Abstract
The development of the vertebral column starts with the formation of a linear array of mesenchymal condensations, forming the blueprint for the eventual alternating pattern of bone and cartilage. Despite growing insight into the molecular mechanisms of morphogenesis, the impact of the physical aspects of the environment is not well understood. We hypothesized that geometric boundary conditions may play a pivotal role in the linear patterning of condensations, as neighbouring tissues provide physical constraints to the cell population. To study the process of condensation and the patterning thereof under tightly controlled geometric constraints, we developed a novel in vitro model that combines micropatterning with the established micromass assay. The spacing and alignment of condensations changed with the width of the cell adhesive patterns, a phenomenon that could not be explained by cell availability alone. Moreover, the extent of chondrogenic commitment was increased on substrates with tighter geometric constraints. When the in vivo pattern of condensations was investigated in the developing vertebral column of chicken embryos, the measurements closely fit into the quantitative relation between geometric constraints and inter-condensation distance found in vitro. Together, these findings suggest a potential role of geometric constraints in skeletal patterning in a cellular process of self-organization.
Keywords: pattern formation, vertebral column, mesenchymal condensation, morphogenesis, geometry, skeletal development
1. Introduction
During the growth and development of the embryo, patterning events give rise to structurally and functionally complex tissues. These morphogenetic events involve an intricate interplay of cellular processes and result in robust outcomes. The role of molecular signalling pathways in achieving these complex outcomes has been studied extensively, however, the impact of physical cues is just starting to be unravelled [1–5]. For example, it was shown recently that the looping pattern of the gut is driven by the forces that arise from differential growth [6], while the pattern of villi lining the gut wall is determined by buckling and folding of the tissue owing to growth under spatial restrictions [7]. Investigations into the role of physical cues in embryogenesis are gaining momentum owing to innovative techniques that allow characterization, modelling and perturbation of developmental processes [1,8–10]. However, the potential role of physical cues in the development of many complex tissue patterns is still to be discovered.
One of the most fundamental and conserved patterns of the vertebrate body plan is the vertebral column. With its alternating pattern of bony elements and cartilaginous hinges, it provides form and stability as well as flexibility to the vertebrate body. Both the bone and cartilage tissue of the vertebral column originate from sclerotomal cells, a mesenchymal cell population originating from the ventral–medial region of the somites [11]. After dissociation from the residual epithelial component of the somites, the sclerotomal cells accumulate around the notochord and subsequently form a robust linear array of mesenchymal condensations [11,12]. Mesenchymal condensation is the first stage in bone development [13–16] and an essential step in skeletal patterning since the position, size and number of skeletal elements is established at this stage [17,18].
Mesenchymal condensation primarily involves an increase in local cell density [16,19], which appears to be mainly mediated through local cell movements rather than locally altered proliferation rates [20,21]. It is thought that these local cell rearrangements are predominantly mediated by passive ECM-driven movements and dragging and pushing by neighbouring cells, rather than active cell migration [22–24]. The dominant matrix components in this developmental stage are hyaluronic acid and fibronectin (FN) [25–29]. An increase in specific cell–cell contacts associated with mesenchymal condensation, mediated through N-cadherin and NCAM, facilitates cell–cell communication and is presumably involved in triggering the onset of chondrogenic differentiation [30–32]. This model is in line with the general notion that a high cell density is required for chondrogenesis [33,34].
The fundamental importance of mesenchymal condensation in skeletal development has motivated the development of in vitro models that recapitulate the early stages of skeletogenesis [35,36]. One in vitro model that is often used to study mesenchymal condensation is the micromass assay [37,38]. Typically, pre-chondrogenic cells are isolated from embryonic mouse or chicken limb buds and plated in a high-density drop. Within this high-density culture, condensations spontaneously appear within several days, followed by chondrogenic commitment and concomitant deposition of specific matrix components. Only embryonic cells seem to go through this specific process, as studies using adult or immortalized mesenchymal cells show different results [39,40]. The micromass model has been used to unravel cellular and molecular events involved in condensation as well as to investigate the role of a variety of signalling molecules in skeletal development [13,41,42]. However, physical cues such as the spatial restrictions set by neighbouring tissues are not taken into account in this model.
Micropatterning techniques, whereby cell adhesive islands are created in order to restrict single or multicellular cultures to specific geometries, have facilitated the study of the effect of geometric boundary conditions on a range of cellular events [43]. It has been shown that single-cell shape can control the switch between cell cycle advancement and apoptosis in certain cell types [44], while controlling the geometry of multicellular cultures elucidated emergent patterns of proliferation as well as differentiation [45–47]. Micropatterning, when combined with the micromass culture, could provide a useful tool to test the role of geometric boundary conditions in the supracellular process of condensation and skeletal patterning.
This study addresses the hypothesis that geometric constraints provided to mesenchymal cells determine their condensation pattern. This hypothesis was investigated by combining an in vitro cell patterning technique with the established micromass assay. Freshly isolated mesenchymal cells from chicken embryonic limb buds were used, as this has been proven to be an appropriate cell model for pre-chondrogenic condensation [13,37,38]. We demonstrate that geometric constraints affect linear patterning of condensations as well as subsequent chondrogenic differentiation. Moreover, measurements of the pattern of condensations in the developing vertebral column in the chicken embryo closely fit into the quantitative relation between geometric constraints and inter-condensation distance established by this new in vitro model.
2. Material and methods
2.1. Cell isolation and culture
Pre-chondrogenic limb bud cells were freshly isolated from chicken embryos for each experiment. Chicken eggs (White Leghorn, premium specific pathogen-free) were purchased from Charles River Laboratories (New York, NY, USA) and set to incubate at 37°C and 60% humidity. After 3 days of incubation, the embryos were staged and returned to the incubator. At Hamburger Hamilton stage 21–23 [48], the wing buds were dissected in PBS using a Leica stereomicroscope. Dissected limb buds were incubated in 0.5% trypsin for 12 min at room temperature and transferred to 10% ice-cold chicken serum in PBS. The ectoderm layer was subsequently manually removed from the limb buds. Buds were then suspended in DMEM-F12 culture medium containing 10% FBS and 1% penicillin and streptomycin (referred to as complete medium) and pipetted up and down to create a single-cell suspension. After straining through a 40 μm filter to remove cell clumps, the cells were counted, centrifuged and resuspended in complete medium to obtain a concentration of 6 × 106 cells ml−1. A drop of 60 μl cell suspension was then spread onto the patterned area (approx. 75 mm2) that was obtained as described in the following section (figure 1a(iv)). The cell seeding density was optimized during preliminary experiments which showed that lower cell densities lead to compromised cell survival, while higher cell densities mainly resulted in a larger fraction of cells not attaching to the substrate (data not shown). The plates were incubated at 37°C and 5% CO2 for 1 h to allow the cells to attach to their substrate. Subsequently, the cultures were washed once to remove unattached cells and then 2.5 ml of DMEM-F12 containing 2% FBS and 1% penicillin and streptomycin was added per well. For specific experiments, the culture medium was supplemented with rhTGF-β3, rhBMP2 or rmFGF8 (R&D systems) at the indicated concentrations. The culture medium was refreshed after 24 h and cultures were terminated after 2.5 days.
Figure 1.
