobiosis) or semiaerobically (cultures filled to 50% of the flask volume agitated at 100 rpm in the dark). Antibiotics were used at the following concentrations for E. coli and R. gelatinosus: kanamycin, 50 μg/ml; ampicillin, 25 μg/ml; chloramphenicol, 3 μg/ml; streptomycin, 25 μg/ml; spectinomycin, 25 μg/ml; and tetracycline, 1 μg/ml. Plasmids were maintained in E. coli XL1-Blue or JM109. The bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM-109 | e14−(McrA−) recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 reA1 Δ(lac-proAB) [F′traD36 proAB lacIqZΔM15] | Stratagene |
| XL1-Blue | supE44 hsdR17 recA1 endA1 gyrA46 thi-1 relA1 lac−F′ [proAB+lacIqlacZΔM15 Tn10 (Tcr)] | Stratagene |
| R. sphaeroides 2.4.1. | Wild type | |
| R. gelatinosus strain 1 | Wild type | 30 |
| PPSRK | ppsR disruption strain (ppsR::Km) | This work |
| Plasmids | ||
| Bluescript KS(+) | Cloning vector (Apr) | Stratagene |
| pGEM-T | PCR product cloning vector | Promega |
| pUC4K | Plasmid with Km cartridge | Pharmacia |
| pSO76 | KS(+) + 3-kb SacI fragment containing crtI, bchG, and the 3′ end of ppsR | Unpublished |
| pGE593 | Plasmid bearing promoterless (BamHI-BsaAI) lacZ gene | Unpublished |
| pBBR1MCS-1 | Expression vector (mob+ Cmr) | 14 |
| pBBR1MCS-3 | Expression vector (mob+ Tcr) | 14 |
| pSO3001 | pBBR1MCS-1 + promoterless (BamHI-BsaAI) lacZ gene | This work |
| pA100 | KS(+) + 2.1-kb SacI/HindIII fragment containing pucBA | This work |
| pA410 | pGEM-T + 1.7-kb fragment containing ppsR gene | This work |
| pA411 | pBBR1MCS-3 + ApaI/SacI (from the pGEM-T polylinker) fragment from pA410 containing ppsR gene | This work |
| pA200 | (pGEM-T + ppsR::Km) Km cartridge cloned in the MscI site within ppsR from pA410 | This work |
| pSO3024 | pucB::lacZ (pSO3001 + SacII-BsaAI URS of pucBA) | This work |
| pSO3025 | crtI::lacZ (pSO3001 + XbaI-BamHI URS of crtI) | This work |
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant; URS, upstream regulatory sequence.
Gene transfer and strain selection.
Transformation of R. gelatinosus cells was carried out by electroporation as previously described (19). Transformants were selected on malate plates supplemented with the appropriate antibiotic. Two different antibiotic-resistance markers were used to distinguish a double-crossover event from a single-crossover event, the first (ampicillin) located on the vector and the second located on the cartridge inserted into the gene to be inactivated.
Molecular biology techniques.
Unless otherwise indicated, the standard methods of Sambrook et al. were used (27). Plasmid DNAs were purified using the Bio-Rad Quantum Prep plasmid kit. DNA was treated with restriction enzymes and other nucleic acid-modifying enzymes (Klenow fragment, T4 DNA polymerase, and T4 DNA ligase) according to the manufacturer's specifications. DNA fragments were analyzed on agarose gels, and different restriction fragments were purified using the Geneclean kit (BIO 101). PCRs were carried out using Taq DNA polymerase from Appligene according to the manufacturer's instructions, except that dimethyl sulfoxide was added to a 5% final concentration. Twenty cycles of 30 s at 92°C, 40 s at 60°C, and 60 s at 72°C were performed in a Hybaid thermal cycler. The primers used to clone the ppsR gene were 5′-GCGACGCCGATATCCGGAT-3′ and 5′-CCGTTGCTGCTGCAGAGC-3′.
Membrane protein preparation and spectrophotometric measurements.
The membranes were prepared by cell disruption with a French press in 0.1 M Na phosphate buffer, pH 7.4, containing 1 mM phenylmethylsulfonyl fluoride, followed by differential ultracentrifugation. The membranes were then resuspended in the same buffer. Spectral analysis was carried out on a CARY 500 spectrophotometer. The protein concentration was determined using the Pierce BCA Protein Assay Reagent method with bovine serum albumin as a reference standard, as prescribed by the manufacturer.
Pigment analyses.
Semiaerobically or photosynthetically grown wild-type or PPSRK mutant cells were harvested in early exponential growth phase, resuspended in ice-cold acetone-methanol (7:2 [vol/vol]), and centrifuged to eliminate cell debris. Absorption spectra were recorded using a CARY 500 spectrophotometer. For high-performance liquid chromatography (HPLC), the pooled pigment extracts were vacuum dried and then resolubilized in a small volume of acetone-methanol and concentrated by evaporation. Twenty microliters of pigment preparation was injected onto a 5-μm-particle-diameter Ultrapack C18 column and run with acetonitrile-methanol-dichloromethane (75:15:15 [vol/vol/vol]) at a 1-ml/min solvent flow rate. Detection was performed at 436 nm, and the collected fractions were further analyzed on a CARY 500 spectrophotometer.
Construction of lacZ reporter fusion plasmids.
To construct the transcriptional lacZ fusion plasmid, the promoterless (BamHI-BsaAI) lacZ gene from te pGE593 plasmid was subcloned into pBBR1MCS-1 (BamHI-EcoRV) to generate the plasmid pSO3001. For the different fusions, the upstream regulatory sequences of the pucBA and crtI genes were generated by digestion and cloned into the promoterless lacZ vector pSO3001. To construct a transcriptional pucB::lacZ fusion plasmid, a 0.68-kb SacII-BsaAI DNA fragment containing the putative PpsR-binding site of the pucBA operon was cloned into the SacII-BamHI-digested (blunt-ended) pSO3001, resulting in the plasmid pSO3024. To construct the crtI::lacZ fusion, a 1.2-kb XbaI-BamHI DNA fragment containing the entire crtI promoter region with the putative PpsR-binding site was cloned from pSO76 into plasmid pSO3001 digested with the same enzymes, resulting in plasmid pSO3025. We confirmed the sequences and orientations of the cloned products by restriction analysis before proceeding with transformation into R. gelatinosus.
β-Galactosidase activity assays.
