Abstract
It is not known whether eosinophilic myositis is a specific histopathological feature of limb girdle muscular dystrophy 2A (LGMD2A).
Number and location of eosinophils in skeletal muscle biopsies (n=100) was analysed by Giemsa and modified hematoxylin/eosin staining in patients with genetically confirmed myopathies (LGMD2A, LGMD2B, LGMD2L, facioscapulohumeral muscular dystrophy, dystrophinopathy), histologically confirmed idiopathic inflammatory myopathies (sporadic inclusion body myositis (sIBM), dermatomyositis (DM), polymyositis), amyotrophic lateral sclerosis (neurogenic control), and normal controls.
The number of eosinophils/mm2 was significantly higher in LGMD2A, PM, DM, and sIBM compared to controls but not significantly higher than other myopathies. A large overlap in the number of eosinophils/mm2 between all groups was seen. In all disease groups eosinophils were mainly found endomysially (46- 88%) and intra- and perivascularly (4-37%). There was no correlation between the numbers of eosinophils/mm2 and (i) age at biopsy and (ii) the duration of the disease. The extent of myopathic, fibrotic, and inflammatory changes did not differ in samples with high and low eosinophil count.
Eosinophils seem to represent an unspecific histological finding in hereditary and inflammatory myopathies, but also amyotrophic lateral sclerosis.
Key words: Eosinophil, inflammatory myopathy, hereditary myopathy, limb girdle muscular dystrophy, calpainopathy, Giemsa staining, amyotrophic lateral sclerosis
Introduction
Some hereditary myopathies are known to be variably associated with an unspecific inflammatory response which does not seem to correlate with the extent of myofibre necrosis (1, 2). Eosinophilic myositis is characterized by infiltration of the skeletal muscle tissue (variably an invasion of necrotic muscle fibres) by eosinophils, possibly in association with peripheral blood and/or bone marrow eosinophilia (4-6). It is usually attributed to specific causes (3-5, 7), but if these etiologies are excluded, the diagnosis of idiopathic eosinophilic myositis is made histopathologically. Recently, eosinophilic myositis was suggested to be a pathophysiological component of LGMD2A) (3, 7-10). Later eosinophilic myositis was reported in LGMD2C, Becker muscular dystrophy, and inflammatory myopathies (11-13). This gave rise to the assumption that eosinophil-mediated injury of muscle cells might occur in a wider spectrum of myopathies than previously thought.
In order to get more insight into eosinophilia in skeletal muscle we compared the frequency of eosinophils in patients with five different genetically confirmed myopathies (including LGMD2A), the three idiopathic inflammatory myopathies and amyotrophic lateral sclerosis as a neurogenic disorder.
Patients and methods
Patients
Skeletal muscle biopsy samples from the Department of Neurology, Martin-Luther-University Halle- Wittenberg, Germany, were analysed from patients with genetically confirmed myopathies [LGMD2A (n = 12), LGMD2B (n = 4), LGMD2L (n = 3), facioscapulohumeral muscular dystrophy (FSHD, n = 5), dystrophinopathy (Duchenne (DMD) and Becker (BMD) muscular dystrophy (n = 8)], histologically confirmed idiopathic inflammatory myopathies (IIM) [(sporadic inclusion body myositis (sIBM, n = 10) (14), dermatomyositis (DM, n=11), polymyositis (PM, n = 13) (15)], amyotrophic lateral sclerosis (ALS, neurogenic control (El Escorial Criteria (16), n = 11), and normal controls (n = 24, for characterization see legend of Table I). Biopsies in IIM cases were performed before any steroid or immunosuppressive treatment was started. All patients and tutors had given their informed consent for the analysis of skeletal muscle biopsy samples.
Table 1.