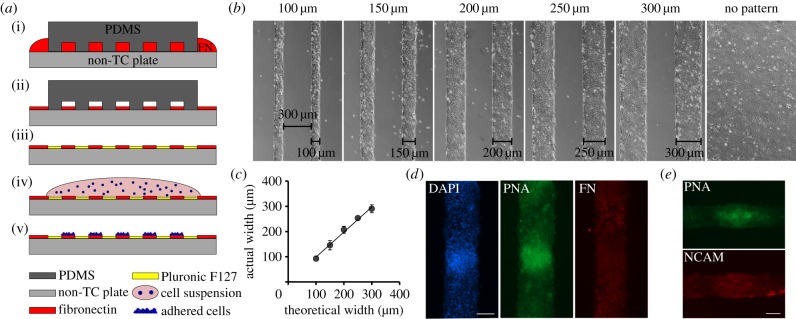
Experimental model. (a) Schematic of cell patterning procedure; (i) PDMS moulds are placed onto a non-tissue culture-treated plate, and an FN solution fills the channels of the mould. (ii) After 1 h of incubation, the FN solution is aspirated. (iii) After drying, the PDMS mould is removed and the plate is treated with Pluronic, which coats the non-patterned surface and leaves it non-adherent. (iv) A drop of cell suspension is spread over the patterned area. (v) The cells adhere to the FN patterns only. (b) Phase contrast micrographs of pre-chondrogenic cells cultured on linear adhesive islands or non-patterned substrates taken after 24 h in culture. The island width varies from 100 to 300 μm, while the non-adhesive space between islands is 300 μm wide for all conditions. (c) Plot indicating the correlation between the theoretical island width and the actual island width measured during cell culture. (d) Fluorescent micrographs of pre-chondrogenic cells isolated from chicken limb buds cultured for 2.5 days on 150 μm wide adhesive islands, showing a representative mesenchymal condensation. Cell nuclei are stained blue (DAPI), green indicates positive staining with PNA lectin, and cell-deposited fibronectin is stained red. Scale bar, 100 μm. (e) Fluorescent micrographs of pre-chondrogenic cells cultured for 2.5 days on 100 μm wide adhesive islands, showing a representative mesenchymal condensation stained with PNA lectin (green), and for NCAM (red). Scale bar, 50 μm. (Online version in colour.)
2.2. Microchannel patterning
Polydimethylsiloxane (PDMS) moulds for creating cell adhesive islands were fabricated by soft lithography. Negative photo resist SU-8 2100 (MicroChem, Newton, MA, USA) was deposited onto clean silicon wafers to a thickness of 130 μm and patterned by exposure to UV light through a transparency photomask (CAD/Art Services, Bandon, OR, USA). PDMS moulds were made by mixing PDMS base and curing agent (Sylgard 184 silicone elastomer kit) in a 10 : 1 ratio and pouring onto the patterned silicon wafer. The PDMS was cured overnight at 65°C after which individual moulds were cut out. Each mould contained raised features 300 μm wide and 8 mm long (figure 1b). The width between raised features was 100, 150, 200, 250 or 300 μm. The moulds were stored in 70% ethanol until use. To create FN islands, the moulds were first sonicated in a water bath for 60 min. They were then dipped in dH2O and placed in a 65°C oven to dry for 30 min. The PDMS moulds were subsequently subjected to O2 plasma treatment for 5 min. Simultaneously, a non-tissue culture six-well plate was UV treated for 7 min. Each mould was then placed features-down in the centre of a well in the six-well plate, creating microchannels. A 50 μl drop of 25 μg ml−1 FN (bovine, Sigma) in dH2O was placed directly next to the moulds and the solution was immediately drawn into the microchannels by capillary forces (figure 1a(i)). To create the non-patterned substrate, a 50 μl drop of FN solution was placed in the centre of the well in the absence of a PDMS mould. The drop was spread to cover the same surface area as the patterned substrates, leading to a similar adhesion ligand density. The FN solution was incubated at room temperature for 1 h. The solution was then carefully aspirated and the plates were placed open at room temperature until dry (figure 1a(ii)). The moulds were removed and 3 ml 1% Pluronic F127 (Sigma) in dH2O was added to each well for 5 min to render the non-coated areas of the well non-adhesive (figure 1a(iii)). The wells were washed three times with PBS and stored at 4°C until use. The actual width of the cell adhesive islands was confirmed by measuring the width of 24-h cultures using ImageJ (figure 1b,c).
2.3. Immunohistochemistry and stainings
After 2.5 days in culture, the samples were fixed in 4% paraformaldehyde in PBS. To identify condensations, the cultures and frozen sections of whole chicken embryos were stained with 50 μg ml−1 peanut agglutinin lectin (PNA lectin) conjugated with Alexa Fluor 488 (Invitrogen) in PBS. Cell nuclei were stained with 2 μg ml−1 Hoechst 33342 (Invitrogen). To probe for newly synthesized FN, the samples were incubated with a mouse-anti-avian FN antibody (Developmental Studies Hybridoma Bank) and stained with an Alexa Fluor 546 conjugated goat-anti-mouse IgG. Since patterned surfaces in the absence of cells only showed very faint FN staining (data not shown), it can be assumed that the strong FN staining after 2.5 days in culture can be attributed to cell-deposited FN. Expression of neural cell adhesion molecule (NCAM) was probed with a mouse-anti-chicken NCAM antibody (Developmental Studies Hybridoma Bank) and stained with an Alexa Fluor 546 conjugated goat-anti-mouse IgG. A rabbit-anti-Sox9 antibody (Millipore) was used to probe for the expression of Sox9. An Alcian Blue staining was performed to stain for deposited glycosaminoglycans (GAGs). The samples were washed in dH2O and subsequently stained in 1% Alcian Blue in 3% acetic acid (Electron Microscopy Sciences) for 30 min.
2.4. Quantitative analysis of condensations
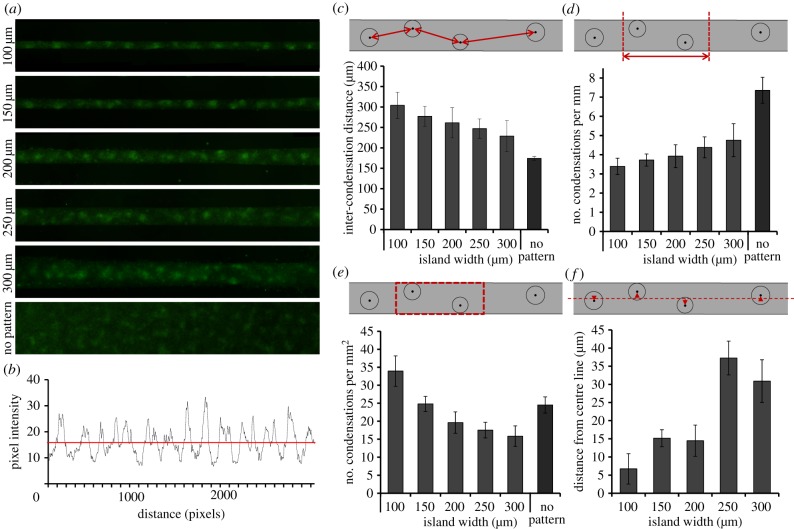
To quantify the condensation patterns, micrographs of PNA lectin staining were analysed using ImageJ (v. 1.46r, NIH) and MATLAB (v. R2011a, MathWorks). First, condensations were identified as high intensity regions in the PNA staining-micrographs by applying a threshold to the images (figure 2b). Secondly, a size limit was applied in order to exclude small high intensity spots caused by staining artefacts. The coordinates of the centre of mass of the identified condensations were then used to calculate the distance between neighbouring condensations (inter-condensation distance), the number of condensations per unit length and the number of condensations per unit area. Neighbouring condensations were determined as nearest neighbours in the x-direction (along the long axis of the island). The ‘distance from centre line’-parameter was measured as the shortest distance from the centre of mass of each condensation to an imaginary line drawn through the centre of the adhesive island along its long axis. For the non-patterned substrates, 300 μm wide strips were imaged and analysed in the same manner as the adhesive islands. All parameters were measured per island and then averaged over a range of replicate islands indicated in the figure legends.
Figure 2.