β-Galactosidase activity was determined on permeabilized cells according to the method of Miller (16). One milliliter of exponential-phase culture was washed with 1 ml of ice-cold 100 mM Na phosphate buffer (pH 7.4). The pellets were resuspended in 1 ml of the same buffer. Cells were permeabilized with a drop of 0.1% sodium dodecyl sulfate (SDS) and chloroform at 37°C for 5 min; 0.9 ml of the samples was then mixed with 0.1 ml of substrate for the β-galactosidase assay (o-nitrophenyl-β-d-galactopyranoside [0.8 mg/ml]) and incubated at 37°C. When a yellow color started to appear, the time was noted, and the reaction was stopped by adding 200 μl of 1 M Na2CO3. Cell debris was removed by centrifugation, and the absorbance of the supernatant at 420 nm was recorded. β-Galactosidase activities were expressed in Miller units, where 1 Miller unit corresponds to 1 μmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per min per unit of optical density of culture at 680 nm. All assays were performed at least three times.
RNA isolation and Northern blot analysis.
RNA was isolated from wild-type R. gelatinosus and the PPSRK mutants in mid-logarithmic-phase culture by the procedure described by Heck et al. (10). Glass beads (0.11-mm diameter; Braun) were added to the lysis buffer to improve cell breakage. After extraction, RNAs were treated with RNase-free DNase (Life Technologies), extracted with acid pH phenol-chloroform, precipitated with ethanol, and resuspended in RNase-free water. RNA (10 μg per lane) was denatured for 10 min at 70°C in morpholinepropanesulfonic acid-formaldehyde (37%)-formamide (50%) buffer (RNA V-buffer 3V). RNA was run on a 1.6% (wt/vol) agarose-formaldehyde gel alongside 3 μg of a 0.24- to 9.5-kb RNA ladder (Bio-Rad Laboratories) per lane at 20 mV for 24 h. Next, the gel was placed for 30 min in 10× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and blotted in 20× SSC with Hybond-N+ membrane (Amersham). After being blotted, the membrane was exposed to a UV Stratalinker (Autocrosslink; 120,000 J/cm2). The blotted membrane was prehybridized in a buffer (5× SSC, 1× Denhardt's solution, 0.05 M phosphate buffer, 0.1% SDS, and 50% formamide) containing denatured sheared salmon sperm DNA for 2 h at 45°C before addition of the denatured labeled probe. The blots were hybridized with the probes for 18 h at 45°C. The 0.4-kb pucBA probe was isolated from pA100 by digestion with HincII and MscI restriction enzymes and labeled with [α-32P]dCTP using the Amersham Pharmacia Oligolabelling kit. After hybridization, the membrane was washed twice for 5 min each time in 2× SSC-0.1% SDS at 45°C and then three times for 10 min each time in 1× SSC-0.1% SDS at 65°C and exposed in a phosphorimager.
Half-life measurement of pucBA mRNA.
Cells of a PPSRK mutant were grown under semiaerobic conditions until logarithmic growth phase, to an optical density of 0.5 to 0.6 at 680 nm. Then, rifampin (200 μg/ml) was added to the cultures and cells were harvested at several time points. The pucBA mRNA half-lives were determined by measuring the radiolabeled bands on the Northern blots using the ImageQuant program. The 16S rRNA of R. gelatinosus was used as an internal-standard probe for the calibration of total RNA.
Immunoblot analyses.
Twenty micrograms of each membrane preparation was resuspended in loading buffer (60 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 0.1% bromphenol blue, and 3% β-mercaptoethanol) and boiled for 3 min. The samples were loaded onto Tris-Tricine-SDS polyacrylamide gels (14% acrylamide). The proteins were transferred onto nitrocellulose membranes by wet electrotransfer in buffer (50 mM Tris, 380 mM glycine, 0.1% SDS, and 20% methanol). The LH2 α polypeptide was detected with an antiserum raised in rabbits (Agro-Bio, Villeny, France) against isolated R. gelatinosus LH2 (at 1:5,000 dilution) and with goat anti-rabbit immunoglobulin conjugated to peroxidase (Bio-Rad) as the secondary antibody (at 1:3,000 dilution). The first antibody is not specific to the LH2 α subunit, since it also revealed nonspecific bands. The LH2 α subunit was detected on the protein blot using the peroxidase color detection system (Bio-Rad).
Nucleotide sequence accession number.
The nucleotide sequence of the ppsR region was deposited in the GenBank Data Library under accession number AY234385.
RESULTS
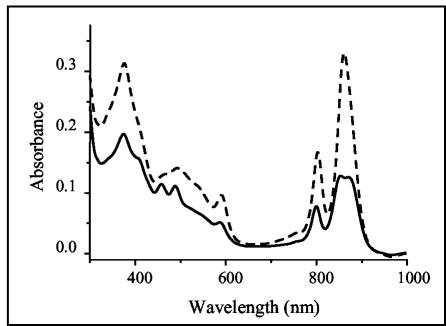
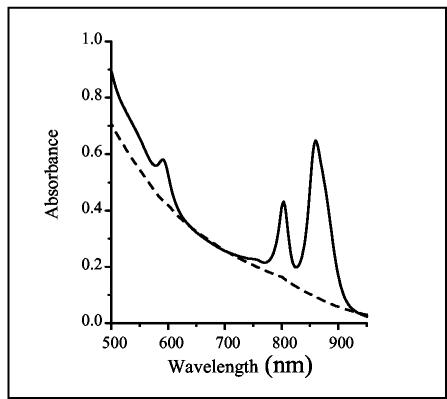
The infrared absorption spectra of R. gelatinosus cells grown under semiaerobic or photosynthetic conditions have shown that photosynthesis genes are oxygen regulated (19), yet semiaerobically grown R. gelatinosus colonies are red rather than the pink color of semiaerobically grown R. sphaeroides. The absorption spectra show that a significant number of the photosynthetic complexes, and particularly the LH2 complexes, were generated under semiaerobiosis in R. gelatinosus, while in R. sphaeroides, a smaller number of these complexes was produced (Fig. 1). In the spectrum of R. gelatinosus, two peaks are observed at 804 and 860 nm, corresponding mainly to LH2 absorption. In R. sphaeroides, three peaks at 804, 850, and 875 nm are observed, but with lower amplitude than in R. gelatinosus. In fact, the amount of LH2 (absorbing at 804 and 850 nm) is small enough for the LH1 absorption peak at 875 nm to be noticed (Fig. 1). This implies that the expression of photosynthesis genes in R. gelatinosus in response to oxygen tension is significantly different from what has been described for R. sphaeroides. Oxygen regulation of photosynthesis is a complex process involving several regulatory proteins (RegA, PpsR, and FnrL). Sequences similar to the PpsR-binding site have been found upstream of photosynthesis genes in R. gelatinosus, prompting us to examine PpsR regulation of these genes.
