Abstract
Gene flow in macroalgal populations can be strongly influenced by spore or gamete dispersal. This, in turn, is influenced by a convolution of the effects of current flow and specific plant reproductive strategies. Although several studies have demonstrated genetic variability in macroalgal populations over a wide range of spatial scales, the associated current data have generally been poorly resolved spatially and temporally. In this study, we used a combination of population genetic analyses and high-resolution hydrodynamic modelling to investigate potential connectivity between populations of the kelp Laminaria digitata in the Strangford Narrows, a narrow channel characterized by strong currents linking the large semi-enclosed sea lough, Strangford Lough, to the Irish Sea. Levels of genetic structuring based on six microsatellite markers were very low, indicating high levels of gene flow and a pattern of isolation-by-distance, where populations are more likely to exchange migrants with geographically proximal populations, but with occasional long-distance dispersal. This was confirmed by the particle tracking model, which showed that, while the majority of spores settle near the release site, there is potential for dispersal over several kilometres. This combined population genetic and modelling approach suggests that the complex hydrodynamic environment at the entrance to Strangford Lough can facilitate dispersal on a scale exceeding that proposed for L. digitata in particular, and the majority of macroalgae in general. The study demonstrates the potential of integrated physical–biological approaches for the prediction of ecological changes resulting from factors such as anthropogenically induced coastal zone changes.
Keywords: dispersal, macroalgae, population genetics, particle tracking modelling, Laminaria digitata, hydrology
1. Introduction
Molecular investigations of gene flow and dispersal offer an opportunity to understand the relationship between life-history traits and population genetic structure. From a biological perspective, macroalgae have often been viewed as poor-dispersing species owing to short-lived spores and gametes [1], complex coastal topography [2], habitat discontinuities [3] and reproductive strategies and intrinsic life-history characteristics [4,5]. Studies investigating genetic structuring of macroalgae have generally demonstrated that gene flow is very limited [3,5–7] and is heavily influenced by their complex life histories [4,5,7]. Gene flow in macroalgae is further influenced by landscape quality between populations in addition to geographical distance and dispersal capabilities [3]. However, macroalgae can achieve long-distance gene flow via the dispersal of whole or partially detached thalli-bearing reproductive structures [8–13]. These may be rare and/or stochastic events, but they are important long-distance dispersal mechanisms connecting potentially isolated populations of macroalgae.
Aside from the importance of reproductive strategies of macroalgae in the control of spore dispersal, the other fundamental influence relates to the ambient current regime of the plants. This will be strongly influenced by the local topography and basic hydrodynamic forces and covers a wide spectrum of spatial scales ranging from the small scale which may affect, for example, the settling pattern of spores to the oceanic scale which may dictate whole or partial plant rafting patterns over large distances [8–13]. In general, however, consideration of the effects of current flow on spore dispersal has depended on rather general descriptors of current flow, although a more concise approach has been attempted by Alberto et al. [14]. Increasing emphasis on the development of highly resolved current models allows the possibility of establishing more rigorous relationships between current patterns and genetic structure in macroalgal populations and, indeed, in a wide range of marine organisms.
The kelp Laminaria digitata (Order Laminariales, Phaeophyta) is an important habitat-forming species and contributes to population and community-level processes. The life cycle of L. digitata alternates between heteromorphic haploid and diploid generations. The dominant sporophyte generation can attain blade lengths of up to approximately 3 m and forms distinctive stands of algae and alternates with the microscopic, dioecious male and female gametophytes. Laminaria digitata sporophytes form sori at the distal end of the thallus and, like many coastal macroalgae, the sori are released into the water directly which can promote spore dispersal (up to 2 km) [3]. However, longevity of spores produced by the sporophyte is believed to be brief (approx. 72 h), thus minimizing dispersal capabilities [3]. For the gamete stage, male gamete dispersal is even more limited as they receive cues (pheromones) from neighbouring female gametophytes to release spermatozoids, where they are retained by female gametophytes and following fertilization grow into sporophyte plants [3].
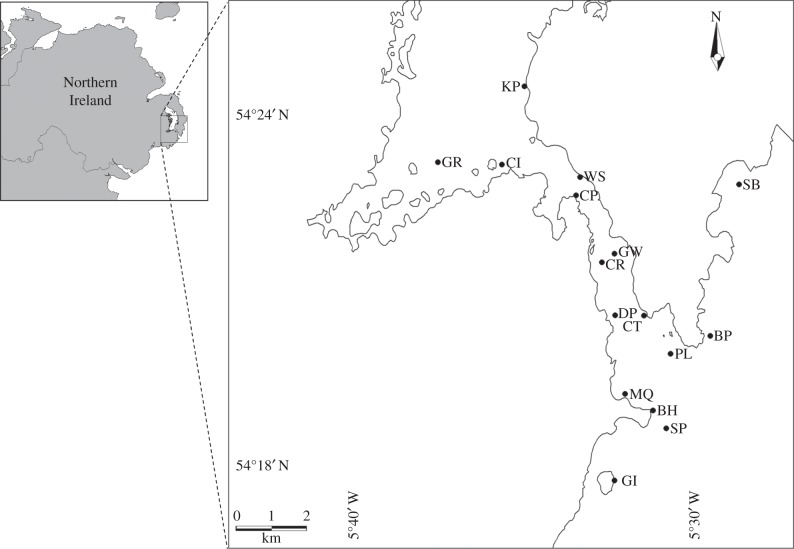
The aim of this study was to examine whether a complex local inshore hydrodynamic regime characterized by strong currents could shape the distribution of genetic variation in natural populations of L. digitata. Such regimes are of increasing significance owing to their potential for the exploitation of the associated currents for energy extraction. The investigation was based on the Strangford Narrows, a narrow channel some 8 km long and 1 km wide linking the main body of the large (150 km2), almost land-locked marine inlet of Strangford Lough on the east coast of Northern Ireland to the Irish Sea (figure 1). The Narrows is known for its strong, tidally driven currents (up to 3.5 m s−1) which led to its choice for the installation of the SeaGen experimental tidal turbine system in 2008. We wanted to investigate whether the complex current regime associated with the Narrows would act as a barrier to dispersal in L. digitata and disrupt the expected distribution of variation under a classic stepping-stone model of gene flow [15]. Thus, the alternative to the null hypothesis of isolation by distance would be that populations along the same shoreline would be more genetically similar to each other than to populations on the opposite shore, irrespective of geographical distance. This premise is based on the assumption that the strong current jet characteristic of the Narrows acts as a major restriction to across-channel flow. This study demonstrates that dispersion processes, both advection and diffusion, are a key driver for spore dispersal.
Figure 1.
Locations of sites sampled in this study.