Demographic and clinical data of patients. Values are given as median (range). (Normal values: CK women < 2.41 mmol / L*s, men <2.81 mmol / L*s, eosinophils <5%.) * Kruskal-Wallis One Way Analysis of Variance on Ranks, post hoc analysis using Dunn's method, n.a. not applicable
| Disease | Number (n) | Gender (F/M) | Age at biopsy (years) | Disase duration (years) | Creatine kinase (μmol/L*s) | Increased eosinophil count in haemogram (n) |
|---|---|---|---|---|---|---|
| Hereditary myopathies | ||||||
| LGMD2A | 12 | 4/8 | 30 (10-72) | 7 (2-49) |
8.3 (2.5-349) |
1/7 |
| LGMD2B | 4 | 2/2 | 43 (34-52) |
20 (6-22) |
67 (38-123) |
0/4 |
| LGMD2L | 3 | 1/2 | 49 (29-69) |
6 (5-24) |
37 (22-74) |
0/3 |
| FSHD | 5 | 5/0 | 39 (21-71) |
8(1-19) | 2.2 (2-12) |
1/3 |
| DMD/BMD | 8 | 0/8 | 11 (3-21) |
3(1-13) | 81 (18-293) |
0/3 |
| Idiopathic inflammatory myopathies | ||||||
| DM | 11 | 5/6 | 54 (15-84) |
0.5 (0.1-10) |
8.6 (0.7-227) |
1/6 |
| PM | 13 | 7/6 | 54 (32-77) |
1.0 (0.3-6) |
15 (0.9-112) |
0/12 |
| sIBM | 10 | 5/5 | 70 (52-77) |
3 (1-10) |
9.4 (2-17) |
1/7 |
| Neurogenic control | ||||||
| ALS | 11 | 6/5 | 47 (35-72) |
1 (0.3-4) |
8.2 (2-24) |
1/7 |
| Normal control# | 24 | 13/11 | 5236 (24-75) |
n.a. | 1.98 (0.6-2.8) |
0/8 |
| p* | <0.0001 | <0.0002 | 0.0023 |
The normal controls were defined as having myalgia and exertion-induced complaints but no paresis or muscle atrophy, normal creatine kinase levels, normal EMG, and only mild unspecific changes or normal features in muscle biopsy. * Kruskal-Wallis One Way Analysis of Variance on Ranks, , post hoc analysis using Dunn's method, n.a. not applicable ALS amyotrophic lateral sclerosis, BMD muscular dystrophy Becker type, DM dermatomyositis, DMD muscular dystrophy Duchenne type, LGMD limb girdle muscular dystrophy, PM polymyositis, sIBM sporadic inclusion body myositis
post hoc analysis:
sIBM, DM, PM vs. DMD/BMD p<0.05, sIBM vs. LGMD2A, controls;
DM vs. LGMD2A, LGMD2B, LGMD2L,
controls vs. DMD/BMD, LGMD2A, LGMD2B, LGMD2L p<0.05.
Histochemical and immunohistochemical of skeletal muscle biopsies
10-μm serial cryosections from skeletal muscle biopsies were stained with (1) Giemsa staining (17, 18) and (B) modified hematoxylin and eosin (H&E) (18) to detect eosinophilic cells (Fig. 1). The modification consisted of a shorter exposure to eosin (modified H&E 3-5s, "common" H&E 60s). The number of eosinophils was related to the cross-sectional area and given as eosinophils /mm2. The location of the eosinophils was classified as intravasal, perivascular, endomysial, perifascicular (Fig. 1). Stained sections were visualized using an Axioplan I microscope (Zeiss, Germany), digitized images were acquired for analysis (Axiovision Rel. 4.8.2, Zeiss, Germany). 4-point semi-quantitative histopathological scales were used to score the degree of degeneration, fibrotic changes, and the amount of CD8 (cytotoxic T cells, dilution 1:80, DAKO M7103) and CD68 (macrophages, dilution 1:250, DAKO M0718) positive cells (19).