FIG. 1.
Absorption spectra of R. gelatinosus (dashed line) and R. sphaeroides 2.4.1 (solid line) crude extracts from cells grown semiaerobically. Cells of R. gelatinosus and R. sphaeroides were grown to optical densities at 680 nm of 0.75 and 0.85, respectively. Membrane samples containing equal amounts of protein were subjected to the spectral analysis.
The ppsR gene and analysis of its deduced amino acid sequence.
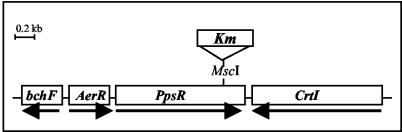
Cloning of the ppsR gene was achieved by PCR using primers based on the published sequence (13). A plasmid (pA410) containing the ppsR gene was obtained and sequenced. The ppsR gene is 1,455 bp long and encodes a 484-amino-acid soluble polypeptide with a predicted molecular mass of ∼52 kDa. The gene is flanked by aerR and crtI genes; it has the same transcriptional orientation as aerR and has a transcriptional orientation opposite to that of the crtI gene (Fig. 2).
FIG. 2.
Physical map of the ppsR region in R. gelatinosus. The putative transcripts are indicated by arrows. Km, kanamycin.
BlastP (3) and clustal analyses of PpsR showed that the polypeptide from R. gelatinosus shows similarity to PpsR from R. sphaeroides (34% identity and 50% similarity) and to CrtJ (33% identity and 50% similarity) from R. capsulatus. Further sequence analyses suggest that R. gelatinosus PpsR possesses a helix-turn-helix DNA binding motif (residues 429 to 469) in its C-terminal domain, as is the case for PpsR from R. sphaeroides and CrtJ from R. capsulatus. The PpsR architecture was determined using the SMART database (28), revealing that R. gelatinosus PpsR contains three well-conserved PAS domains (33) (residues 17 to 83, 159 to 223, and 281 to 350), while Rhodobacter PpsR exhibits only two PAS domains.
PPSRK mutant construction and characterization.
The ppsR gene was disrupted with a kanamycin cassette introduced at the MscI site of ppsR to generate plasmid pA200. In this construction, the kanamycin cartridge was inserted before the region coding for the helix-turn-helix DNA binding domain contained in the last 135 amino acids. This plasmid was used to transform R. gelatinosus. Kanamycin-resistant and ampicillin-sensitive colonies were isolated, and Southern blot analyses were performed and confirmed inactivation by a double-crossover event (data not shown). Compared to the wild type, the isolated PPSRK mutant was dark red and showed a high level of pigmentation when grown semiaerobically. However, when grown photosynthetically, no difference in pigmentation could be observed between the wild type and the PPSRK mutant, suggesting that ppsR is involved in regulation by oxygen in R. gelatinosus. The PPSRK mutant grows with the same generation time as the wild type under photosynthetic and semiaerobic conditions. As reported for R. sphaeroides (9), the PPSRK mutant of R. gelatinosus was genetically unstable. A variety of secondary mutants with altered pigmentation appear in the culture with a high frequency of ∼10−2, probably because of the excess of bacteriochlorophylls and photosynthetic complexes accumulated in the presence of oxygen. Under these conditions, the excited states of bacteriochlorophylls may sensitize oxygen to produce singlet molecular oxygen, a powerful oxidant that damages DNA and induces mutagenesis.
Decrease of LH2 production in the PPSRK mutant.
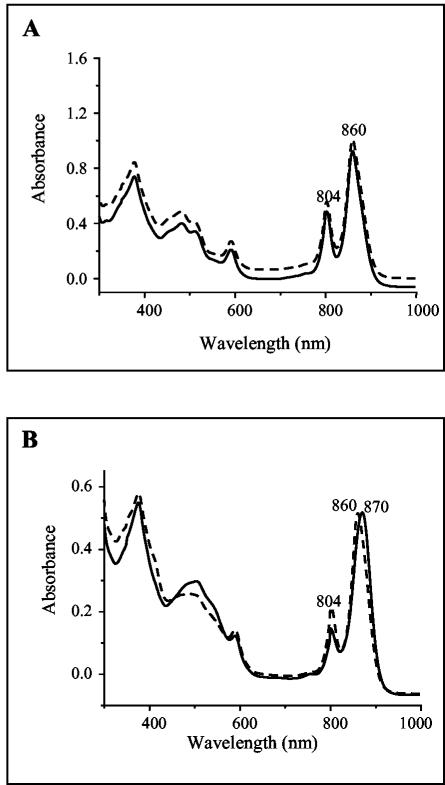
Because of its dark-red color under semiaerobic conditions, one could expect an increase in the expression of photopigments and photopigment binding proteins (LH and RC) in the PPSRK mutant strain. To examine this hypothesis, we compared equal amounts of membranes from wild-type and PPSRK cells grown under the same conditions, i.e., both under photosynthesis or both under semiaerobiosis. As shown in Fig. 3A, no difference was observed between the two spectra when cells were grown photosynthetically, indicating that there is no difference in the levels of production of the photopigment and photopigment binding proteins (LH1, LH2, and RC). Intriguingly, when grown semiaerobically, LH2 complex production was reduced in the PPSRK mutant compared to the wild type. Indeed, in the wild type, two peaks are observed at 804 and 860 nm, corresponding mainly to LH2 absorption, while in the PPSRK mutant a shift of the 860-nm peak to 870 nm and a reduction of ∼50% in the absorbance at 804 nm were observed (Fig. 3B), indicating a substantial decrease in the production of LH2. These results suggest that for full production of LH2, PpsR protein is required under semiaerobic conditions, and that this factor is an activator of LH2 production under semiaerobiosis. Moreover, an increase in carotenoid production (broad carotenoid absorption region, 420 to 550 nm) after growth under semiaerobic conditions was observed in the PPSRK mutant (Fig. 3B). This observation suggests that PpsR acts as a repressor of the carotenoid biosynthesis pathway.
FIG. 3.
(A) Absorption spectra of chromatophores of the wild type (dashed line) and of the PPSRK mutant (solid line) grown photosynthetically (A) or semiaerobically (B). (The zero level of the upper spectrum in panel A was shifted for better viewing.) (B) In the PPSRK mutant, a shift of the 860-nm peak to 870 nm, a reduction in the absorbance at 804 nm, and an increase in the absorbance at 420 to 550 nm are apparent.