To validate the findings of the genetic analysis, we used a hydrodynamic modelling approach to determine the spatial and temporal variability in spore dispersal to identify whether populations could be connected, thus facilitating gene flow. The relevant tools to predict advection and diffusion on this scale in the ocean and coastal environments are now readily available and frequently applied by ocean scientists and coastal engineers. Therefore, it is argued that more usage should be made of these tools given the importance of dispersion processes to nutrient availability and mixing, pollution dispersion and aggregation and reproductive processes in the marine environment. By contrast, it is advocated that, in equivalent terrestrial processes such as air pollution, the dispersion is less deterministic, because in the marine environment there is strong directionality owing to the inherent nature of tidal, topography and density-driven currents.
2. Material and methods
2.1. Site selection and DNA extraction
Samples were obtained from 16 discrete populations of L. digitata located around the Strangford Narrows of Strangford Lough, Northern Ireland (table 1 and figure 1), in June 2010. Although it is recognized that outside the lough wave action as well as currents can influence spore dispersal, in this study we only investigated current action as the greatest area of interest was within the Narrows, where wind influence would be minimal. At each site, 40 adult sporophytes with a blade length of more than 1 m were haphazardly selected and a small disc 0.7 cm in diameter was hole punched approximately 4 cm from the stipe/laminar junction and stored at −20°C on return to the laboratory. DNA was extracted using a CTAB (cetyltrimethylammonium bromide) procedure [16] and quantified visually on 1% agarose gels stained with ethidium bromide and diluted to a concentration of 50 ng μl−1 for subsequent polymerase chain reaction (PCR). Between 14 and 32 individuals per population were successfully genotyped.
Table 1.
Laminaria digitata populations studied and summary diversity statistics: N, number of individuals; AR, alleleic richness; HO, observed heterozygosity; HE, expected heterozygosity; FIS, inbreeding coefficient. n.s., not significant.
| location | code | latitude (N) | longitude (W) | N | AR | HO | HE | FIS |
|---|---|---|---|---|---|---|---|---|
| Guns Island | GI | 54.29495 | 5.54242 | 31 | 4.360 | 0.495 | 0.650 | 0.243*** |
| St Patrick's Rock | SP | 54.30925 | 5.51632 | 29 | 3.830 | 0.513 | 0.590 | 0.134** |
| Ballyhornan | BH | 54.31448 | 5.52252 | 30 | 4.240 | 0.496 | 0.629 | 0.216*** |
| Mill Quarter Bay | MQ | 54.31947 | 5.53582 | 22 | 4.413 | 0.410 | 0.661 | 0.387*** |
| Dick's Point | DP | 54.34202 | 5.53933 | 30 | 3.990 | 0.516 | 0.623 | 0.174*** |
| Cloghy Rocks | CR | 54.35403 | 5.53988 | 19 | 4.071 | 0.474 | 0.624 | 0.247** |
| Church Point | CP | 54.37717 | 5.55423 | 31 | 4.301 | 0.594 | 0.645 | 0.081* |
| Chapel Island | CI | 54.38607 | 5.59210 | 32 | 3.639 | 0.501 | 0.576 | 0.134** |
| Gate Rock | GR | 54.38740 | 5.62325 | 32 | 4.080 | 0.594 | 0.656 | 0.096* |
| Kate's Pladdy | KP | 54.40818 | 5.57980 | 32 | 3.321 | 0.497 | 0.507 | 0.019n.s. |
| Walter Shore | WS | 54.38172 | 5.55402 | 31 | 4.513 | 0.511 | 0.658 | 0.226*** |
| Gowland Rocks | GW | 54.35955 | 5.53850 | 14 | 4.705 | 0.612 | 0.692 | 0.119* |
| Carrstown | CT | 54.34162 | 5.52522 | 21 | 3.888 | 0.412 | 0.552 | 0.259*** |
| Pladdy Lug | PL | 54.33043 | 5.51292 | 32 | 4.474 | 0.482 | 0.688 | 0.304*** |
| Ballyquintin Point | BP | 54.33520 | 5.49318 | 31 | 3.691 | 0.469 | 0.601 | 0.223*** |
| South Bay | SB | 54.42823 | 5.47955 | 21 | 3.582 | 0.324 | 0.563 | 0.437*** |
*p < 0.05; **p < 0.01; ***p < 0.001.
2.2. Microsatellite genotyping
Microsatellite markers were developed by searching L. digitata-expressed sequence tag (EST) sequences in the GenBank database for all trinucleotide repeat motifs as described in [17]. Of 10 loci tested, only three (AW401303, CN466672 and CN467658) gave consistently scorable, polymorphic products (table 2). Consequently, we also used three trinucleotide loci (Ld2/167, Ld2/371 and Ld2/531) described in [18]. Forward primers included a 19 bp M13 tail (CACGACGTTGTAAAACGAC) and reverse primers included a 7 bp tail (GTGTCTT). PCR was carried out in a total volume of 10 μl containing 100 ng genomic DNA, 10 pmol of HEX-labelled M13 primer, 1 pmol of tailed forward primer, 10 pmol reverse primer, 1× PCR reaction buffer, 200 µM each dNTP, 2.5 mM MgCl2 and 0.25 U GoTaq Flexi DNA polymerase (Promega). PCR was carried out on a MWG Primus thermal cycler using the following parameters: initial denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 30 s and a final extension at 72°C for 5 min. Genotyping was carried out on an AB3730xl capillary genotyping system. Allele sizes were scored using LIZ size standards and were checked by comparison with previously sized control samples. Microsatellite data are available upon request.
Table 2.
Laminaria digitata microsatellite loci developed in this study. Forward tailed with CACGACGTTGTAAAACGAC. Reverse tailed with GTGTCTT.
| locus | repeat | primers | size range (bp) | source |
|---|---|---|---|---|
| AW401303 | complex (TGC) | TTCGAACCCAGTATCTACCTCGT CCGAGTACAGGCGGCAAC |
278–335 | this study |
| CN466672 | (AAC)18 | CTCCAACGATCCGCCTTG GGTCGCTTTCTTTCGTTGC |
99–135 | this study |
| CN467658 | (AGC)11 | CTCTTCCGTCGACCTTGTTC GGTCGAGGCGATTTTTCATA |
228–276 | this study |
| Ld2/167 | (CAA)2CGA(CAA)3CAG(CAA)6 | CGGACTCGATTTTAGCGATGGG TCGGAAGCACGTGTTCTGTAT |
169–205 | [18] |
| Ld2/371 | (TTG)TG(TTG)13TTT(TTG)2 | TACATGCCTCGTGTCTTTTGTGCG AGGAAAAAGCGGTGCAGTATTA |
138–204 | [18] |
| Ld2/531 | (ATT)8(CTT)3T(CTT) | TACTAACCCATTGTTTGTTGTGC CGACGTGGTCCTTGTTCTCATC |
251–287 | [18] |
2.3. Genetic data analysis
As three of the microsatellite markers were derived from ESTs, we carried out an analysis to detect whether any of the loci used were potentially under selection, which could skew values of within-population diversity and between-population differentiation calculated under the assumption of neutrality. We used the software package BayeScan (v. 1.0), which implements the Bayesian method described in [19] to identify outlier loci based on population-specific and locus-specific FST. The program was run using the default parameters, and loci were classified as potentially under selection where log10(BF) > 1, which corresponds to ‘strong’ evidence based on Jeffreys criteria [20].