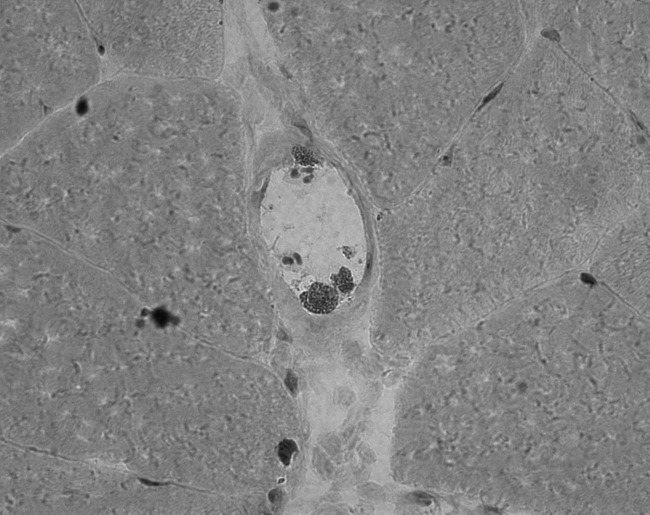
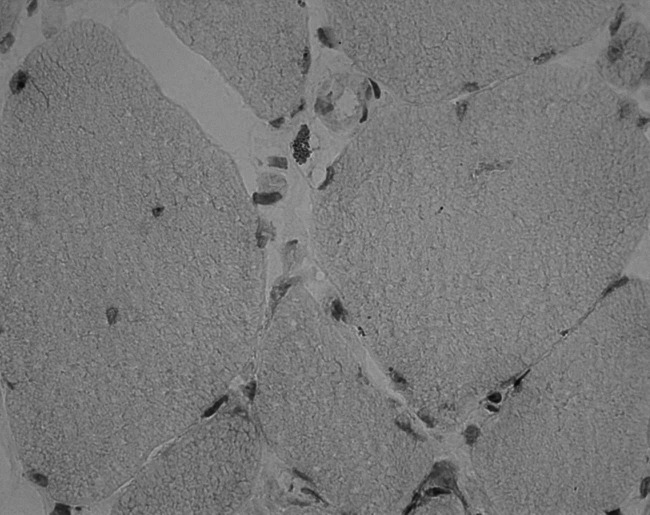
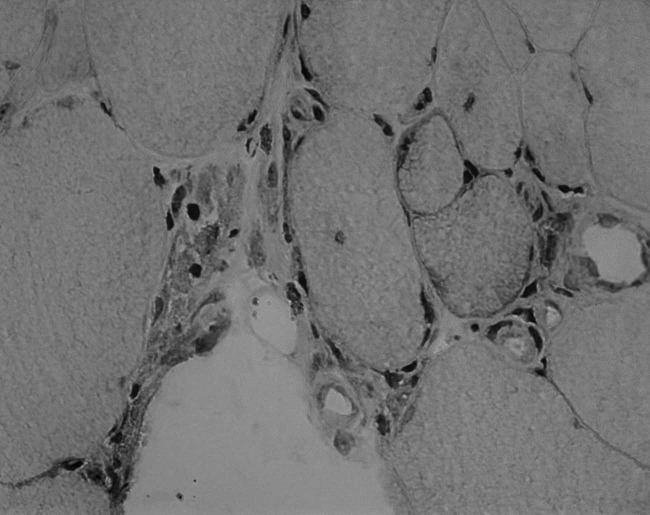
Figure 1.
Intravasal (A), perivascular (B), and perifascicular (C) presence of eosinophils in Giemsa staining (20-fold magnification).
Statistical analysis
All data are given as median + 1 SD. All statistical tests were considered to be significant if p < 0.05. Statistical analyses comprised the Kruskal-Wallis One Way Analysis of Variance on Ranks with Dunn's method posthoc and the Spearman rank order correlation (Sigma Stat Version 9.0, San Jose, USA).
Results
Demographic and clinical data are summarized in Table 1. The genetic data of patients with LGMD2A, LGMD2B, LGMD2L, and DMD/BMD are given in supplementary Table 1.