Increase in pigment production in the PPSRK mutant.
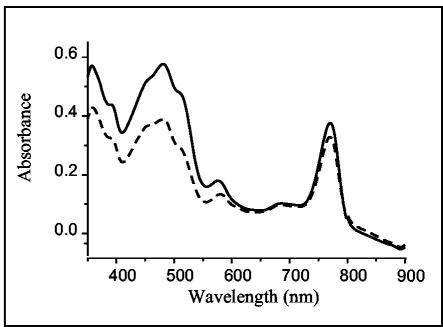
To further characterize the PPSRK mutant, carotenoid analyses were carried out by HPLC and spectral absorption on cells grown semiaerobically. Increases in the amount and in the color of the pigment extract from the PPSRK mutant were observed. Spectral analyses showed that for approximately equivalent bacteriochlorophyll-containing samples, the amount of carotenoid was significantly larger in the PPSRK mutant than in the wild type (Fig. 4). In the wild-type cells grown under semiaerobic conditions, carotenoids, such as neurosporene, OH-neurosporene, spheroidene, OH-spheroidene, and ketocarotenoids (spheroidenone, OH-spheroidenone, and diketospirilloxanthin), were identified (23). In the PPSRK mutant, the HPLC analysis (not shown) revealed that the most abundant species of carotenoids accumulating in the cells under semiaerobiosis corresponded to spheroidenone and OH-spheroidenone.
FIG. 4.
Absorption spectra of pigments from wild-type (dashed line) and PPSRK mutant (solid line) cells grown semiaerobically. For equal amounts of bacteriochlorophyll, the amount of carotenoid in the PPSRK mutant is dramatically increased (absorption between 420 and 550 nm).
The HPLC analysis has also shown that in the PPSRK mutant, bacteriochlorophyll a was produced, and it revealed the presence of a bacteriochlorophyll a intermediate. This intermediate, eluted before bacteriochlorophyll a, has the same absorbance characteristics as bacteriochlorophyll a (i.e., a 770-nm peak); thus, we propose that this intermediate corresponds to bacteriochlorophyllide a. Bacteriochlorophyllide a accumulation in the absence of PpsR suggests that the PpsR protein should regulate the expression of at least one bacteriochlorophyll biosynthesis enzyme under semiaerobic conditions.
In summary, the spectral and HPLC analyses have shown that disruption of ppsR results in an increase of carotenoid biosynthesis, in a change of the bacteriochlorophyll a intermediate balance, and in a pronounced reduction in the amount of LH2.
Overexpression of ppsR in the wild type.
To obtain further evidence that R. gelatinosus PpsR represses pigment and photosystem biosynthesis, we examined the consequences of the overexpression of ppsR on photosystem complex production. With that aim, we subcloned a DNA fragment containing the ppsR gene from pA410 into the pBBR1MCS-3 replicative plasmid to give the pA411 plasmid. In pA411, the ppsR gene is under the lacZ constitutive promoter. The plasmid was introduced into the wild type. All of the selected transformants carrying extra copies of ppsR were no longer pigmented but whitish. Transformants were grown under semiaerobic conditions and compared to the wild type containing the pBBR1-MCS3 (control) grown under the same conditions. Unlike the control strain, the strain overexpressing ppsR did not produce any photosynthetic complex (Fig. 5). When shifted to photosynthesis conditions, after a lag time, the photosynthetic complexes were produced and allowed photosynthetic growth (data not shown). We conclude that the addition of several ppsR copies leads to the total repression of photosynthetic-complex biogenesis under semiaerobic conditions. This severe phenotype is probably due to the repression of carotenoid and bacteriochlorophyll biosynthesis.
FIG. 5.
Spectral analysis of wild-type cells containing either pBBR1MCS-3 (control; solid line) or pA411 (dashed line) and grown semiaerobically.
Transcriptional regulation of LH2 and pigments by PpsR.
The difference observed in the production of photopigments and photopigment binding proteins in the PPSRK mutant could be a consequence of transcriptional control of puc and crt genes by ppsR. To test this hypothesis, fusions to the lacZ reporter gene were constructed, and the β-galactosidase activities were determined in the wild type and the PPSRK mutant grown under different conditions.
Regulation of the pucBA operon.
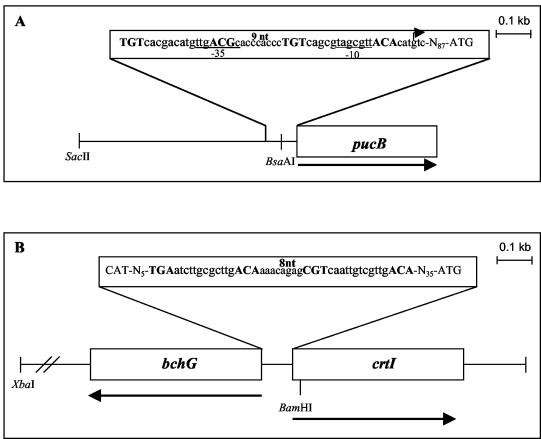
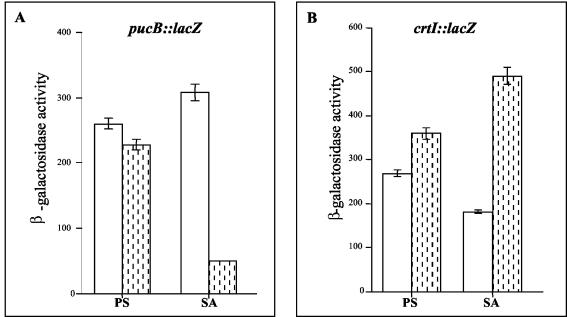
The presence of a putative PpsR-binding motif upstream of the pucBA operon (Fig. 6A) prompted us to examine whether this operon is regulated by the transcriptional regulator PpsR. The wild type and the PPSRK mutant, containing the pucBA::lacZ fusion, were grown photosynthetically or semiaerobically and assayed for β-galactosidase activity. As shown in Fig. 7A, under photosynthesis conditions, the expressions of the pucB::lacZ fusion in the mutant and the wild type were not different, suggesting that PpsR is not required. In contrast, under semiaerobiosis, in the PPSRK mutant strain the level of pucB::lacZ expression decreased by a factor of 6 compared to the wild type. This shows that PpsR acts as an aerobic activating transcriptional factor and supports the requirement for PpsR for the induction of pucBA expression in response to an increase in oxygen tension, consistent with the spectral analyses.