Tests for linkage disequilibrium between pairs of loci in each population were carried out in the program FSTAT (v. 2.9.3.2) [21]. Levels of polymorphism measured as allelic richness (AR), and observed (HO) and expected (HE) heterozygosity averaged over loci were calculated using the FSTAT and Arlequin (v. 3.5.1.2) [22] software packages, respectively. Inbreeding coefficients (FIS) were estimated using FSTAT. Overall levels of interpopulation differentiation were estimated using Φ-statistics, which give an analogue of F-statistics [23] calculated within the analysis of molecular variance (AMOVA) framework [24], also using the Arlequin software package. Population-pairwise FST values were calculated using the Genepop software package (v. 4.1) [25]. Population-pairwise estimates of gene flow (Nm) were also calculated using the private alleles method [26,27] as implemented in Genepop.
To further identify possible spatial patterns of gene flow, a principal coordinate analysis (PCA) was carried out in GenAlEx (v. 6.1; [28]). Inter-individual genetic distances were calculated as described in [29], and the PCA was carried out using the standard covariance approach.
A test for isolation-by-distance (IBD) [30] was carried out to test the null hypothesis of a stepping-stone model of gene flow between populations of L. digitata, independent of the effects of currents. The Isolde test implemented in the Genepop software package was used to assess the relationship between genetic distance, measured as FST/(1 − FST), and geographical distance between population pairs. Geographical distances were measured as coastal distance between pairs occupying the same coastline and coastal distances plus direct distance across water between pairs occupying opposite sides of the Strangford Narrows: thus, linear not log-transformed distances were used for the analysis. One thousand permutations were used for the Mantel test.
2.4. Hydrodynamic and particle tracking modelling
To numerically predict the spore dispersal of each of the L. digitata populations, a particle tracking model was coupled to the Strangford Lough current model [31] using MIKE21 modelling software (DHI Water and Environment software package: www.dhisoftware.com). The Strangford Lough hydrodynamic model uses a finite volume method to determine the current field by solving a depth averaged shallow water approximation [31]. Simulations were carried out separately for each site. Fertile L. digitata can be found throughout April to November, but as current flow is driven by tidal elevation which is predictable and varies little over a year, each model was run for a 56 day period (October/November 2010), which incorporated four spring and four neap tides, representative of the flow velocities that L. digitata spores would experience during the expected reproductive period.
It is unknown during which phase of the tide L. digitata releases spores, so a trickle release approach was adopted whereby, at each site, 200 particles were released every 5 min during the time period (56 days): this assumed that spores (particles) could be released from the plants at all states of the tide. Particles were released 0.5 m above the seabed, representative of the average stipe length in the lough (L. Kregting 2010, unpublished data). The particle model simulates the movement of particles in the horizontal and vertical dimensions using the Langevin equation to simulate the spore dispersal by diffusion and advection [32]. The scaled eddy viscosity formulation was used for the horizontal dispersion, whereas the dispersion is assumed to be proportional to the scaled eddy viscosity (Smagorinsky formulation) [33]. In the absence of any dispersion information, we assumed the dispersion to be of the same magnitude as the viscous mixing and used a constant value of 1.0. For the vertical dispersion, a constant dispersion coefficient value of 0.01 m2 s−1 was used for the vertical dispersion of particles in the water column. To estimate the change in velocity with water depth, a logarithmic velocity profile was calculated based on the bed friction velocity which can be directly calculated in the hydrodynamic model. While the model is capable of simulating processes such as settling and migration of the spores, there are no data to provide input for these parameters. Therefore, we took the approach to simulate the spore dispersal and settlement using neutrally buoyant inert particles and assumed the endpoint to be where the particles reached the substrate.
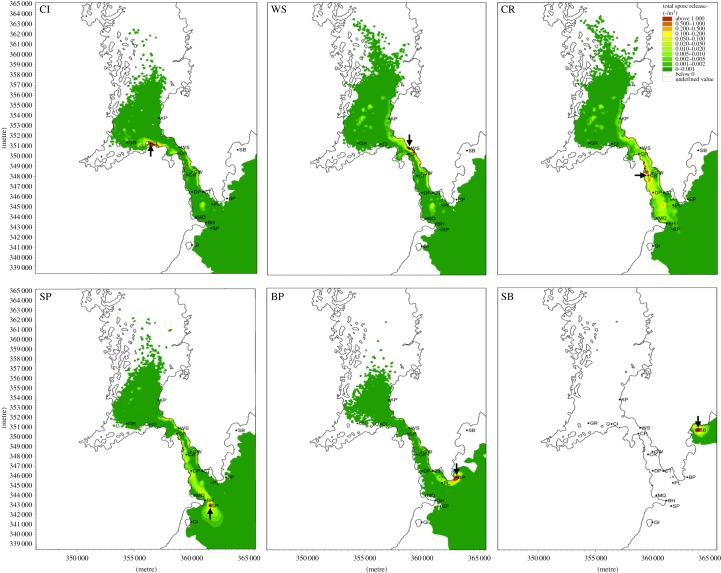
Prior to running the model, the dispersion pattern of the particles is unknown and therefore a reasonable time period is required to allow the simulation to reach a stationary pattern for the settled particles. Consequently, only the last fortnight of the simulation period was used to derive the figures in the paper (figure 2) still allowing for spring and neap tides to be incorporated, thus ensuring a good representation of overall flow conditions. To derive an average density of particles reaching the substrate per square metre, at each time step the number of particles on the bed per square metre was represented in relation to the total number of particles released (1 million particles) in the model to this point.
Figure 2.
Predicted particle distribution after 56 days from six representative release sites (arrowed) at Chapel Island (CI), Walter Shore (WS), Cloghy Rocks (CR), St Patrick's Rock (SP), Ballyquintin Point (BP) and South Bay (SB). Release of particles is constant simulating trickle spawning; time step 5 min, 200 particles per time step.