Patterning of condensations under geometric constraints. (a) Fluorescent micrographs of PNA lectin staining (green) indicating mesenchymal condensations on linear islands of varying width and on non-patterned substrates after 2.5 days in culture, as used for quantitative analysis. (b) Representative intensity plot of PNA lectin staining of cells cultured on a 150 μm wide island for 2.5 days, indicating the pixel intensity along a line drawn along the long axis of the island. The horizontal red line indicates the threshold used for quantitative image analysis to identify the location of the condensations. (c) The inter-condensation distance, the distance between neighbouring condensations, is plotted against the width of the adhesive islands. Linear regression analysis shows a statistically significant trend between the inter-condensation distance and the island width ((F = 34.74) > (Fcritical = 7), p < 0.0125). Data represent mean ± s.d. from n ≥ 7 8-mm long islands from two independent experiments. (d) Plot of the number of condensations per unit length of adhesive island as a function of the island width. Linear regression analysis shows a statistically significant trend between the number of condensations per unit length and the island width ((F = 27.22) > (Fcritical = 7.09), p < 0.0125). Data represent mean ± s.d. from n ≥ 6 8-mm long islands from two independent experiments. (e) The number of condensations per unit area of adhesive island, obtained by dividing the number of condensations per unit length by the island width, is plotted against island width. Linear regression analysis shows a statistically significant trend between the number of condensations per unit area and the island width ((F = 103.47) > (Fcritical = 7.09), p < 0.0125). Data represent mean ± s.d. from n ≥ 6 8-mm long islands from two independent experiments. (f) The distance to the centre line is the average distance of the centre of mass of the condensations to a line drawn through the middle of the adhesive island along its long axis. The distance to the centre line is plotted against the island width. Linear regression analysis shows a statistically significant trend between the distance to the centre line and the island width ((F = 63.77) > (Fcritical = 7.4), p < 0.0125). Data represent mean ±s.d. from n ≥ 5 8-mm long islands from two independent experiments. (Online version in colour.)
2.5. Proliferation assay
To assess proliferation, a ClickiT EdU Alexa Fluor 555 kit (Invitrogen) was used. Samples were incubated with 5 μM EdU for 4 h. For imaging, the samples were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and stained following the manufacturer's protocol for 30 min at room temperature. Nuclei were counterstained with 2 μg ml−1 Hoechst 33342. The percentage of proliferating cells was quantified by flow cytometry. After incubation with EdU, the cells were trypsinized to create a single-cell suspension, fixed in 4% paraformaldehyde in PBS and stained following the manufacturer's protocol for 30 min at room temperature. Flow analysis was performed on a BD LSR II Analyzer.
2.6. Glycosaminoglycan quantification
To quantify the deposition of GAGs, a colourimetric dimethyl methylene blue (DMB) assay was used. GAG deposition was normalized to DNA content to obtain GAG/DNA ratios. After 2.5 days in culture, the samples were digested in papain digestion buffer containing 10 mM l-cysteine and 0.125 mg ml−1 papain in PBE buffer (100 mM sodium phosphate buffer, 10 mM Na2EDTA, pH 6.5) and incubated for 24 h at 65°C. Directly after digestion, the DNA content of the samples was determined using a PicoGreen dsDNA assay kit (Invitrogen) using calf thymus DNA as a standard. A 200× 1,9-dimethyl methylene blue stock solution (3.2 mg ml−1) in 100% ethanol was made to enhance solubility. GAG content was determined by mixing 10 μl sample with 250 μl DMB staining solution (2.37 mg ml−1 NaCl, 3.04 mg ml−1 glycine, 16 μg ml−1 1,9-dimethyl methylene blue, pH 3) and reading the absorbance at 590 and 525 nm using a BioTek Synergy HT plate reader.
2.7. Sox9 expression
Expression of Sox9 was analysed by flow cytometry. After fixation and permeabilization, cells were probed with a rabbit-anti-Sox9 polyclonal antibody (Millipore) at 5 μg ml−1. Cells were subsequently stained with an Alexa Fluor 647 conjugated goat-anti-rabbit IgG (Invitrogen). Flow analysis was performed on a BD LSR II Analyzer. Mean fluorescent intensities of cell populations retrieved at different time points, as well as freshly isolated cells (‘day 0’) were used as a measure of Sox9 expression.
2.8. Whole embryo sectioning and analysis
Chicken embryos at Hamburger Hamilton stage 22 were euthanized, washed in PBS and fixed in 4% paraformaldehyde in PBS at 4°C overnight. After multiple PBS washes, the embryos were incubated in 30% sucrose in PBS at 4°C overnight and then in a 50–50 mixture of 30% sucrose in PBS and OCT (Optimal Cutting Temperature, Tissue Tek) at 4°C overnight. Embryos were subsequently embedded in OCT and frozen on dry ice. A Leica cryotome was used to cut 10 μm frozen sections in either the sagittal or the transverse plane of the embryo. Sections were stained with PNA lectin and Hoechst 33342 as described above, and imaged using a Nikon E800 upright microscope. Image analysis was performed using ImageJ to measure the geometric constraint provided to the mesenchymal cells in transverse sections, as well as the inter-condensation distance in sagittal sections.
2.9. Statistics
Linear regression analysis and ANOVA tests were performed using Stata v. 13.0. Linear regression analysis was performed on data for the inter-condensation distance, the number of condensations per unit length, number of condensations per unit area, distance from centre line, Sox9 mean fluorescent intensity, Sox9 pixel intensity and GAG deposition, to test whether a statistical trend was observed between these variables and the island width, by testing whether the slope of the predicted curve was statistically different from zero. The F statistic from each regression analysis was compared with the critical F-value at a significance level of α = 0.05. Since the inter-condensation distance, the number of condensation per unit length, the number of condensation per unit area and the distance from centreline are derived from the same set of images, a multiple comparison correction is required. For these datasets, the critical F-value was therefore determined at a significance level of α = 0.05/4 = 0.0125 (Bonferroni correction). One-way ANOVA tests were performed on the data for initial cell density and proliferation on the different island widths to test for significant differences between any of the groups using a significance level of α = 0.05. One-tailed unpaired Student's t-tests were performed to test whether the Sox9 mean fluorescent intensity, Sox9 pixel intensity and the GAG deposition on 100 μm adhesive islands was significantly different from that on non-patterned substrates.
3. Results
3.1. Experimental model system
In order to study the role of geometric boundary conditions in the formation and patterning of mesenchymal condensations, a microchannel patterning technique was employed to create adhesive islands of specific dimensions (figure 1a). The adhesive islands were designed to be 8 mm long and 100, 150, 200, 250 or 300 μm wide, while a continuous FN-coated substrate was used as a negative control (figure 1b). The microchannel technique resulted in robust and reproducible patterns, as it was confirmed that the actual island widths closely resembled the desired dimensions (figure 1c). Pre-chondrogenic cells were freshly isolated from chicken embryonic limb buds, as it has been demonstrated that these cells are able to form mesenchymal condensations in vitro in micromass culture [37]. We adapted the classical micromass culture to conform to our patterned substrates and it was confirmed that typical condensation took place on the adhesive islands (figure 1d,e). A typical mesenchymal condensation is indicated by a local increase in cell density, positive staining with PNA lectin [15], the deposition of abundant FN matrix and increased expression of the adhesion molecule NCAM [32].
3.2. Patterning of condensations
To investigate the effect of geometric constraints on patterning of condensations, pre-chondrogenic cells were cultured on linear adhesive islands of varying width or on non-patterned substrates. PNA lectin staining revealed mesenchymal condensations (figure 2a) and the pixel intensity landscapes were used to identify the locations of individual condensations (figure 2b). The distance between the centre of mass of neighbouring condensations, the inter-condensation distance, was measured and compared between the different island sizes. A significant trend was observed, where the inter-condensation distance decreased with increasing island width (figure 2c). The inter-condensation distance on non-patterned substrates was smaller than for any of the linear islands. As the inter-condensation distance becomes shorter and the condensations have more room to disperse across the width of the island, as the islands become wider, the number of condensations per millimetre length of adhesive island increased significantly with increasing island width (figure 2d). As wider islands contain more cells per millimetre length, the number of condensations per unit area was also determined. Interestingly, a significant downward trend was observed between the number of condensations per unit area and the island width (figure 2e). The number of condensations per unit area on the non-patterned substrates was higher than the trend would predict, possibly because cells can be drawn in from outside the quantified region, as it is a continuous culture, resulting in a higher number of condensations. Additionally, the alignment of condensations was evaluated by measuring the distance of the centre of mass of each condensation from the centre line of the island; a line through the middle of the island along its long axis (figure 2f). The average distance from the centre line increased significantly with increasing island width; the condensations on the thinnest island (100 μm) had the highest degree of alignment.