FIG. 6.
pucBA (A) and crtI-bchG (B) promoter regions used to construct lacZ fusions. The restriction sites used for cloning are indicated. The PpsR-binding sites are in boldface uppercase letters. The spacing between the two palindromes is 9 bp in the case of pucBA and 8 bp in the case of crtI-bchG. The σ70-type −10 and −35 promoter regions are underlined. The pucBA transcriptional start site (+1) located at −90 nucleotides (nt) from the pucB initiation codon start site is indicated (29).
FIG. 7.
Promoter activities of lacZ fusions in the wild type (open bars) and in the PPSRK mutant strains (shaded bars) grown either under photosynthetic (PS) or under semiaerobic (SA) conditions. The strains contain either the pucB::lacZ transcriptional fusion (A) or the crtI::lacZ transcriptional fusion (B). Experiments were performed in triplicate, and the error bars indicate standard deviations. Units of β-galactosidase activity represent micromoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per unit of optical density of culture at 680 nm.
Regulation of the crtI gene.
The increase in the amount of carotenoid in the PPSRK mutant suggested an increase in the expression of carotenoid biosynthesis genes. In particular, the increase in the amount of spheroidenone and OH-spheroidenone in the mutant suggested the induction of the crtA gene encoding the hydroxyspheroidene oxygenase. However, sequence analysis upstream of the start codon of crtA did not reveal any putative PpsR-binding motif. The increase in carotenoid could, alternatively, be a consequence of induction of enzymes involved earlier in the biosynthetic pathway, such as CrtB and CrtI. Inspection of the crtI gene sequence revealed the existence of a PpsR-binding motif (Fig. 6B). To address whether crtI is regulated by PpsR, we measured the promoter activities of crtI in the PPSRK background, as well as in the wild-type strain, using a crtI::lacZ transcriptional fusion. Under photosynthetic conditions, no significant difference between the promoter activities was observed in the PpsR strain and the wild type. However, the induction was substantial under semiaerobic conditions. Indeed, the crtI promoter activity was 2.7 times greater in the PPSRK mutant than in the wild type (Fig. 7B). We conclude that PpsR represses the activity of the crtI promoter under semiaerobic conditions. The increase in the amount of hydroxyspheroidenone in the PPSRK mutant may be explained by the fact that induction of expression of crtI in this mutant will provide the cells with more substrate for the CrtA enzyme, thus increasing the amount of hydroxyspheroidenone.
Promoter activities determined by the lacZ fusion assays clearly demonstrate the involvement of PpsR as a transcriptional factor exhibiting positive control over pucBA expression and negative control over the carotenoid crtI gene.
Analysis of pucBA transcripts.
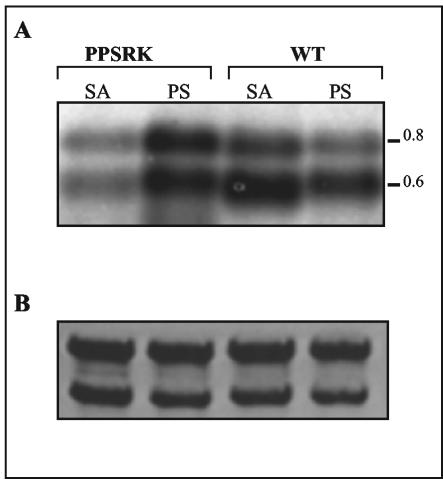
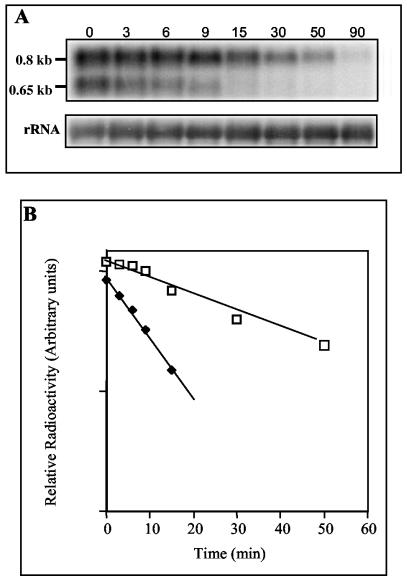
Northern hybridization was performed to correlate the abundance of pucBA transcripts with the results obtained for pucB::lacZ fusion expression. Total RNA was extracted from the wild type and the PPSRK mutant grown either photosynthetically or semiaerobically and then hybridized with the pucBA probe. Two pucBA hybridizing transcripts (0.8 and 0.65 kb) were detected (Fig. 8). Both transcripts initiate at the same promoter identified by primer extension but end at two distinct terminator structures (29).
FIG. 8.
Analysis of pucBA mRNA levels in the wild type (WT) and in the PPSRK mutant strains grown under photosynthetic (PS) or under semiaerobic (SA) conditions. (A) Total RNA was extracted from the wild type and the PPSRK mutant and then hybridized with a pucBA-specific probe. Approximately 10 μg of total RNA was loaded in each lane. (B) Values were normalized by the levels of 23S and 16S rRNAs.
Under photosynthetic conditions, the pucBA transcripts seem to be more abundant in the PPSRK mutant than in the wild type. However, when grown semiaerobically, the amount of pucBA in the PPSRK mutant is obviously reduced compared to that in the wild type (Fig. 8). The reduction of pucBA transcripts in the PPSRK mutant is in agreement with the β-galactosidase activity data. However, corroborating the β-galactosidase activity, the amount of transcripts detected in the wild type grown semiaerobically is higher than that in the photosynthetically grown wild-type cells (Fig. 8). This result is explained by the fact that under semiaerobiosis, PpsR activates pucBA transcription. Thus, pucBA Northern blot analysis supports the conclusions derived from the pucB::lacZ fusion studies and confirms that pucBA transcription is strongly activated by PpsR under semiaerobic conditions. However, in disagreement with the spectral and Western blot analyses, the amount of transcripts detected in the wild type grown photosynthetically is reduced compared to the semiaerobically grown wild-type cells. This may be explained by posttranscriptional events (availability of pigments to assemble LH2). For the crtI gene, the abundance of the transcript should be very low, and we were unable to detect any transcript. The low abundance of the crtI transcripts in spite of the high promoter activity might be explained by instability of the transcripts.
Half-life measurement of pucBA mRNA.