A Mantel test was carried out to determine whether there was any correlation between spore dispersal between populations based on the particle tracking model and estimates of population-pairwise gene flow (Nm) calculated from the microsatellite data.
3. Results
3.1. Levels of within-population diversity
Between 14 and 32 individuals were genotyped per population (total = 438; mean number of individuals per population = 27.375). No evidence of selection on any of the six loci was observed, and no significant linkage disequilibrium was detected between pairs of loci after sequential Bonferroni correction. The six microsatellite loci were highly polymorphic, exhibiting 14 (Ld2/531) to 29 (Ld2/371) alleles per locus (mean = 20.833; electronic supplementary material, table S1). Mean levels of within-population allelic richness (AR) averaged across the six loci ranged from 3.321 (KP) to 4.705 (GR). Mean levels of expected heterozygosity (HE) ranged from 0.507 (KP) to 0.692 (GW). Significant levels of inbreeding were detected in all but one of the populations (table 1).
3.2. Population genetic structuring
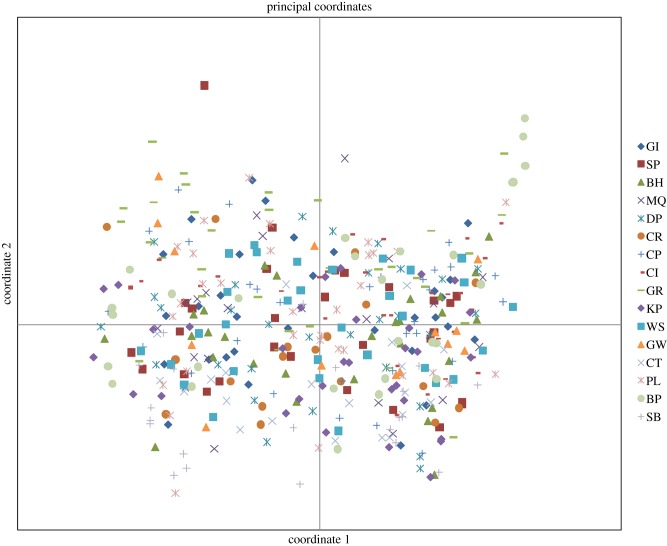
The AMOVA analysis indicated an overall small but significant level of differentiation across all populations (ΦST = 0.024, p < 0.001; table 3). Values of ΦST calculated separately for EST-based markers and non-genic markers (0.023 and 0.025, respectively) were almost identical, further suggesting neutrality of the markers employed in the study. Population-pairwise FST values (table 4 and figure 3) ranged from non-significant (GW/MQ, SP/MQ, PL/GI, CP/GW, SP/GW) to 0.156 (SB/GR, the most open site, located outside the narrows, and the site located furthest inside the lough). Population-pairwise Nm values calculated from private alleles (table 4) ranged from 1.04 (GW/SB) to 6.67 (BP/CP). The low global ΦST value was reflected in the PCA, which showed no evidence of geographical clustering of individuals (figure 4).
Table 3.
Analysis of molecular variance.
| source of variation | d.f. | variance | % variation |
|---|---|---|---|
| among populations | 15 | 0.0385 | 2.42 |
| within populations | 860 | 1.5548 | 97.58 |
Table 4.
Above diagonal: population-pairwise estimates of gene flow (Nm) calculated from nuclear microsatellite data using the private alleles method [26]. Below diagonal: population-pairwise FST values calculated from nuclear microsatellite data. Values between populations located on the same shoreline (with the exception of the outlying SB population) are shaded. n.s., non-significant FST value.
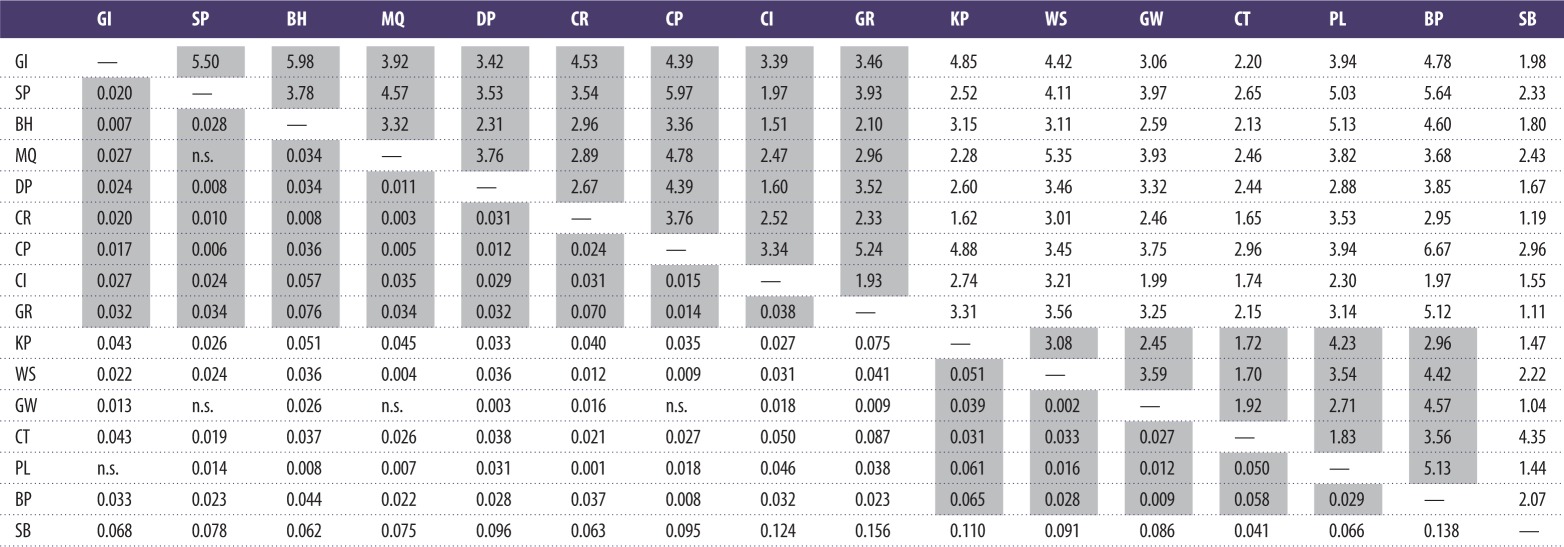 |
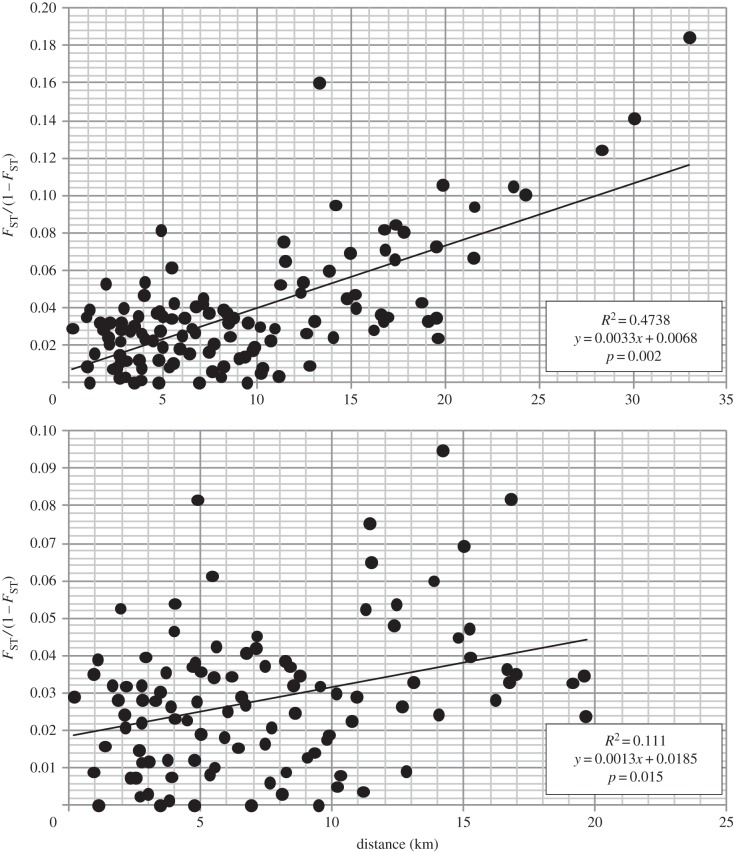
Figure 3.