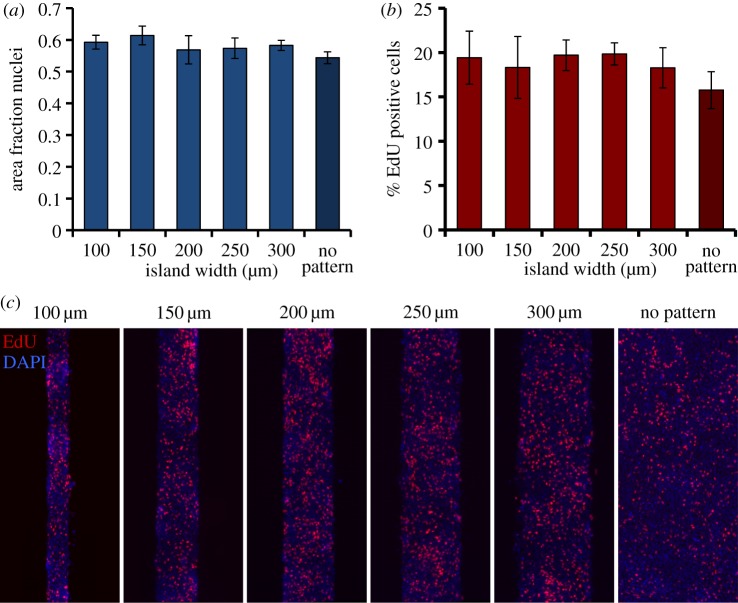
3.3. Cell density and proliferation
As the process of condensation is highly dependent on cell density, the initial cell densities on the differently sized islands was quantified to determine whether the observed differences in patterning were simply caused by unequal cell attachment. Initial cell densities on the patterned and non-patterned substrates were found to be not significantly different (figure 3a). Furthermore, we investigated the potential effect of the geometric constraints on the proliferation rate as this could result in differences in cell density over time between the differently sized islands, or inhomogeneous cell densities within the islands. Proliferation was assessed after 48 h of culture, while the cells were in the process of condensation. There was no statistically significant difference in proliferation rate between any of the various island sizes (figure 3b). Additionally, no regional differences in proliferation were observed on any of the island geometries (figure 3c). Apoptosis is assumed not to play a considerable role, as previous studies have shown that apoptosis rates at early time points of in vitro condensation are negligible [49].
Figure 3.
Cell density and proliferation. (a) The area fraction occupied by cell nuclei after 1 h in culture, as a measure of initial cell density, is plotted against the island width. None of the differences between conditions is statistically significant. Data represent mean ± s.d., n = 3. (b) The percentage of proliferating cells at 48 h of culture on adhesive islands, as identified by incorporation of EdU during 4 h of culture, is plotted against the island width. None of the differences between conditions is statistically significant. Data represent mean ± s.d., n = 4. (c) Fluorescent micrographs of cells cultured on adhesive islands for 48 h, stained for incorporation of EdU (red) and cell nuclei (blue) after 4 h of incubation with EdU. (Online version in colour.)
3.4. Chondrogenic differentiation
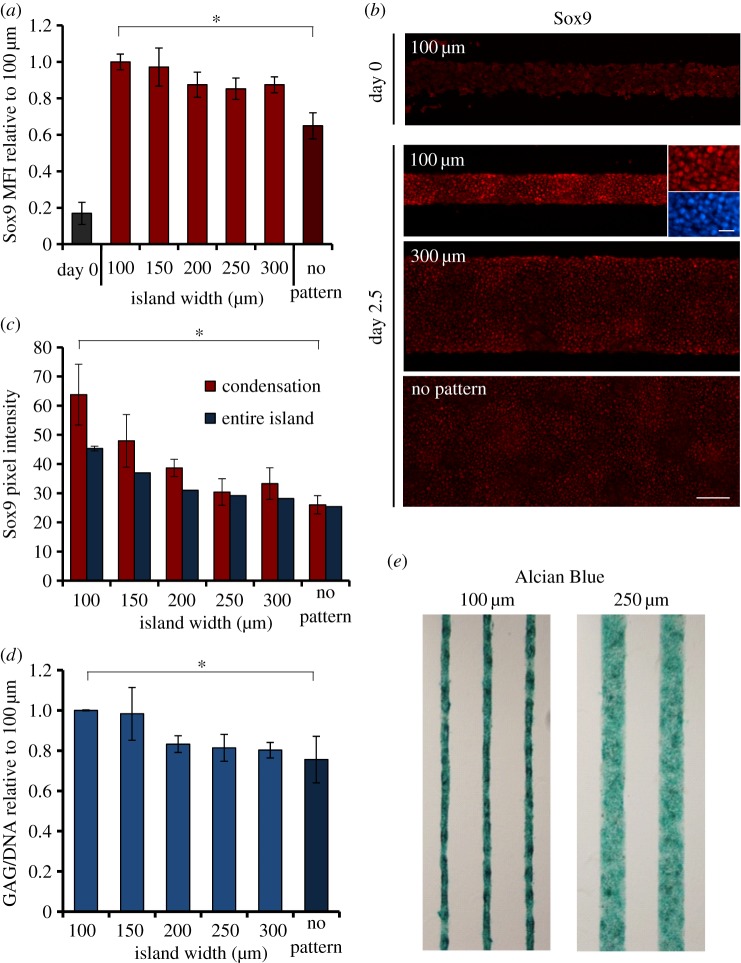
Mesenchymal condensation is believed to be the initial step in skeletal development, followed by chondrogenic commitment of the involved cells. We thus investigated whether the geometric constraints imposed on the pre-chondrogenic cells affected their chondrogenic differentiation. Firstly, the expression of the transcription factor Sox9 was investigated, as it is known to play a major role in chondrogenesis during embryonic development and is often used as an early indicator of chondrogenic commitment [50]. Expression of Sox9 at 2.5 days of culture, under all conditions, was increased compared with day 0 (figure 4a,b). Additionally, at 2.5 days a significant difference was observed between the non-patterned control substrates and the 100 μm islands. When expression levels were compared between the different island sizes, it was found that Sox9 expression decreased when the cells were cultured on wider islands, showing a statistically significant downward trend. Immunohistochemistry revealed that at 2.5 days Sox9 was located in the nucleus of the cells (figure 4b), as expected. The average intensity of the staining of the entire island, as a measure of Sox9 expression, decreased with increasing island width (figure 4b,c), consistent with the trend observed by flow cytometry (figure 4a). While the intensity within the condensations was higher than the average of the entire island on thinner islands, this difference diminished with increasing island width (figure 4c). In addition to the early transcription factor Sox9, the deposition of GAGs was quantified. Similar to Sox9 expression, GAG deposition decreased significantly with increasing island width and also the difference between the 100 μm island and the non-patterned substrates was statistically significant (figure 4d,e).
Figure 4.