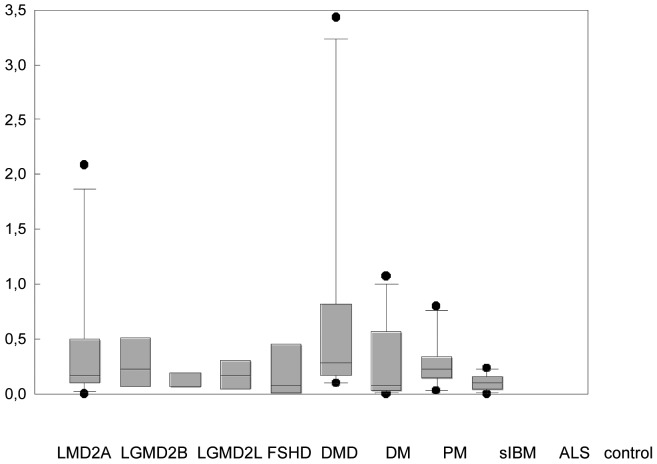
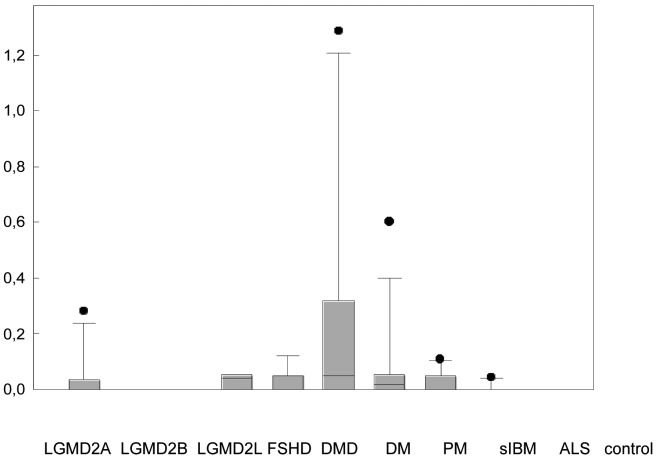
The patients with a considerably higher number of eosinophils/mm2 (LGMD2A n = 2, DM n = 2, cut-off: > 1.0 eosinophils/mm2) did not differ in their clinical, genetic or histopathological features compared to other patients with LGMD2A or DM. The number of eosinophils/mm2 in LGMD2A and all three IIM (DM, PM, sIBM) was significantly higher than normal controls, but not significantly higher than the other disease groups, using Giemsa staining (Fig. 2A). There was a large overlap in the number of eosinophils/mm2 in LGMD2A, the IIM, and the other myopathies. Eosinophils were detected in 72 cases using Giemsa staining, but in only 31 cases using modified H&E staining (Fig. 2). In controls only one eosinophil was detected in 1 case with both stainings. There was a moderate correlation between the number of eosinophils/mm2 in Giemsa and modified H&E staining (r2 0.58, standard error 0.438, p < 0.001, power 1.0). Using the cut-off value of 0.3 eosinophils/mm2 (20), 18/100 patients had values > 0.3 eosinophils/mm2 (DM n = 5, PM n = 3, DMD n = 3, LGMD2A n = 3, LGMD2B n = 3, FSHD n = 1).
Figure 2.
Eosinophils/mm2 in different myopathies with Giemsa staining (A) and modified H&E staining (B). Data are given as median, 5%, 25%, 75%, 95% CI.
*Kruskal-Wallis One Way Analysis of Variance on Ranks revealed a statistically significant difference of p < 0.001 (Giemsa staining) and p = 0.021 (mod. H&E). Pairwise multiple comparison procedures using Dunn's method showed a statistically significant difference (p < 0.05) between LGMD2A, DM, PM, and sIBM with normal controls with Giemsa stain but not with mod. H&E.
There was no correlation between age at biopsy, duration of the disease or myopathy score with the number of eosinophils/mm2 in patients with (i) LGMD2A, (ii) the group of the IIM, or (iii) total group of myopathies together (data not shown). In addition, there was no difference between the extent of myopathic, fibrotic, and inflammatory changes using the semi-quantitative scores between samples with low and high (i.e. mean eosinophils/ mm2+1SD) eosinophil count (data not shown).