Mantel test for IBD between populations including (above) and excluding (below) the SB population.
Figure 4.
Results of the PCA. The first two axes accounted for 22.87% and 19.85%, respectively, of the total variation (42.72%).
3.3. Particle dispersal
Connectivity of spores between sites was evident for all sites except SB (table 5 and figure 2; electronic supplementary material, figure S1). Greatest dispersal of particles was evident for sites within the Narrows where the highest flow rates occur. For example, populations at WS and CR (figure 2) that experience flow rates of more than 2 m s−1 show high levels of dispersal throughout the Narrows and into the main body of the lough and also, importantly, into the main body of the Irish Sea close to the entrance to the Narrows. High levels of dispersal were also apparent at sites within the lough (e.g. CI, figure 2) and sites adjacent to the entrance of the Narrows on the outer coast, that is, sites SP, GI and BP (table 5 and figure 2), with particles being dispersed throughout the length of the main tidal channel. For example, figure 2 shows in detail that particles released from SP at the entrance to the Narrows were predominantly dispersed along the western shores until approximately the mid-point of the Narrows after which the particles were transported across the channel to the eastern shores. In this instance, the across-channel transport is assumed to reflect the control of the direction of flow of the current in the Narrows by changes in the alignment of the shores. The only exception where connectivity between the outer sites and the Narrows was not evident was SB, located to the north of the entrance to the Narrows from where particles were predicted to advect away from the ambit of the lough.
Table 5.
Indication of the connectivity between sites. Values represent the likelihood of number of particles (×10−9 m−2) to be found at each of the source release sites (horizontal axis across the top). Values between populations located on the same shoreline (with the exception of the outlying SB population) are shaded. Values on the diagonal (black boxes) represent numbers of particles remaining at the source release sites.
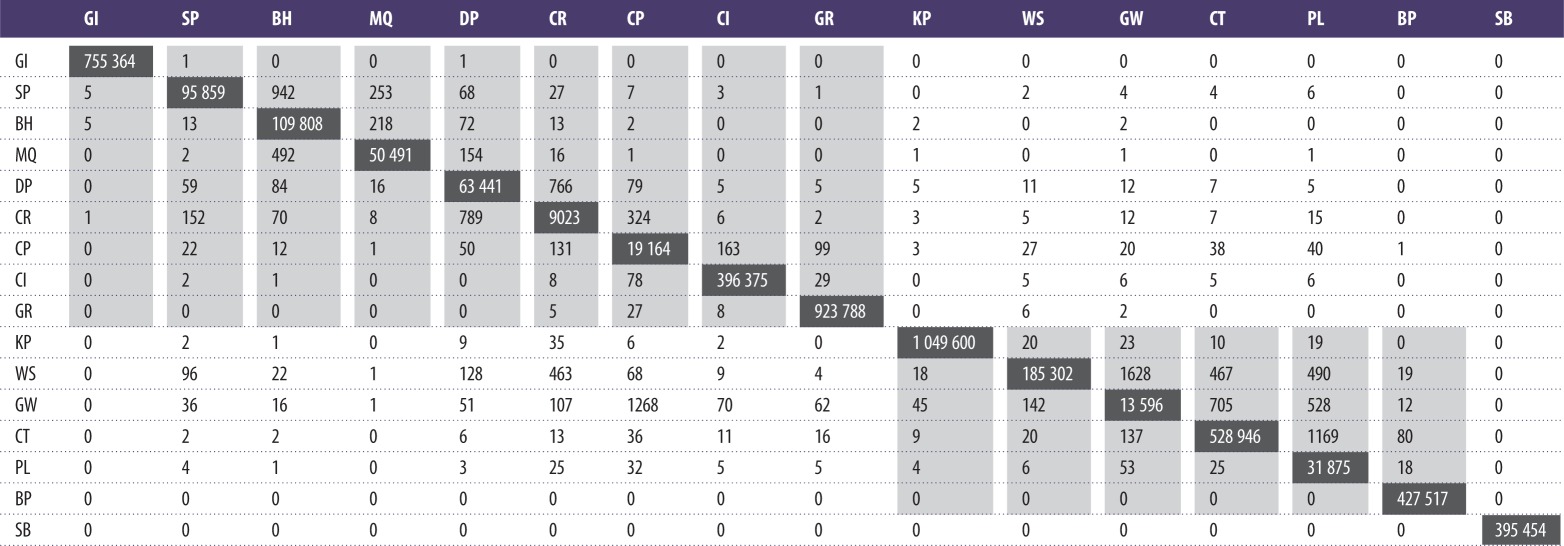 |
Particles released from sites in the Narrows generally demonstrated limited exchange between the east and west shores with relatively high densities of particles being recorded downstream of the release locations as for sites DP, CR and GW (table 5). This is likely to reflect the strength and bidirectional flow of the tidally induced jet aligned parallel to the axis of the main channel restricting the across-channel component of flow. Some exchange of spores between shores was suggested for a few release sites, for example SP, presumably reflecting localized flow patterns.