Chondrogenic differentiation under geometric constraints. (a) The mean fluorescent intensity of Sox9-stained cells was measured by flow analysis at day 0 (directly after isolation; grey column) and after 2.5 days in culture on adhesive islands of a range of widths and on non-patterned substrates. Data are normalized to the ‘100 μm’ condition. Linear regression analysis shows a statistically significant trend of Sox9 expression at 2.5 days with the island width ((F = 11.67) > (Fcritical = 4.41), p < 0.05). Data represent mean ± s.d., n = 4, *p < 0.0005. (b) Fluorescent micrographs of immunohistochemistry of Sox9 (red) at day 0 (100 μm, top) and day 2.5 (100, 300 μm and no pattern). A higher magnification of Sox9 (red) and DAPI (nuclei, blue) of the 100 μm island shows the nuclear localization of Sox9 at day 2.5. For the lower magnification, scale bar is 100 μm. For the higher magnification, scale bar is 20 μm. (c) The intensity of Sox9 staining was quantified on the entire island and in the condensations and plotted against the islands width. Linear regression analysis shows a statistically significant trend of Sox9 immonufluorescent intensity on the entire island, with the island width ((F = 35.03) > (Fcritical = 7.71), p < 0.05). Data represent mean ± s.d., n = 10, *p < 0.0005. (d) GAG content was quantified for 2.5-day cultures and normalized to DNA content. The data are presented relative to the ‘100 μm’ condition. Linear regression analysis shows a statistically significant trend of GAG/DNA content with the island width ((F = 16.74) > (Fcritical = 4.75), p < 0.05). Data represent mean ± s.d., n = 3, *p < 0.05. (e) Colour micrographs of 2.5-day cultures stained with Alcian Blue, indicating the deposition of GAGs on 100 versus 250 μm islands. (Online version in colour.)
3.5. Time course of proliferation, condensation and differentiation
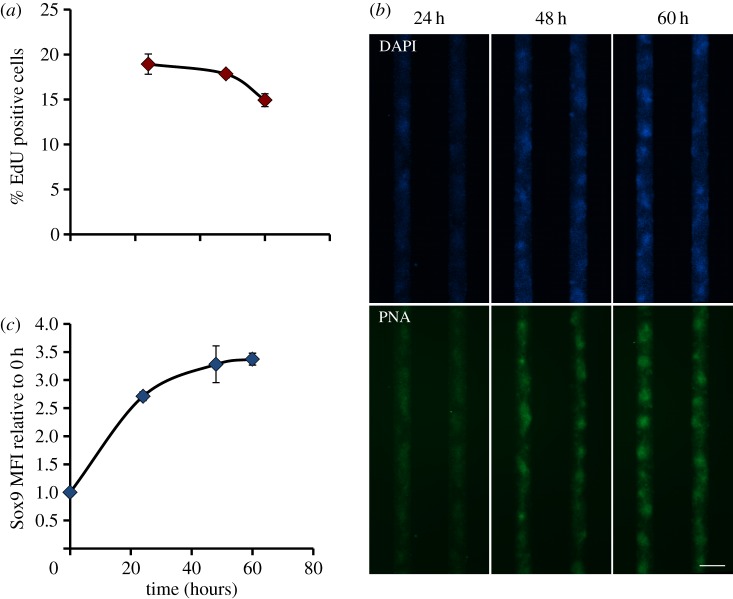
In order to gain a better understanding of the temporal sequence of the processes of proliferation, condensation and differentiation on the adhesive islands, these processes were investigated at different time points during the 2.5-day culture. First, it was found that the rate of proliferation was high initially and decreased over time (figure 5a). At 24 h, areas of increased cell density started to appear, however typical condensations—indicated by PNA staining—were still absent (figure 5b). At 48 h, condensations were visible and they subsequently became more defined over time. In line with the decreasing rate of proliferation and the appearance of condensations, differentiation—indicated by the relative expression of Sox9—increased over time (figure 5c).
Figure 5.
Time course of proliferation, condensation and differentiation. (a) The percentage of proliferating cells on 150 μm wide adhesive islands, as identified by incorporation of EdU during 4 h of culture, is plotted against time. Data represent mean ± s.d., n = 3. (b) Fluorescent micrographs of pre-chondrogenic cells cultured on 150 μm wide adhesive islands stained with DAPI (blue, cell nuclei), and PNA lectin (green, indicating condensations) at 24, 48 and 60 h. Scale bar is 200 μm. (c) The Sox9 mean fluorescent intensity of cells retrieved from 150 μm wide adhesive islands, as measured by flow cytometry, is plotted against time. Data represent mean ± s.d., n = 3. (Online version in colour.)
3.6. Impact of added morphogens
In vivo, tissue development takes place in the presence of various morphogens. To test the effect of specific morphogens on the process of condensation under geometric constraints, cultures were supplemented with varying concentrations and combinations of TGF-β3, BMP2 and FGF8, factors that are known to be involved in the development of the vertebral column [51–54]. In control cultures, after 2.5 days of culture, condensations can be readily identified by increased cell density and positive PNA staining (electronic supplementary material, figure S1). In the presence of a range of concentrations of TGF-β3 or BMP2 however, both the overall cell density and the extent of condensation were reduced. In the presence of FGF8, distinct local increases in cell density are discernable, however PNA staining is more faint. The combination of TGF-β3 and BMP2 resulted in a culture similar to the control condition, while TGF-β3 and FGF8 together lead to distinctive areas of highly increased cell density and considerable PNA staining.
3.7. Patterning of condensations in the chicken embryo
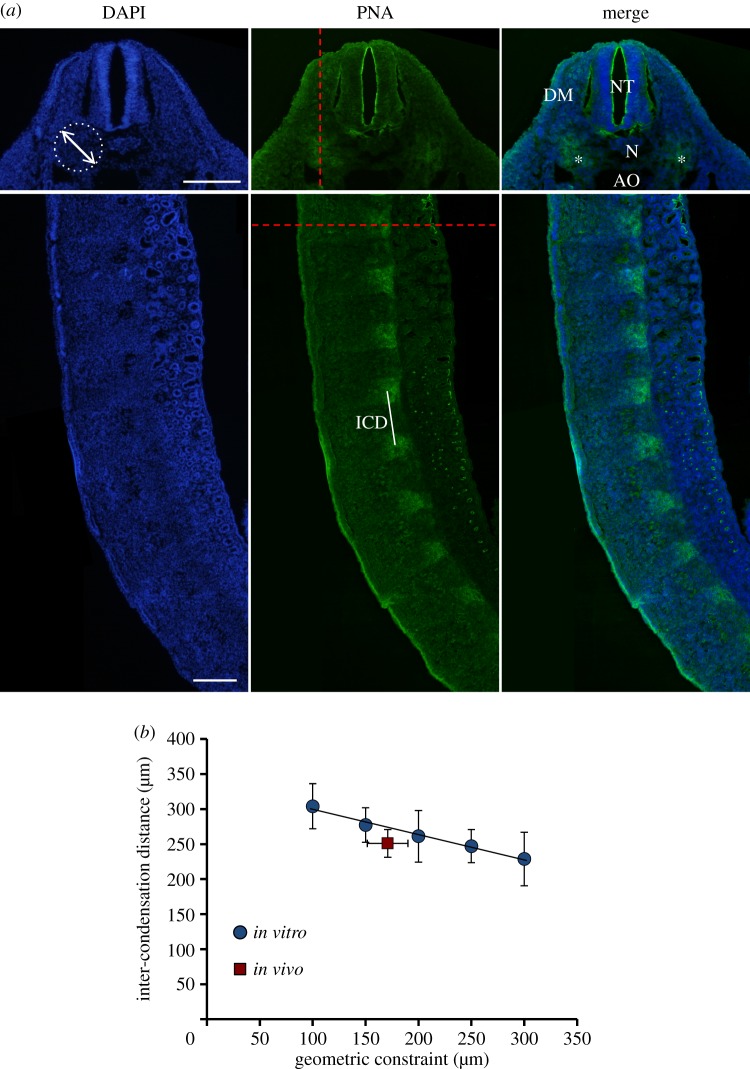
Finally, in order to relate the observed patterning of condensations in our in vitro model to patterning in vivo, the linear array of condensations of the developing vertebral column was investigated in chicken embryos (figure 6). At Hamburger Hamilton stage 22, the sclerotomal cells that will eventually give rise to the vertebral column have formed a linear pattern of condensations along the body axis, as indicated by PNA lectin staining (figure 6a). The neighbouring tissues that provide geometric constraints to this population of cells are the neural tube on the dorsal side, the epithelial dermamyotome on the dorsal–lateral sides and the notochord and aorta medially (figure 6a). At this stage, the width of the sclerotomal cell population constrained within the transverse plane was measured to be 171 ± 19 μm. The inter-condensation distance was measured in the sagittal plane and had a mean value of 251 ± 20 μm (figure 6b).