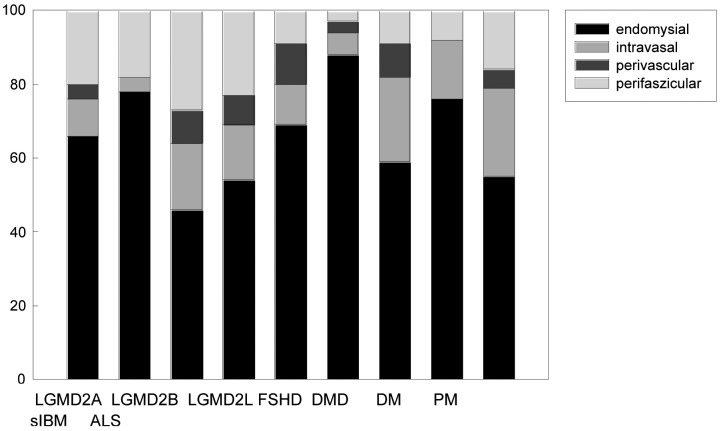
46% (LGMD2L) to 88% (DM) of all eosinophils were located in the endomysium (Fig. 3). The number of eosinophils located in the vessels, on the passage through the vessel wall and perivascularly ranged from 4% in LGMD2B to 37% in LGMD2L (mostly 15-35%, Fig. 1). Necrotic fibres were not infiltrated by eosinophils.
Figure 3.
Distribution of eosinophils in endomysial (black), intravascular (light grey), perivascular (dark grey), perifascicular (white) locations in the different myopathies and ALS.
Absolute number of eosinophils: LGMD2A (n = 205), LGMD2B (n = 23), LGMD2L (n = 11), FSHD (n = 13), DMD (n = 45), DM (n = 169), PM (n = 106), sIBM (n = 60), ALS (n = 37), normal controls (n = 1).
Five patients with five different diagnoses had an increased eosinophil count in the haemogram (Table 1).
None of these patients had a high number of eosinophils/mm2 in the muscle biopsy with Giemsa staining.
Discussion
This is the first systematical survey comparing the frequency of eosinophils in a wide variety of inflammatory and hereditary myopathies and ALS. There was a wide range in the number of eosinophils/mm2 in all disorders including ALS. Eosinophilia was neither specific for LGMD2A nor idiopathic inflammatory myopathies. However, we showed that eosinophils are no feature in normal skeletal muscle. The eosinophil density in idiopathic inflammatory myopathies in the present study was similar to the density in a previous study (13). We found (i) no dominance of eosinophils among the monocytes composing the infiltrates, (ii) no association between high numbers of eosinophils in skeletal muscle and hypereosinophilia in peripheral blood, (iii) no infiltration of necrotic muscle fibres by eosinophils, and (iv) no correlation between the number of eosinophils and the extent of myopathic changes or the duration of the disease. This might be due to the low sample size of these rare disorders and its varying pathological background, but could also reflect the unspecific nature of eosinophils in skeletal muscle.
In the present study, eosinophils were frequently located either within the lumen of the vessel or perivascularly, which might be related to the mechanisms of chemotaxis. It has been suggested that eosinophilic proteins (e.g. eosinophil cation protein) have a specific role in skeletal muscle protein degradation (21, 23) and that eosinophils secrete cytokines, and attract inflammatory cells. In addition, T-lymphocytes, macrophages, monocytes and dendritic cells are able to activate eosinophils (24).
Eosinophils and fibrosis
Eosinophils were frequently seen in the endomysium in the present study, which might be due to mechanisms related to extracellular matrix proteins and their metabolism, finally leading to fibrosis. Eosinophilia in areas of active fibrosis (25) has been described in a variety of inflammatory diseases (25-27). Eosinophils themselves contain granule proteins that might participate in fibrosis (25, 28, 29). Fibroblasts are a natural source of the chemokine eotaxin, which is able to attract eosinophils (30-32). Fibrosis-related mechanisms might explain why eosinophils were seen in early disease stages in previous reports (3, 8) rather than highly dystrophic muscle in later stages of disease.