High levels of particle retention in the Narrows were also observed for some of the sites after 56 days of continuous release (figure 2 and table 5). Sites with the highest particle retention were generally associated with a low-flow regime (e.g. SB, CI, MQ and KP), whereas, conversely, sites with the highest flows had the lowest particle retention (e.g. CR, CP and GW) (table 5).
No significant correlation was observed between spore dispersal between populations based on the particle tracking model and estimates of population-pairwise gene flow (Nm) calculated from the microsatellite data (p = 0.07).
4. Discussion
To date, we are aware of only a single other study that has used a combination of hydrodynamic modelling and genetic analyses to gain insights into spore dispersal in macroalgae [14]. Although the results of both approaches in this study were not completely congruent, they did indicate that currents are an important mechanism in spore dispersal, and that the strong, predominantly jet-like currents in the Narrows at the entrance to Strangford Lough do not necessarily present an insurmountable barrier to dispersal between L. digitata populations on opposite sides of the channel. Furthermore, results revealed that, while the vast majority of spores are predicted to settle close to the source population, there is potential for long-distance dispersal, although the general pattern is one of IBD.
Observed levels of within-population genetic diversity were similar to those observed in previous microsatellite-based studies in L. digitata [3,34,35]. Levels of inbreeding, as measured by FIS, however, were far higher than those detected previously. Analysis of the data using the Micro-checker software package (v. 2.2.3) [36] indicated the possibility of null alleles at loci Ld2/167 and Ld2/531, which would also lead to increased homozygosity, but no evidence for null alleles was reported in previous studies using the same markers [3,34,35]. The limited dispersal implied by high inbreeding coefficients is generally inconsistent with the low levels of population differentiation observed in this study (global ΦST = 0.024), but a similar scenario was also reported in the kelp Macrocystis pyrifera [37].
Both the global and population-pairwise ΦST values as well as the results of the PCA indicate extensive gene flow between populations of L. digitata in the vicinity of the Narrows at the entrance to Strangford Lough. Nevertheless, a pattern of IBD was observed, indicating that the populations are more likely to exchange migrants with geographically proximal populations, irrespective of their position on the same or opposite shores of the Narrows. The particle tracking model also demonstrated that, while the majority of particles settled near the site of release, there was potential for long-distance dispersal within the ambit of the Narrows and also low-level transport of the particles out of the Narrows into the adjacent Irish Sea coastal waters.
The modelling approach also provides insight into the possible distribution and fate of the particles leaving the Narrows. Although the data in figure 2 are of necessity somewhat spatially limited outside the Narrows, there is a suggestion that the low-level transport of the particles to the south of the entrance to the Narrows is restricted compared with the offshore and northwards transport: this may reflect a mean northwards drift in this sector of the Irish Sea. A consistent feature of the distribution predictions is the minimal influence of particles from the Narrows in the close inshore region to the north of the Narrows entrance and is particularly noteworthy in the case of BP where it may have been expected that particles would have remained close inshore immediately to the north of the site. This feature would tend to accentuate the potential isolation of the SB population situated outside the lough several kilometres north of the entrance to the Narrows. Both the genetic data and the hydrodynamic modelling predictions confirmed that this was the most isolated population sampled; removal of the data for this population from the IBD analysis, however, still resulted in a clear pattern of IBD, albeit not as pronounced. Although the GI site was located some 3 km from the entrance to the Narrows, particles from this location were, in contrast to those from SB, predicted to enter the Narrows. This may again reflect the effects of the possible overall northwards drift, as suggested above, for this inshore sector of the Irish Sea.
The global level of population differentiation estimated in this study (ΦST = 0.024) was lower than those reported in three previous studies on L. digitata (global FST = 0.068 and 0.070 in [3] and [34], respectively, and mean population-pairwise FST = 0.02–0.12 in [35]), and far lower than those reported in other kelps, with the exception of M. pyrifera (FST = 0.021; summarized in [37]). The more continuous dispersal afforded to subtidal macroalgae generally results in lower levels of population differentiation than those observed for intertidal species [34], and the particularly low levels in this study compared with earlier studies on L. digitata are most likely due to the complex and dynamic hydrodynamic environment in Strangford Lough which results in high levels of both horizontal and vertical mixing.
The particle tracking model provides a visual representation of the extent that particles can travel throughout a dynamic system such as the Strangford Narrows and provides a powerful tool to predict the dispersal of spores accurately. High levels of recruitment near parent plants of macroalgae have been shown experimentally [38–40]; this observation has been confirmed from the observed results where dispersal distance was reduced as evidenced by the high retention of particle numbers near the release sites, thus providing support to the self-sustaining ‘closed’ population scenario [34]. By contrast, results from the model do, however, suggest that spores can be transported many kilometres (more than 8 km) away from the source population, although this is likely to be spatially and temporally specific. For example, spores released on the flood or ebb tide of the Narrows such as at CR will travel further during a 72 h period than spores released at BP.
In order to improve the resolution of the modelling approach, it would be necessary to determine the detailed track that spores would take during their life cycle. However, such an approach requires knowledge of when spores are released by macroalgae during the course of the tidal cycle. Depending on tidal elevation as controlled by the spring–neap tidal cycle and the consequent varying location of the algae in relation to the actual low-tide level, different spore release scenarios will emerge that may be simulated in the model. The most likely scenario appears to be that spores are released during slack low tide, explaining the high levels of recruitment near the parent plants [1]. This has been observed in fucoid algae, where gametes are released principally at low tide in rock pools or under calm conditions (flows less than 0.02 m s−1) [41,42]. Further, culturing techniques to stimulate the release of zoospores require algal tissue to be exposed to air for 18–24 h, suggesting that a stress response, as may be provided by exposure to air at low tide, may be required for the release of the spores [43]. This factor may explain why intertidal species are more genetically structured than subtidal species [34]. As ambient flow rates increase with the inflowing tide, the heterogeneous bathymetry resulting from the presence of local features, such as the plants themselves, rocks, boulders and changes in depth, give rise to increased turbulence which can effectively transport the newly released spores into the overlying water column [44]. Once the spores become buoyant, because of their small size (approx. 10–100 µm) [45] and hence low settling velocities, they will be transported solely by advective flow processes many kilometres downstream. It is recognized that adopting a trickle spawn approach, as in the present model, where particles are released continuously throughout the tidal cycle, only provides an approximation of the distance spores can travel via current flow alone.