The decrease in LH2 in the PPSRK mutant under semiaerobic conditions could also be a consequence of a reduced stability of the pucBA transcripts in this mutant. The half-lives of the pucBA transcripts have been determined in the wild type (29). For the evaluation of pucBA mRNA decay kinetics, the Northern blot was normalized to rRNA. The half lives of the small 0.65-kb and large 0.8-kb pucBA transcripts in PPSRK were determined under semiaerobic growth conditions by Northern hybridization with the pucBA radiolabeled probe. The results are shown in Fig. 9. The transcript of 0.65 kb presents a short half-life (6 min), while the transcript of 0.8 kb presents a half-life of 22.5 ± 3 min. The results were similar to those obtained for the wild type (6 and 25 ± 3 min), indicating that the stability of pucBA mRNA was not affected in the PPSRK mutant (29).
FIG. 9.
pucBA mRNA decay kinetics. Total RNA was isolated at different times after the addition of rifampin to the cultures grown under semiaerobic conditions. (A) Northern blot analysis of pucBA mRNAs from the PPSRK mutant. The pucBA mRNAs were revealed with 32P-labeled pucBA-specific probe. The values were normalized by the levels of 16S rRNA. (B) Quantification of radiolabeled pucBA transcript from the PPSRK mutant blot. Squares, 0.8-kb transcript; diamonds, 0.65-kb transcript.
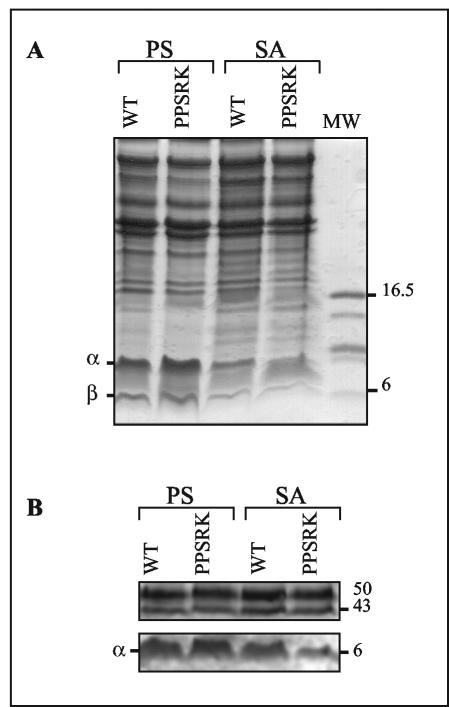
Immunoblot analysis of LH2 proteins.
Western blot analysis of R. gelatinosus membranes was performed to visualize a possible posttranscriptional regulation of LH2 and to confirm the spectral analysis. For that purpose, cultures of wild-type and PPSRK mutants grown under photosynthetic or semiaerobic conditions were harvested, and the membrane fraction of each culture was purified. Equivalent amounts of membrane were subjected to Tris-Tricine-SDS-polyacrylamide gel electrophoresis (Fig. 10A). After transfer onto nitrocellulose membranes, the fractions were probed with antiserum raised against R. gelatinosus LH2 proteins. The 6-kDa α subunit of LH2 was detected in all strains and under all conditions, although at different concentrations. The largest amounts were present in photosynthetically grown cells (Fig. 10B). In semiaerobically grown wild-type cells, the amount of α subunit detected was slightly reduced compared to that in the photosynthetically grown cells, suggesting once again a posttranscriptional step in the regulation of LH2 under semiaerobic conditions. In the PPSRK mutant membranes, the amount of the α subunit detected was drastically reduced, in agreement with the spectroscopic study and the transcriptional-fusion analyses. In the membranes, other polypeptides with high molecular masses (43 to 50 kDa) reacted with the LH2 antiserum. The nonspecific signals from these proteins were the same irrespective of the strain and the growth conditions. This signal served as a positive control. Taken together, the results of the Northern and Western blot analyses confirm the requirement for PpsR in the expression of the LH2 antenna under semiaerobic conditions.
FIG. 10.
Western blot analysis of R. gelatinosus membranes from the wild type (WT) and PPSRK mutant strains grown under photosynthetic (PS) or semiaerobic (SA) conditions. (A) Coomassie staining. Equal amounts of membrane protein (20 μg) were loaded on a Tris-Tricine-SDS-14% polyacrylamide gel. (B) Immunoreaction with anti-LH2 antibodies. MW, molecular weight (thousands).
DISCUSSION
In this work, we investigated the role of PpsR, a repressor of gene expression in response to oxygen in Rhodobacter species, in photosystem production in R. gelatinosus. R. gelatinosus produces a substantial number of photosystems even under semiaerobic conditions. To inactivate ppsR, the kanamycin cartridge was inserted in the gene before the region coding for the helix-turn-helix DNA binding domain. In this construct, the last 135 amino acids of PpsR should be deleted, resulting in an inactive protein. In R. sphaeroides, deletion of only the last 33 residues, including the helix-turn-helix, yields an inactive PpsR (22). Other experiments indicated that deletion of the C terminus of PpsR completely abolished its repressor activity (8).
The PPSRK mutant was dark red, and spectral analyses showed that PpsR is effectively involved in photosystem production. We observed a decrease in LH2 and a change in the pigment content in the mutant under semiaerobic conditions in darkness. Furthermore, we recorded the spectra of wild-type and PPSRK cells grown semiaerobically with light (data not shown). The results show that the PPSRK mutant synthesizes amounts of LH2 complexes comparable to the wild type under semiaerobiosis with light, strongly supporting the idea that PpsR detects only the oxygen in R. gelatinosus and is not essential for light sensing. The decrease in LH2 production could be due to a reduction in pucBA transcription, as well as to postranscriptional events, or simply a consequence of the change in the pigment content of the cells. To clarify the mechanism involved in LH2 decrease in the PPSRK mutant, pucBA and crtI promoter activities were measured using lacZ transcriptional fusions. We showed that PpsR possesses both positive and negative transcriptional activities, as this factor simultaneously induces pucBA transcription and represses crtI expression under semiaerobic conditions. pucBA Northern blot analysis corroborates the data from the pucB::lacZ fusion experiments and confirms the fact that pucBA transcription is induced by the PpsR factor under semiaerobic conditions without affecting the stability of the transcripts. While a correlation between transcription of pucBA and LH2 production could be observed in photosynthetically grown cells, a correlation between high pucBA promoter activity, high RNA stability, and the reduced number of LH2 complexes in semiaerobically grown wild-type cells is not obvious. Posttranscriptional events determined by the availability of carotenoid and bacteriochlorophylls may restrict the amount of LH2 antenna in the membranes under such conditions. In spite of high expression of pucBA, if the amounts of pigments (crt and bch) are small under semiaerobiosis, it may affect the assembly of LH2. Indeed, mutants of R. sphaeroides and R. gelatinosus lacking carotenoid do not assemble any LH2 complex (15, 20).