Eosinophils in neurogenic disorders
This is the first report to mention eosinophils in skeletal muscle in ALS. Neurotoxic substances generated by eosinophils were suggested to be involved in the development of an axonal neuropathy in Churg-Strauss syndrome (33). Peripheral nerves have been shown to actively recruit eosinophils via the release of chemotactic factors and expression of adhesion molecules (34, 35). Eosinophil-nerve cell interactions led to generation of neuronal reactive oxygen species and the release of eosinophil proteins (36). In vivo eosinophils induced neurite retraction and prevented neurite outgrowth during differentiation (37). Some of these mechanisms might contribute to the axonal damage in motor neurons in motor neuron disease.
In conclusion, the present study suggests that the occurrence of eosinophils in hereditary and idiopathic inflammatory myopathies might not be associated with specific genetic causes (such as mutations in the CAPN3 gene), but rather related to mechanisms related to extracellular matrix proteins and their metabolism, finally leading to fibrosis in skeletal muscle tissue and the degeneration or denervation of skeletal muscle cells.
Acknowledgements
We thank Kathleen Zietz and Thekla Wangemann (Neurological Department, Halle (Saale)) for their skilful technical assistance, Hermann Herbst (Department of Pathology, Vivantes Klinikum, Neukölln, Berlin) for his kind collaboration, Marcus Deschauer, Pushpa R. Joshi (Neurological Department, Halle (Saale)) and Tiemo Grimm (Institute of Human Genetics, Würzburg) for genetic analysis, Stephan Zierz (Head of the Neurological Department, Halle (Saale)) for support, and Kathryn Birch for proof-reading the manuscript.
Supplementary
Table 1.
Genetic data of patients with hereditary myopathies. *In 5 of 8 cases of dystrophinopathy no deletion of the dystrophin gene could be identified, and a point mutation was not investigated so these cases were diagnosed by western blot and immunochemistry for dystrophin. ** Heterozygous mutation without detection of another mutation. Cases were diagnosed by western blot.
| Disease / Gene | patient | 1st allele | 2nd allele |
|---|---|---|---|
| LGMD2A / CAPN3 | 1 2 3 4 5 6 7 8 9 10 11 12 |
c.200-204delFWSAL c.550delA c.550delA c.550delA c.550delA c.550delA c.550delA c.550delA c.550delA p.Arg490Trp c.550delA p.Gly445Arg |
p.A705H c.550delA n.d.** n.d.** p.V509P n.d.** p.G565Stop p.A355T Intron13 1746-20C>G p.Arg489Trp c.550delA n.d.** |
| LGMD2B / DYSF | 1 2 3 4 |
c.2779delG c.763delC c.1930G>T in exon 20 c.247dupG |
n.d.** c.3059insC n.d.** c.C757T |
| LGMD2L / ANO5 | 1 2 3 |
c.191dupA c.191dupA c.191dupA |
p.R758C c.191dupA c.1898+1G>A |
| DMD/BMD / DMD | 1 2 3 4 5 6 7 8 |
n.d.* del exons 2-7 (in frame) n.d.* n.d.* n.d.* del exons 53-55 (frameshift) del exons 19-44 n.d.* |
n. a. n.a. n.a. n.a. n.a. n.a. n.a. n.a. |
BMD muscular dystrophy Becker type, DMD muscular dystrophy Duchenne type, LGMD limb girdle muscular dystrophy
Footnotes
F.H. received lecturer honoraria and travel feels from Astellas, Genzyme, and Biomarin Incorp.