It is also necessary to consider that currents are only one aspect of coastal processes that play a critical role in determining the distribution, connectivity and genetic structure of marine populations. Wind is also an integral part of coastal transport processes through its influence on both wind-generated currents and waves (wind and swell waves): both features provide additional significant mechanisms for spore dispersal. Wind-driven dispersal will be especially important on outer coastlines and non-inclusion of this factor could explain the apparently restricted dispersal of the population on the outer coastline at SB compared with populations within the Narrows. However, because of the sheltered nature of the Narrows, most of the L. digitata populations sampled in the investigation experience predominantly current-dominated flow [46] and it is hence considered that the results from the current-based model in the study are an ccurate representation of spore dispersal in the Narrows.
This study has highlighted the potential strengths of recent developments in high-resolution hydrodynamic modelling to powerfully assist the interpretation of the genetic properties of macroalgae and also their distribution in relation to current flow. In particular, we have demonstrated the influence of local coastal morphology on the distribution of macroalgal spores. This has obvious application on a more general basis for coastal zone management by allowing the prediction of possible changes in a wide range of marine fauna and flora characterized by planktonic dispersal stages arising from coastal engineering or naturally occurring changes or events. A major strength of the developments in current modelling and particle transport lies in their ability to describe ecological processes over a wide range of spatial scales ranging from the sub-metre to the oceanic scale while also incorporating the effects of turbulence. Thus, for example, these approaches have been applied to consider the effect of current flow on benthic distributions in coastal waters [47] and more specifically the effect on the distribution of Modiolus modiolus [48] and also in relation to the influence of current flow perturbations on the benthos adjacent to SeaGen, a full-scale, grid-connected tidal turbine [49]. Although not immediately relevant to the spatial scales considered in this study, the modelling approaches could also conceivably be applied in open ocean situations where they will have more relevance to evolutionary time scales.
Acknowledgements
The authors are grateful to J. Rodgers and P. Johnston for providing technical assistance in the field.
Funding statement
Aspects of this research were financially supported as part of the EPSRC SuperGen Marine Energy Research Consortium II (grant no. EP/E040136/1) and EPSRC grant EP/J010065/1.
References
- 1.Santelices B. 1990. Patterns of reproduction, dispersal and recruitment in seaweeds. Oceanogr. Mar. Biol. Annual Rev. 28, 177–276. [Google Scholar]
- 2.Muhlin JF, Engel R, Stessel R, Weatherbee RA, Brawley SH. 2008. The influence of coastal topography, circulation patterns, and rafting in structuring populations of an intertidal alga. Mol. Ecol. 17, 1198–1210. ( 10.1111/j.1365-294X.2007.03624.x) [DOI] [PubMed] [Google Scholar]
- 3.Billot C, Engel CR, Rousvoal S, Kloareg B, Valereo M. 2003. Current patterns, habitat discontinuities and population genetic structure: the case of the kelp Laminaria digitata in the English Channel. Mar. Ecol. Prog. Ser. 253, 111–121. ( 10.3354/meps253111) [DOI] [Google Scholar]
- 4.Lu TT, Williams SL. 1994. Genetic diversity and genetic structure in the brown alga Halidrys dioica (Fucales: Cystoseiraceae) in Southern California. Mar. Biol. 121, 363–371. ( 10.1007/BF00346746) [DOI] [Google Scholar]
- 5.Williams SL, Di Fiori RE. 1996. Genetic diversity and structure in Pelvetia fastigiata (Phaeophyta: Fucales): does a small effective neighbourhood size explain fine-scale genetic structure? Mar. Biol. 126, 371–382. ( 10.1007/BF00354619) [DOI] [Google Scholar]
- 6.Van der Strate HJ, Van de Zande L, Stam WT, Olsen JL. 2002. The contribution of haploids, diploids and clones to fine-scale population structure in the seaweed Cladophoropsis membranacea (Chlorophyta). Mol. Ecol. 11, 329–345. ( 10.1046/j.1365-294X.2002.01448.x) [DOI] [PubMed] [Google Scholar]
- 7.Krueger-Hadfield SA, Collen J, Daguin-Thiebault C, Valero M. 2011. Genetic population structure and mating system in Chondrus crispus (Rhodophyta). J. Phycol. 47, 440–450. ( 10.1111/j.1529-8817.2011.00995.x) [DOI] [PubMed] [Google Scholar]
- 8.Norton TA. 1992. Dispersal by macroalage. Eur. J. Phycol. 27, 293–301. [Google Scholar]
- 9.Valero M, Engel C, Billot C, Kloareg B, Destombe C. 2001. Concepts and issues in population genetics in seaweeds. Cah. Biol. Mar. 42, 53–62. [Google Scholar]
- 10.McKenzie PF, Bellgrove A. 2008. Dispersal of Hormosira banksii (Phaeophyceae) via detached fragments: reproductive viability and longevity. J. Phycol. 44, 1108–1115. ( 10.1111/j.1529-8817.2008.00563.x) [DOI] [PubMed] [Google Scholar]
- 11.Fraser CI, Hay CH, Spencer HG, Waters JM. 2009. Genetic and morphological analyses of the southern bull kelp Durvillaea Antarctica (Phaeophyceae: Durvillaeales) in New Zealand reveal cryptic species. J. Phycol. 45, 436–443. ( 10.1111/j.1529-8817.2009.00658.x) [DOI] [PubMed] [Google Scholar]
- 12.Buchanan J, Zuccarello GC. 2012. Decoupling of short- and long-distance dispersal pathways in the endemic New Zealand seaweed Carpophyllum maschalocarpum (Phaeophyceae, Fucales). J. Phycol. 48, 518–529. ( 10.1111/j.1529-8817.2012.01167.x) [DOI] [PubMed] [Google Scholar]
- 13.Provan J, Glendinning K, Kelly R, Maggs CA. 2013. Levels and patterns of population genetic diversity in the red seaweed Chondrus crispus (Florideophyceae): a direct comparison of single nucleotide polymorphisms (SNPs) and microsatellites. Biol. J. Linn. Soc. 108, 251–262. ( 10.1111/j.1095-8312.2012.02010.x) [DOI] [Google Scholar]
- 14.Alberto F, Raimondi PT, Reed DC, Watson JR, Siegel DA, Mitarai S, Coelho NC, Serrão E. 2011. Isolation by oceanographic distance explains genetic structure for Macrocystis pyrifera in the Santa Barbara Channel. Mol. Ecol. 20, 2543–2554. ( 10.1111/j.1365-294X.2011.05117.x) [DOI] [PubMed] [Google Scholar]
- 15.Kimura M, Weiss GH. 1964. The stepping-stone model of population structure and the decrease of genetics correlation with distance. Genetics 49, 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem. Bull. 19, 11–15. [Google Scholar]
- 17.Provan J, Beatty GE, Maggs CA, Savidge G. 2007. Expressed sequence tag-derived microsatellites for the cool-water marine copepod Calanus finmarchicus. Mol. Ecol. Notes 7, 1369–1371. ( 10.1111/j.1471-8286.2007.01889.x) [DOI] [Google Scholar]
- 18.Billot C, Rousvoal S, Estoup A, Epplen JT, Saumitou-Laprade P, Valereo M, Kloareg B. 1998. Isolation and characterization of microsatellite markers in the nuclear genome of the brown alga Laminaria digitata (Phaeophyceae). Mol. Ecol. 7, 1778–1780. [DOI] [PubMed] [Google Scholar]
- 19.Foll M, Gaggiotti O. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180, 977–995. ( 10.1534/genetics.108.092221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffreys H. 1961. Theory of probability. Oxford, UK: Clarendon Press. [Google Scholar]
- 21.Goudet J. 2002. FSTAT, v. 2.9.3: a program to estimate and test gene diversities and fixation indices. See http://www2.unil.ch/popgen/softwares/fstat.htm.