Figure 6.
Patterning of condensations in vivo. (a) Fluorescent micrographs of transverse (top) and sagittal (bottom) sections of a chicken embryo at HH stage 22. Cell nuclei are stained blue (left) and PNA lectin staining is indicated in green (middle). Red dashed line in transverse section indicates the cross-sectional plane of the sagittal section and vice-versa. The white dashed circle indicates the estimated geometric constraint of the population of sclerotomal cells. Neighbouring tissues that provide the spatial constraints are the neural tube (NT), the dermamyotome (DM), the notochord (N) and the aorta (AO). Mesenchymal condensations in the transverse plane are indicated with an asterisk (*). The inter-condensation distance (ICD) is indicated in the sagittal section. Scale bars, 200 μm. (b) The inter-condensation distance is plotted against the geometric constraint for the in vitro model (also in figure 2c) as well as the chicken embryo (in vivo). For the in vitro model, the geometric constraint on the x-axis indicates the island width, while for the in vivo case it indicates the measured width of the population of sclerotomal cells (indicated by the white dashed circle in (a)). Data represent mean ± s.d. from 20 measurements from three embryos. (Online version in colour.)
4. Discussion
The role of physical cues in the creation of the characteristic pattern of the vertebral column has not been widely explored. This study addresses the hypothesis that geometric constraints play a role in determining the linear pattern of condensations at the onset of the development of the vertebral column. By employing a novel in vitro model, we found that the spacing and alignment of condensations, as well as the number of condensation per unit area, were influenced by the geometry of the cell adhesive pattern. Interestingly, measurements of the spatial constraints of the developing vertebral column in the chicken embryo and its pattern of condensations were shown to closely fit into the quantitative relation between geometric constraints and inter-condensation distance found in vitro.
The distance between neighbouring condensations, the inter-condensation distance, was shown to increase significantly when skeletal precursor cells were cultured on linear adhesive islands of decreasing width (figure 2c). Condensation is defined by an increase in local cell density [16,19]. A minimum number of cells must be drawn together to create a condensation [55] and the process is thus dependent on cell availability. On the thinner islands, fewer cells are available along the short axis, so this could explain why the inter-condensation distance increases and the number of condensations per unit island length decreases with decreasing island width. If the spacing of condensations was only a function of cell availability, the number of condensations per unit area would presumably be constant. We found a different trend, however, implying that the mechanism that governs the patterning of condensations is more complex. The process of condensation involves a fine balance between the cohesiveness of cells versus their interaction with the extracellular matrix. As varying geometric constraints will alter the ratio between perimeter and surface area, this might shift the balance between cell–cell versus cell–matrix contacts and the cellular forces applied at those contacts, as indicated by several investigations on force generation by multicellular cultures [46,47,56]. Theoretical models have indicated these forces as key players in the condensation process [24,57,58]. Further investigations are needed to unravel the mechanisms that cause the difference in patterning of condensations under varying geometric constraints.
Interestingly, the in vivo inter-condensation distance and geometric constraints of the developing vertebral column, measured in chicken embryos, were found to closely fit into the quantitative relation described by the in vitro model (figure 6). The in vitro model is limited in its capacity to capture all aspects of the in vivo condensation process, as it does not recapitulate the full three-dimensional environment or possible signalling events from nearby epithelium implied to be involved in the process of condensation [59]. Remarkably, despite these discrepancies, the isolated mesenchymal cells are able to form condensations on the provided adhesive islands in specific patterns that match the condensation pattern in the developing vertebral column in vivo. This can thus be seen as a process of self-organization, as in vitro this is not orchestrated by signalling from neighbouring tissues.
In the developing vertebral column, the condensations are not only consistently spaced, but they are also accurately aligned along the body axis, which is critical for the eventual structure of the spine. The distance of the centre of mass of each condensation to the centre line of the island was quantified in this study (figure 2f) as a measure of alignment. A significant trend was found, with the distance diminishing for the thinnest islands. The enhanced alignment of condensations on thinner islands reflects the more limited space for the condensations to disperse. Similarly, if the population of cells in the in vivo environment of the developing spine is spatially restricted, the condensations may also be forced to align.
Geometric boundary conditions have been previously shown to play a role in a variety of cellular events. In the context of the developing embryo, it was shown that elongation of the body axis in Xenopus embryos is a process of self-organization controlled by physical alignment cues from the edges of the cell population [60]. Additionally, anterior–posterior axis polarization in mouse embryos is indicated to be triggered by the boundary conditions that the uterine wall provides to the early embryo [61]. In this study, we show the effect of geometric constraints on the supracellular process of patterning of mesenchymal condensations.
Morphogens are soluble factors that are thought to play an important role in pattern formation and tissue development in vivo. The effect of three relevant morphogens—TGF-β3, BMP2 and FGF8—on the condensation process was tested on 171 μm adhesive islands, based on the measured tissue width in vivo (figure 6). The addition of TGF-β3 or BMP2 had an inhibiting effect on proliferation and condensation, as indicated by the decreased cell density and faint PNA staining when compared with control cultures (electronic supplementary material, figure S1). FGF8, either alone or in combination with the other morphogens, caused a strong increase in local cell densities, however the faint PNA staining in the majority of these conditions indicates that these areas of increased cell density might not represent characteristic condensations. Possibly, FGF8 reduces cell–matrix adhesion, causing the cells to cluster in a non-specific manner. In contrast to the static presence of morphogens in this study, during tissue development in vivo morphogens are secreted at specific locations at specific times, resulting in complex spatial and temporal gradients [53,62]. More complex model systems are required to further elucidate the interplay between geometric constraints and morphogen gradients in patterning and differentiation.
To test whether chondrogenic lineage commitment was affected by the imposed geometric constraints in our model system, the expression of Sox9 and extent of GAG deposition were also investigated (figure 4). Both Sox9 expression and GAG deposition decreased significantly as the islands became wider, indicating that the overall chondrogenic differentiation was enhanced on substrates with tighter geometric constraints. For both differentiation markers, a significant difference was observed between the non-patterned control substrates and the 100 μm islands. These findings correspond with the observation that the number of condensations per unit area is increased on thinner islands. Moreover, on the thinner islands the difference in intensity of Sox9 expression between condensations and the rest of the island was more pronounced than on wider islands. This might indicate that the condensations are more mature, in terms of chondrogenic differentiation, under tighter geometric constraints. It has been shown previously that the extent of chondrogenic differentiation from condensations can be modulated by various soluble factors [63,64], but the observed effect of geometric constraints provides new insight into the role of physical cues in the differentiation process.
Taken together, this study suggests a role for geometric boundary conditions in regulating skeletal patterning during embryonic development in a process of self-organization. The pattern of condensations in the chicken embryo was shown to closely fit the relationship found in vitro. Different species with varying dimensions could be studied in the future to investigate whether this relationship extends to other vertebrates as well. Moreover, further experimentation could provide more insight into the mechanisms involved and the possible impact of geometric constraints on additional developmental processes. This line of investigations is likely to provide a more profound understanding of the processes of tissue patterning and morphogenesis.
Acknowledgements
The authors would like to thank Alan Rodrigues and Clifford Tabin for sharing their expertise on chicken embryo limb bud cell isolations, Cristiana Cunha for help with flow cytometry experiments and Max Darnell for help with statistical analysis.
Funding statement
This work was funded by National Institute of Health (R37 DE013033), ZonMW-VICI grant no. 918.11.635 (The Netherlands), and Research Institute MOVE at the VU University, Amsterdam. A.S.M. acknowledges funding from the National Science Foundation Graduate Research Fellowships Program, and T.H.S. was supported by a visiting scholarship from the Wyss Institute for Biologically Inspired Engineering at Harvard University.