References
- 1.Choi JH, Park YE, Kim SI, et al. Differential immunohistological features of inflammatory myopathies and dysferlinopathy. J Korean Med Sci. 2009;24:1015–1023. doi: 10.3346/jkms.2009.24.6.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benveniste O, Romero NB. Myositis or dystrophy? Traps and pitfalls. Presse Med. 2011;40:e249–e255. doi: 10.1016/j.lpm.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 3.Krahn M, Lopez de Munain A, Streichenberger N, et al. CAPN3 mutations in patients with idiopathic eosinophilic myositis. Ann Neurol. 2006;59:905–911. doi: 10.1002/ana.20833. [DOI] [PubMed] [Google Scholar]
- 4.Hall FC, Krausz T, Walport MJ. Idiopathic eosinophilic myositis. QJM. 1995;88:581–586. [PubMed] [Google Scholar]
- 5.Pickering MC, Walport MJ. Eosinophilic myopathic syndromes. Curr Opin Rheumatol. 1998;10:504–510. doi: 10.1097/00002281-199811000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Krahn M, Bartoli M, Levy N. In: Eosinophils in health and disease. Lee JJ, Rosenberg HF, editors. Elsevier: 2013. pp. 533–536. Chapter 13.4. [Google Scholar]
- 7.Titlić M, Kodzoman K, Loncar D. Neurologic manifestations of hypereosinophilic syndrome – review of the literature. Acta Clin Croat. 2012;51:65–69. [PubMed] [Google Scholar]
- 8.Krahn M, Goicoechea M, Hanisch F, et al. Eosinophilic infiltration related to CAPN3 mutations: a pathophysiological component of primary calpainopathy? Clin Genet. 2010;80:308–402. doi: 10.1111/j.1399-0004.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 9.Amato AA. Adults with eosinophilic myositis and calpain-3 mutations. Neurology. 2008;70:730–731. doi: 10.1212/01.wnl.0000287138.89373.6b. [DOI] [PubMed] [Google Scholar]
- 10.Oflazer PS, Gundesli H, Zorludemir S, et al. Eosinophilic myositis in calpainopathy: Could immunosuppression of the eosinophilic myositis alter the early natural course of the dystrophic disease? Neuromuscular Disord. 2009;19:261–263. doi: 10.1016/j.nmd.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Baumeister SK, Todorovic S, Milić-Rasić V, et al. Eosinophilic myositis as presenting symptom in gamma-sarcoglycanopathy. Neuromuscul Disord. 2009;19:167–171. doi: 10.1016/j.nmd.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Weinstock A, Green C, Cohen BH, et al. Becker muscular dystrophy presenting as eosinophilic inflammatory myopathy in an infant. J Child Neurol. 1997;12:146–147. doi: 10.1177/088307389701200214. [DOI] [PubMed] [Google Scholar]
- 13.Cantarini L, Volpi N, Carbotti P, et al. Eosinophilia-associated muscle disorders: an immunohistological study with tissue localisation of major basic protein in distinct clinicopathological forms. J Clin Pathol. 2009;62:442–447. doi: 10.1136/jcp.2008.060616. [DOI] [PubMed] [Google Scholar]
- 14.Brady S, Squier W, Hilton-Jones D. Clinical assessment determines the diagnosis of inclusion body myositis independently of pathological features. J Neurol Neurosurg Psychiatry. 2013;84:1240–1246. doi: 10.1136/jnnp-2013-305690. [DOI] [PubMed] [Google Scholar]
- 15.Dalakas MC, Hohlfeld R. Polymyositis and dermatomyositis. Lancet. 2003;362:971–982. doi: 10.1016/S0140-6736(03)14368-1. [DOI] [PubMed] [Google Scholar]
- 16.Brooks BR, Miller RG, Swash M, et al. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 17.Jankowski R, Persoons M, Foliguet B, et al. Eosinophil count in nasal secretions of subjects with and without nasal symptoms. Rhinology. 2000;38:23–32. [PubMed] [Google Scholar]