- 22.Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. ( 10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 23.Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. ( 10.2307/2408641) [DOI] [PubMed] [Google Scholar]
- 24.Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raymond M, Rousset F. 1995. GENEPOP (v. 1.2): population genetic software for exact tests and ecumenicism. J. Hered. 86, 248–249. [Google Scholar]
- 26.Slatkin M. 1985. Rare alleles as indicators of gene flow. Evolution 39, 53–65. ( 10.2307/2408516) [DOI] [PubMed] [Google Scholar]
- 27.Barton NH, Slatkin M. 1986. A quasi-equilibrium theory of the distribution of rare alleles in a subdivided populations. Heredity 56, 409–415. ( 10.1038/hdy.1986.63) [DOI] [PubMed] [Google Scholar]
- 28.Peakall R, Smouse PE. 2006. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. ( 10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smouse PE, Peakall R. 1999. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82, 561–573. ( 10.1038/sj.hdy.6885180) [DOI] [PubMed] [Google Scholar]
- 30.Rousset F. 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145, 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kregting L, Elsäßer B. 2014. A hydrodynamic modelling framework for Strangford Lough part 1: tidal model . J. Mar. Sci. Eng. 2, 46–65. ( 10.3390/jmse2010046) [DOI] [Google Scholar]
- 32.Pope SB. 2002. A stochastic Lagrangian model for acceleration in turbulent flows. Phys. Fluids 14, 2360–2375. ( 10.1063/1.1483876) [DOI] [Google Scholar]
- 33.Rodi W. 1980. Turbulence models and their application in hydraulics: a state of the art review. Special IAHR Publication. Leiden, The Netherlands: AA Balkema. [Google Scholar]
- 34.Valero M, Destombe C, Mauger S, Ribout C, Engel CR, Daguin-Thiebaut C, Tellier F. 2011. Using genetic tools for sustainable management of kelps: a literature review and the example of Laminaria digitata. Cah. Biol. Mar. 52, 467–483. [Google Scholar]
- 35.Couceiro L, Robuchon M, Destombe C, Valero M. 2013. Management and conservation of the kelp species Laminaria digitata: using genetic tools to explore the potential exporting role of the MPA “Parc naturel marin d'Iroise”. Aquat. Living Resour. 26, 197–205. ( 10.1051/alr/2012027) [DOI] [Google Scholar]
- 36.van Oosterhout C, Hutchinson WF, Willis DP, Shipley P. 2004. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. ( 10.1111/j.1471-8286.2004.00684.x) [DOI] [Google Scholar]
- 37.Alberto F, Raimondi PT, Reed DC, Coelho NC, Leblois R, Whitmer A, Serrão E. 2010. Habitat continuity and geographic distance predict population genetic differentiation in giant kelp. Ecology 91, 49–56. ( 10.1890/09-0050.1) [DOI] [PubMed] [Google Scholar]
- 38.Sundene O. 1962. The implications of transplant and culture experiments on the growth and distribution of Alaria esculenta. Nytt Mag. Bot. 9, 155–174. [Google Scholar]
- 39.Dayton PK. 1973. Dispersion, dispersal, and persistence of the annual intertidal alga, Postelsia palmaeformis Ruprecht. Ecology 54, 433–438. ( 10.2307/1934353) [DOI] [Google Scholar]
- 40.Kendrick GA, Walker DI. 1991. Dispersal distances for propagules of Sargassum spinuligerum (Sargassaceae, Phaeophyta) measured directly by vital staining and venturi suction sampling. Mar. Ecol. Prog. Ser. 79, 133–138. ( 10.3354/meps079133) [DOI] [Google Scholar]
- 41.Pearson GA, Brawley SH. 1996. Reproductive ecology of Fucus distichus (Phaeophyceae): an intertidal alga with successful external fertilization. Mar. Ecol. Prog. Ser. 143, 211–223. ( 10.3354/meps143211) [DOI] [Google Scholar]
- 42.Serrão EA, Pearson G, Kautsky L, Brawley SH. 1996. Successful external fertilization in turbulent environments. Proc. Natl Acad. Sci. USA 93, 5286–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edwards M, Watson L. 2011. Aquaculture explained: no. 26, Cultivating Laminaria digitata. Dunlaoghaire, Ireland: Irish Sea Fisheries Board. [Google Scholar]
- 44.Kregting LT, Bass Al, Òscar G, Yund PO, Thomas FIM. 2013. Effects of oscillatory flow on fertilization in the green sea urchin Strongylocentrotus droebachiensis. PLoS ONE 8, e76082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clayton MN. 1992. Propagules of marine macroalgae: structure and development. Br. Phycol. J. 27, 219–23. ( 10.1080/00071619200650231) [DOI] [Google Scholar]
- 46.Kregting LT, Blight A, Elsäßer B, Savidge G. Waves and currents influence the growth of the kelp. Laminaria digitata. Submitted. [Google Scholar]
- 47.Kennedy R, O'Carroll J, Kregting LT, Elsäßer B, Savidge G. In preparation The influence of current velocity as predicted by fine-scale modelling on benthic distributions in the Strangford Narrows and adjacent waters. [Google Scholar]
- 48.Elsäßer B, Fariñas-Franco J, Wilson CD, Kregting L, Roberts D. 2013. Identifying optimal sites for natural recovery and restoration of impacted biogenic habitats in a special area of conservation using hydrodynamic and habitat suitability. J. Sea Res. 77, 11–21. ( 10.1016/j.seares.2012.12.006) [DOI] [Google Scholar]
- 49.O'Carroll J, Creech A, Savidge G, Kennedy R. In preparation The effect of the SeaGen tidal turbine on local benthic community structure: an integrated metre-scale modelling and observational study. [Google Scholar]