Because CrtJ, the PpsR homologue in R. capsulatus, was shown to bind to DNA, we believe that the effect of ppsR on the amount of LH2 could be interpreted as a direct effect on pucBA transcription. This was sustained by the pucBA promoter activities and the Northern blots. However, other possibilities should be considered. Indeed, the natures and the relative amounts of carotenoids may affect the LH1/LH2 ratio, as reported for R. sphaeroides (31). In that case, the level of LH2 would be the result of both the transcriptional activation of pucBA by PpsR and the carotenoid composition of the membranes.
In regard to carotenoid biosynthesis in the PPSRK mutant, the data we obtained suggest that PpsR represses the expression of crtI in semiaerobiosis. The large amounts of hydroxyspheroidenone and spheroidenone accumulated in the PPSRK mutant could result from overexpression of crtI, as mentioned above, but also from an induction of crtA gene expression. Because crtA is part of the puf-crt operon in R. gelatinosus (23) and putative PpsR-binding sites were found in the puf promoter region, a puf::lacZ fusion will be soon available to investigate the involvement of PpsR in puf-crtA regulation.
In addition, we should keep in mind that genetic and molecular data suggest that puf is cotranscribed with crtEF and/or bchCXYZ (21; S. Liotenberg, unpublished data). Both the crtE and bchC promoters exhibit well-conserved PpsR-binding sites. Hence, we cannot exclude at present possible PpsR regulation of the crtEF-bchCXYZ-pufBALMCcrtADC superoperon in R. gelatinosus.
The accumulation of bacteriochlorophyllide a in the PPSRK mutant suggests that PpsR regulates the expression of at least one bacteriochlorophyll biosynthesis enzyme. In the bacteriochlorophyll a biosynthesis pathway, bacteriochlorophyllide a is the product of BchC and the substrate of BchG (2). As both bchC and bchG promoters possess the putative PpsR-binding sites, it will be informative to study their expression.
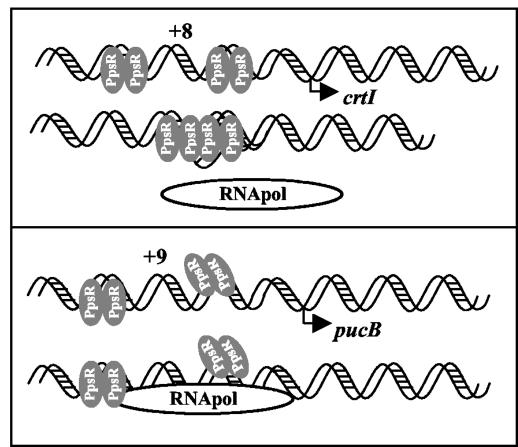
In Rhodobacter, it was shown that PpsR/CrtJ acts as a repressor by binding to the well-conserved TGT-N12-ACA consensus sequences positioned either upstream of or overlapping the RNA polymerase binding sites of the puc, bch, and crt genes (7, 8, 25). The results described above for R. gelatinosus show that PpsR acts as an activator of pucBA expression and as a repressor of crtI gene expression in this bacterium. A plausible explanation of the difference from Rhodobacter could be the difference in the sequence promoters of the genes studied, especially the sequence of PpsR-binding sites. As shown in Fig. 6, inspection of the putative PpsR-binding consensus sequences from the pucBA and crtI genes revealed significant divergences. Compared to the PpsR-binding sequence consensus (TGT-N12-ACA), the two sites from R. gelatinosus are not well conserved. In addition, compared to each other, the spacing between the two PpsR consensus sequences in the two promoters varies. Nine nucleotides were found in the spacing between the two consensus sequences in the pucBA promoter, while 8 nucleotides were found in the spacing sequence for the crtI promoter. It was reported that both the sequence and the spacing between the two PpsR consensus sequences are important for PpsR/CrtJ binding in Rhodobacter species (26). We assume that R. gelatinosus PpsR behavior (activator-repressor) should be related to the sequence divergences in the PpsR-binding site. Many reports describe transcriptional regulators exhibiting both positive and negative control, i.e., the TyrR factor in E. coli (4, 24). Like PpsR, TyrR recognizes the palindrome TGT-N12-ACA. Depending on the spacing sequence between the two TGT-N12-ACA sequences, the activity of TyrR could be modified. The maximum repression occurs when the two consensus sequences are positioned on the same face of the DNA helix, promoting cooperative interaction of the two TyrR dimers (4, 12). If the cooperative interaction is abolished (+3 or +12 bp between the two consensus sequences), the activation occurs through the interaction between one TyrR dimer and the RNA polymerase (4, 12). In addition, Ponnampalam et al. have reported that +6 and +11 nucleotide additions between the two consensus sequences abolished CrtJ binding to the bchC promoter, and they suggest that the 8-bp spacing between the two palindrome sites is important for proper cooperative binding of two CrtJ dimers to the promoter (26).
How could PpsR exhibit both positive and negative control in R. gelatinosus? A possible mechanism could be the fact that expression of the pucBA operon requires the presence of PpsR to facilitate the recruitment and the binding of trans-acting factors or the recruitment of the RNA polymerase, which in turn will induce transcription of the pucBA operon under semiaerobic conditions. In the case of crtI, the two PpsR dimers should bind to the same face of the DNA double helix (when the spacing is 8 bp) and may form the tetramer that prevents binding of the RNA polymerase (Fig. 11). In the case of pucBA, when the spacing is 9 bp, the formation of the tetramer is no longer possible, and the two PpsR dimers will increase transcription initiation. Site-directed mutagenesis has been undertaken to test this model. The results of this work show that the differences in gene expression between the β proteobacterium R. gelatinosus and the α proteobacterial species have real consequences for the biosynthesis of the photosynthetic complexes.
FIG. 11.
Model representing binding of PpsR to the crtI promoter (the PpsR binding sites are separated by 8 bp) and to the pucBA promoter (the PpsR binding sites are separated by 9 bp). The difference in spacing between the two PpsR consensus sequences would affect the formation of the PpsR tetramer. The tetramer would prevent binding of the RNA polymerase (26).
Acknowledgments
We thank Françoise Reiss-Husson and Sylvie Elsen for fruitful discussions and Linda Sperling for hepful comments on the manuscript.