References
- 1.Gjorevski N, Nelson CM. 2010. The mechanics of development: models and methods for tissue morphogenesis. Birth Defects Res. C Embryo Today 90, 193–202. ( 10.1002/bdrc.20185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingber DE. 2006. Mechanical control of tissue morphogenesis during embryological development. Int. J. Dev. Biol. 50, 255–266. ( 10.1387/ijdb.052044di) [DOI] [PubMed] [Google Scholar]
- 3.Mammoto T, Ingber DE. 2010. Mechanical control of tissue and organ development. Development 137, 1407–1420. ( 10.1242/dev.024166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowlan NC, Murphy P, Prendergast PJ. 2007. Mechanobiology of embryonic limb development. Ann. N Y Acad. Sci. 1101, 389–411. ( 10.1196/annals.1389.003) [DOI] [PubMed] [Google Scholar]
- 5.Wozniak MA, Chen CS. 2009. Mechanotransduction in development: a growing role for contractility. Nat. Rev. Mol. Cell Biol. 10, 34–43. ( 10.1038/nrm2592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savin T, Kurpios NA, Shyer AE, Florescu P, Liang H, Mahadevan L, Tabin CJ. 2011. On the growth and form of the gut. Nature 476, 57–62. ( 10.1038/nature10277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shyer AE, Tallinen T, Nerurkar NL, Wei Z, Gil ES, Kaplan DL, Tabin CJ, Mahadevan L. 2013. Villification: how the gut gets its villi. Science 342, 212–218. ( 10.1126/science.1238842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beloussov LV, Grabovsky VI. 2006. Morphomechanics: goals, basic experiments and models. Int. J. Dev. Biol. 50, 81–92. ( 10.1387/ijdb.052056lb) [DOI] [PubMed] [Google Scholar]
- 9.Brodland GW, et al. 2010. Video force microscopy reveals the mechanics of ventral furrow invagination in Drosophila. Proc. Natl Acad. Sci. USA 107, 22 111–22 116. ( 10.1073/pnas.1006591107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gjorevski N, Nelson CM. 2010. Endogenous patterns of mechanical stress are required for branching morphogenesis. Integr. Biol. (Camb) 2, 424–434. ( 10.1039/c0ib00040j) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christ B, Huang R, Wilting J. 2000. The development of the avian vertebral column. Anat. Embryol. (Berl) 202, 179–194. ( 10.1007/s004290000114) [DOI] [PubMed] [Google Scholar]
- 12.Smith LJ, Nerurkar NL, Choi KS, Harfe BD, Elliott DM. 2011. Degeneration and regeneration of the intervertebral disc: lessons from development. Dis. Model Mech. 4, 31–41. ( 10.1242/dmm.006403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLise AM, Fischer L, Tuan RS. 2000. Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage 8, 309–334. ( 10.1053/joca.1999.0306) [DOI] [PubMed] [Google Scholar]
- 14.Hall BK. 1987. Earliest evidence of cartilage and bone development in embryonic life. Clin. Orthop. Relat. Res. 255, 255–272. [PubMed] [Google Scholar]
- 15.Hall BK, Miyake T. 1992. The membranous skeleton: the role of cell condensations in vertebrate skeletogenesis. Anat. Embryol. (Berl) 186, 107–124. ( 10.1007/BF00174948) [DOI] [PubMed] [Google Scholar]
- 16.Thorogood PV, Hinchliffe JR. 1975. An analysis of the condensation process during chondrogenesis in the embryonic chick hind limb. J. Embryol. Exp. Morphol. 33, 581–606. [PubMed] [Google Scholar]
- 17.Grüneberg H. 1963. The pathology of development: a study of inhereted skeletal disorders in animals. Oxford, UK: Blackwells Scientific Publications. [Google Scholar]
- 18.Hall BK, Miyake T. 2000. All for one and one for all: condensations and the initiation of skeletal development. Bioessays 22, 138–147. () [DOI] [PubMed] [Google Scholar]
- 19.Hall BK, Miyake T. 1995. Divide, accumulate, differentiate: cell condensation in skeletal development revisited. Int. J. Dev. Biol. 39, 881–893. [PubMed] [Google Scholar]
- 20.Janners MY, Searls RL. 1970. Changes in rate of cellular proliferation during the differentiation of cartilage and muscle in the mesenchyme of the embryonic chick wing. Dev. Biol. 23, 136–165. ( 10.1016/S0012-1606(70)80011-2) [DOI] [PubMed] [Google Scholar]
- 21.Summerbell D, Wolpert L. 1972. Cell density and cell division in the early morphogenesis of the chick wing. Nat. New Biol. 239, 24–26. ( 10.1038/newbio239024a0) [DOI] [PubMed] [Google Scholar]
- 22.Cui C. 2005. Dynamics of cell movement and tissue motion in gastrulation and micromass cell culture. Bloomington, IN: Indiana University. [Google Scholar]
- 23.Newman SA, Frenz DA, Tomasek JJ, Rabuzzi DD. 1985. Matrix-driven translocation of cells and nonliving particles. Science 228, 885–889. ( 10.1126/science.4001925) [DOI] [PubMed] [Google Scholar]
- 24.Oster GF, Murray JD, Maini PK. 1985. A model for chondrogenic condensations in the developing limb: the role of extracellular matrix and cell tractions. J. Embryol. Exp. Morphol. 89, 93–112. [PubMed] [Google Scholar]
- 25.Dessau W, von der Mark H, von der Mark K, Fischer S. 1980. Changes in the patterns of collagens and fibronectin during limb-bud chondrogenesis. J. Embryol. Exp. Morphol. 57, 51–60. [PubMed] [Google Scholar]
- 26.Knudson CB, Toole BP. 1985. Changes in the pericellular matrix during differentiation of limb bud mesoderm. Dev. Biol. 112, 308–318. ( 10.1016/0012-1606(85)90401-4) [DOI] [PubMed] [Google Scholar]
- 27.Kulyk WM, Upholt WB, Kosher RA. 1989. Fibronectin gene expression during limb cartilage differentiation. Development 106, 449–455. [DOI] [PubMed] [Google Scholar]
- 28.Stern CD. 1984. Mini-review: hyaluronidases in early embryonic development. Cell Biol. Int. Rep. 8, 703–717. ( 10.1016/0309-1651(84)90108-5) [DOI] [PubMed] [Google Scholar]
- 29.Tomasek JJ, Mazurkiewicz JE, Newman SA. 1982. Nonuniform distribution of fibronectin during avian limb development. Dev. Biol. 90, 118–126. ( 10.1016/0012-1606(82)90217-2) [DOI] [PubMed] [Google Scholar]
- 30.Oberlender SA, Tuan RS. 1994. Expression and functional involvement of N-cadherin in embryonic limb chondrogenesis. Development 120, 177–187. [DOI] [PubMed] [Google Scholar]
- 31.Tavella S, Raffo P, Tacchetti C, Cancedda R, Castagnola P. 1994. N-CAM and N-cadherin expression during in vitro chondrogenesis. Exp. Cell Res. 215, 354–362. ( 10.1006/excr.1994.1352) [DOI] [PubMed] [Google Scholar]
- 32.Widelitz RB, Jiang TX, Murray BA, Chuong CM. 1993. Adhesion molecules in skeletogenesis: II. Neural cell adhesion molecules mediate precartilaginous mesenchymal condensations and enhance chondrogenesis. J. Cell Physiol. 156, 399–411. ( 10.1002/jcp.1041560224) [DOI] [PubMed] [Google Scholar]
- 33.Hui TY, Cheung KM, Cheung WL, Chan D, Chan BP. 2008. In vitro chondrogenic differentiation of human mesenchymal stem cells in collagen microspheres: influence of cell seeding density and collagen concentration. Biomaterials 29, 3201–3212. ( 10.1016/j.biomaterials.2008.04.001) [DOI] [PubMed] [Google Scholar]