REFERENCES
- 1.Agalidis, I., E. Rivas, and F. Reiss-Husson. 1990. Reaction center-light harvesting B875 complexes from Rhodocyclus gelatinosus, characterisation and identification of quinones. Photosynth. Res. 23:249-255. [DOI] [PubMed] [Google Scholar]
- 2.Alberti, M., D. H. Burke, and J. E. Hearst. 1995. Structure and sequence of the photosynthesis gene cluster, p. 1083-1106. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, A. E., B. Dickson, B. Lawley, C. Cobbett, and A. J. Pittard. 1991. Importance of the position of TYR R boxes for repression and activation of the tyrP and aroF genes in Escherichia coli. J. Bacteriol. 173:5079-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer, C., S. Elsen, L. R. Swem, D. L. Swem, and S. Masuda. 2003. Redox and light regulation of gene expression in photosynthetic prokaryotes. Phil. Trans. R. Soc. Lond. B 358:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen-Bazire, G. W., W. R. Sistorm, and R. Y. Stanier. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell. Comp. Physiol. 49:25-68. [DOI] [PubMed] [Google Scholar]
- 7.Elsen, S., S. N. Ponnampalam, and C. E. Bauer. 1998. CrtJ found in distant binding sites interacts cooperatively to aerobically repress photopigment biosynthesis and light harvesting II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 273:30762-30769. [DOI] [PubMed] [Google Scholar]
- 8.Gomelsky, M., and S. Kaplan. 1995. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J. Bacteriol. 177:1634-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomelsky, M., and S. Kaplan. 1997. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 179:128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heck, C., R. Rothfuchs, A. Jager, R. Rauhut, and G. Klug. 1996. Effect of the pufQ-pufB intercistronic region on puf mRNA stability in Rhodobacter capsulatus. Mol. Microbiol. 20:1165-1178. [DOI] [PubMed] [Google Scholar]
- 11.Hu, X., A. Damjanovic, T. Ritz, and K. Schulten. 1998. Architecture and mechanism of the light-harvesting apparatus of purple bacteria. Proc. Natl. Acad. Sci. USA 95:5935-5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang, J. S., J. Yang, and A. J. Pittard. 1997. Critical base pairs and amino acid residues for protein-DNA interaction between the TyrR protein and tyrP operator of Escherichia coli. J. Bacteriol. 179:1051-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Igarashi, N., J. Harada, S. Nagashima, K. Matsuura, K. Shimada, and K. V. Nagashima. 2001. Horizontal transfer of the photosynthesis gene cluster and operon rearrangement in purple bacteria. J. Mol. Evol. 52:333-341. [DOI] [PubMed] [Google Scholar]
- 14.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 15.Lang, H. P., and C. N. Hunter. 1994. The relationship between carotenoid biosynthesis and the assembly of the light-harvesting LH2 complex in Rhodobacter sphaeroides. Biochem. J. 298:197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Nagashima, K. V. P., K. Matsuura, S. Ohyama, and K. Shimada. 1994. Primary structure and transcription of genes encoding B870 and photosynthetic reaction center apoproteins from Rubrivivax gelatinosus. J. Biol. Chem. 269:1-8. [PubMed] [Google Scholar]
- 18.Ouchane, S., I. Agalidis, and C. Astier. 2002. Natural resistance to inhibitors of the ubiquinol-cytochrome c oxidoreductase from Rubrivivax gelatinosus. Sequence and functional analysis of the bc1 complex. J. Bacteriol. 184:3815-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouchane, S., M. Picaud, F. Reiss-Husson, C. Vernotte, and C. Astier. 1996. Development of genetic transfers for Rubrivivax gelatinosus S1. Construction, characterization and complementation of a puf operon deletion strain. Mol. Gen. Genet. 252:379-385. [DOI] [PubMed] [Google Scholar]
- 20.Ouchane, S., M. Picaud, C. Vernotte, and C. Astier. 1997. Photooxidative stress stimulates illegitimate recombination and mutability in carotenoid-less mutants of Rubrivivax gelatinosus. EMBO J. 16:4777-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouchane, S., M. Picaud, C. Vernotte, F. Reiss-Husson, and C. Astier. 1997. Pleiotropic effects of puf interposon mutagenesis on carotenoid biosynthesis in Rubrivivax gelatinosus. A new gene organization in purple bacteria. J. Biol. Chem. 272:1670-1676. [DOI] [PubMed] [Google Scholar]
- 22.Penfold, R. J., and J. M. Pemberton. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J. Bacteriol. 176:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinta, V., S. Ouchane, M. Picaud, S. Takaichi, C. Astier, and F. Reiss-Husson. 2003. Characterization of unusual hydroxy- and ketocarotenoids in Rubrivivax gelatinosus: involvement of enzyme CrtF or CrtA. Arch. Microbiol. 179:354-362. [DOI] [PubMed] [Google Scholar]
- 24.Pittard, A. J., and B. E. Davidson. 1991. TyrR protein of Escherichia coli and its role as repressor and activator. Mol. Microbiol. 5:1585-1592. [DOI] [PubMed] [Google Scholar]
- 25.Ponnampalam, S. N., J. J. Buggy, and C. E. Bauer. 1995. Characterization of an aerobic repressor that coordinately regulates bacteriochlorophyll, carotenoid, and light harvesting-II expression in Rhodobacter capsulatus. J. Bacteriol. 177:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponnampalam, S. N., S. Elsen, and C. E. Bauer. 1998. Aerobic repression of the Rhodobacter capsulatus bchC promoter involves cooperative interactions between CrtJ bound to neighboring palindromes. J. Biol. Chem. 273:30757-30761. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steunou, A. S., S. Ouchane, F. Reiss-Husson, and C. Astier. 2003. Involvement of the C-terminal extension of the α polypeptide and of the PucC protein in LH2 complex biosynthesis in Rubrivivax gelatinosus. J. Bacteriol. 186:3143-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uffen, R. L. 1976. Anaerobic growth of a Rhodopseudomonas species in the dark with carbon monoxide as sole carbon and energy substrate. Proc. Natl. Acad. Sci. USA 73:3298-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeliseev, A. A., J. M. Eraso, and S. Kaplan. 1996. Differential carotenoid composition of the B875 and B800-850 photosynthetic antenna complexes in Rhodobacter sphaeroides 2.4.1: involvement of spheroidene and spheroidenone in adaptation to changes in light intensity and oxygen availability. J. Bacteriol. 178:5877-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeilstra-Ryalls, J. H., M. Gomelsky, A. A. Yeliseev, J. M. Eraso, and S. Kaplan. 1998. Transcriptional regulation of photosynthesis operons in Rhodobacter sphaeroides 2.4.1. Methods Enzymol. 297:151-166. [DOI] [PubMed] [Google Scholar]
- 33.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]