- 34.Mauck RL, Seyhan SL, Ateshian GA, Hung CT. 2002. Influence of seeding density and dynamic deformational loading on the developing structure/function relationships of chondrocyte-seeded agarose hydrogels. Ann. Biomed. Eng. 30, 1046–1056. ( 10.1114/1.1512676) [DOI] [PubMed] [Google Scholar]
- 35.Centola M, Tonnarelli B, Scharen S, Glaser N, Barbero A, Martin I. 2013. Priming 3D cultures of human mesenchymal stromal cells toward cartilage formation via developmental pathways. Stem Cells Dev. 22, 2849–2858. ( 10.1089/scd.2013.0216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ten Berge D, Brugmann SA, Helms JA, Nusse R. 2008. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development 135, 3247–3257. ( 10.1242/dev.023176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahrens PB, Solursh M, Reiter RS. 1977. Stage-related capacity for limb chondrogenesis in cell culture. Dev. Biol. 60, 69–82. ( 10.1016/0012-1606(77)90110-5) [DOI] [PubMed] [Google Scholar]
- 38.DeLise AM, Stringa E, Woodward WA, Mello MA, Tuan RS. 2000. Embryonic limb mesenchyme micromass culture as an in vitro model for chondrogenesis and cartilage maturation. Methods Mol. Biol. 137, 359–375. ( 10.1385/1-59259-066-7:359) [DOI] [PubMed] [Google Scholar]
- 39.Denker AE, Haas AR, Nicoll SB, Tuan RS. 1999. Chondrogenic differentiation of murine C3H10T1/2 multipotential mesenchymal cells: I. Stimulation by bone morphogenetic protein-2 in high-density micromass cultures. Differentiation 64, 67–76. ( 10.1046/j.1432-0436.1999.6420067.x) [DOI] [PubMed] [Google Scholar]
- 40.Denker AE, Nicoll SB, Tuan RS. 1995. Formation of cartilage-like spheroids by micromass cultures of murine C3H10T1/2 cells upon treatment with transforming growth factor-beta 1. Differentiation 59, 25–34. ( 10.1046/j.1432-0436.1995.5910025.x) [DOI] [PubMed] [Google Scholar]
- 41.Bhat R, Lerea KM, Peng H, Kaltner H, Gabius HJ, Newman SA. 2011. A regulatory network of two galectins mediates the earliest steps of avian limb skeletal morphogenesis. BMC Dev. Biol. 11, 6 ( 10.1186/1471-213X-11-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woods A, Wang G, Dupuis H, Shao Z, Beier F. 2007. Rac1 signaling stimulates N-cadherin expression, mesenchymal condensation, and chondrogenesis. J. Biol. Chem. 282, 23 500–23 508. ( 10.1074/jbc.M700680200) [DOI] [PubMed] [Google Scholar]
- 43.Thery M. 2010. Micropatterning as a tool to decipher cell morphogenesis and functions. J. Cell Sci. 123, 4201–4213. ( 10.1242/jcs.075150) [DOI] [PubMed] [Google Scholar]
- 44.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. 1997. Geometric control of cell life and death. Science 276, 1425–1428. ( 10.1126/science.276.5317.1425) [DOI] [PubMed] [Google Scholar]
- 45.Li B, Li F, Puskar KM, Wang JH. 2009. Spatial patterning of cell proliferation and differentiation depends on mechanical stress magnitude. J. Biomech. 42, 1622–1627. ( 10.1016/j.jbiomech.2009.04.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. 2005. Emergent patterns of growth controlled by multicellular form and mechanics. Proc. Natl Acad. Sci. USA 102, 11 594–11 599. ( 10.1073/pnas.0502575102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruiz SA, Chen CS. 2008. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells 26, 2921–2927. ( 10.1634/stemcells.2008-0432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamburger V, Hamilton H. 1951. A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49–92. ( 10.1002/jmor.1050880104) [DOI] [PubMed] [Google Scholar]
- 49.Coleman CM, Tuan RS. 2003. Growth/differentiation factor 5 enhances chondrocyte maturation. Dev. Dyn. 228, 208–216. ( 10.1002/dvdy.10369) [DOI] [PubMed] [Google Scholar]
- 50.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. 1999. Sox9 is required for cartilage formation. Nat. Genet. 22, 85–89. ( 10.1038/8792) [DOI] [PubMed] [Google Scholar]
- 51.Brent AE, Tabin CJ. 2004. FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development 131, 3885–3896. ( 10.1242/dev.01275) [DOI] [PubMed] [Google Scholar]
- 52.Huang R, Stolte D, Kurz H, Ehehalt F, Cann GM, Stockdale FE, Patel K, Christ B. 2003. Ventral axial organs regulate expression of myotomal Fgf-8 that influences rib development. Dev. Biol. 255, 30–47. ( 10.1016/S0012-1606(02)00051-9) [DOI] [PubMed] [Google Scholar]
- 53.Roberts AB, Sporn MB. 1992. Differential expression of the TGF-beta isoforms in embryogenesis suggests specific roles in developing and adult tissues. Mol. Reprod. Dev. 32, 91–98. ( 10.1002/mrd.1080320203) [DOI] [PubMed] [Google Scholar]
- 54.Wan M, Cao X. 2005. BMP signaling in skeletal development. Biochem. Biophys. Res. Commun. 328, 651–657. ( 10.1016/j.bbrc.2004.11.067) [DOI] [PubMed] [Google Scholar]
- 55.Cottrill CP, Archer CW, Wolpert L. 1987. Cell sorting and chondrogenic aggregate formation in micromass culture. Dev. Biol. 122, 503–515. ( 10.1016/0012-1606(87)90314-9) [DOI] [PubMed] [Google Scholar]
- 56.Kim JH, et al. 2013. Propulsion and navigation within the advancing monolayer sheet. Nat. Mater. 12, 856–863. ( 10.1038/nmat3689) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bard JB. 1990. Traction and the formation of mesenchymal condensations in vivo. Bioessays 12, 389–395. ( 10.1002/bies.950120809) [DOI] [PubMed] [Google Scholar]
- 58.Murray JD, Oster GF. 1984. Cell traction models for generating pattern and form in morphogenesis. J. Math. Biol. 19, 265–279. ( 10.1007/BF00277099) [DOI] [PubMed] [Google Scholar]
- 59.Mammoto T, et al. 2011. Mechanochemical control of mesenchymal condensation and embryonic tooth organ formation. Dev. Cell 21, 758–769. ( 10.1016/j.devcel.2011.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Green JB, Dominguez I, Davidson LA. 2004. Self-organization of vertebrate mesoderm based on simple boundary conditions. Dev. Dyn. 231, 576–581. ( 10.1002/dvdy.20163) [DOI] [PubMed] [Google Scholar]
- 61.Hiramatsu R, Matsuoka T, Kimura-Yoshida C, Han SW, Mochida K, Adachi T, Takayama S, Matsuo I. 2013. External mechanical cues trigger the establishment of the anterior–posterior axis in early mouse embryos. Dev. Cell 27, 131–144. ( 10.1016/j.devcel.2013.09.026) [DOI] [PubMed] [Google Scholar]
- 62.Tabata T, Takei Y. 2004. Morphogens, their identification and regulation. Development 131, 703–712. ( 10.1242/dev.01043) [DOI] [PubMed] [Google Scholar]
- 63.Hatakeyama Y, Tuan RS, Shum L. 2004. Distinct functions of BMP4 and GDF5 in the regulation of chondrogenesis. J. Cell Biochem. 91, 1204–1217. ( 10.1002/jcb.20019) [DOI] [PubMed] [Google Scholar]
- 64.Sohn P, Cox M, Chen D, Serra R. 2010. Molecular profiling of the developing mouse axial skeleton: a role for Tgfbr2 in the development of the intervertebral disc. BMC Dev. Biol. 10, 29 ( 10.1186/1471-213X-10-29) [DOI] [PMC free article] [PubMed] [Google Scholar]