- 18.Meyerholz DK, Griffin MA, Castilow EM, et al. Comparison of histochemical methods for murine eosinophil detection in an RSV vaccine-enhanced inflammation model. Toxicol Pathol. 2009;37:249–255. doi: 10.1177/0192623308329342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fanin M, Nardetto L, Nascembeni AC. Correlations between clinical severity, genotype and muscle pathology in limb gridle muscular dystrophy type 2A. J Med Genet. 2007;44:609–614. doi: 10.1136/jmg.2007.050328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumamoto T, Ueyama H, Fujimoto S, et al. Clinicopathologic characteristics of polymyositis patients with numerous tissue eosinophils. Acta Neurol Scand. 1996;94:110–114. doi: 10.1111/j.1600-0404.1996.tb07039.x. [DOI] [PubMed] [Google Scholar]
- 21.Cai B, Spencer MJ, Nakamura G, et al. Eosinophilia of dystrophindeficient muscle is promoted by perforin-mediated cytotoxicity by T cell effectors. Am J Pathol. 2000;156:1789–1789. doi: 10.1016/S0002-9440(10)65050-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanisch F, Müller CR, Grimm D, et al. Frequency of calpain-3 c.550delA mutation in limb girdle muscular dystrophy type 2 and isolated hyperCKemia in German patients. Clin Neuropathol. 2007;26:157–163. doi: 10.5414/npp26157. [DOI] [PubMed] [Google Scholar]
- 23.Sugihara R, Kumamoto T, Ito T, et al. Human muscle protein degradation in vitro by eosinophil cationic protein (ECP) Muscle Nerve. 2001;24:1627–1634. doi: 10.1002/mus.1198. [DOI] [PubMed] [Google Scholar]
- 24.Murata K, Sugie K, Takamure M, et al. Eosinophilic major basic protein and interleukin-5 in eosinophilic myositis. Eur J Neurol. 2003;10:35–38. doi: 10.1046/j.1468-1331.2003.00481.x. [DOI] [PubMed] [Google Scholar]
- 25.Noguchi H, Kephart GM, Colby TV, et al. Tissue eosinophilia and eosinophil degranulation in syndromes associated with fibrosis. Am J Pathol. 1992;140:521–528. [PMC free article] [PubMed] [Google Scholar]
- 26.Janin A. Eosinophilic myocarditis and fibrosis. Hum Pathol. 2005;36:592–593. doi: 10.1016/j.humpath.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Xu X, Rivkind A, Pikarsky A, Pappo O, Bischoff SC, Levi-Schaffer F, et al. Mast cells and eosinophils have a potential profibrogenic role in Crohn disease. Scand J Gastroenterol. 2004;9:440–447. doi: 10.1080/00365520310008566. [DOI] [PubMed] [Google Scholar]
- 28.Furuta GT, Ackerman SJ, Varga J, et al. Eosinophil granule-derived major basic protein induces IL-8 expression in human intestinal myofibroblasts. Clin Exp Immunol. 2000;122:35–40. doi: 10.1046/j.1365-2249.2000.01337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letuve S, Pretolani M. Potential role of eosinophilgranule proteins in tissue remodeling and fibrosis. In: Lee JJ, Rosenberg HF, editors. Eosinophils in health and disease. Elsevier: 2013. pp. 393–398. Chapter 12.2. [Google Scholar]
- 30.Wehling-Henricks M, Sokolow S, Lee JJ, et al. Major basic protein- 1 promotes fibrosis of dystrophic muscle and attenuates the cellular immune response in muscular dystrophy. Hum Mol Genet. 2008;17:2280–2292. doi: 10.1093/hmg/ddn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salvadori C, Peters IR, Day MJ, et al. Muscle regeneration, inflammation, and connective tissue expansion in canine inflammatory myopathy. Muscle Nerve. 2005;31:192–198. doi: 10.1002/mus.20252. [DOI] [PubMed] [Google Scholar]
- 32.Kohan M, Puxeddu I, Reich R, et al. Eotaxin-2/CCL24 and eotaxin- 3/CCL26 exert differential profibrogenic effects on human lung fibroblasts. Ann Allergy Asthma Immunol. 2010;104:66–72. doi: 10.1016/j.anai.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Chao CC, Hsieh ST, Shun CT, et al. Skin denervation and cutaneous vasculitis in eosinophilia-associated neuropathy. Arch Neurol. 2007;64:959–960. doi: 10.1001/archneur.64.7.959. [DOI] [PubMed] [Google Scholar]
- 34.Sunohara N, Furukawa S, Nishio T, et al. Neurotoxicity of human eosinophils towards peripheral nerves. J Neurol Sci. 1989;92:1–7. doi: 10.1016/0022-510x(89)90170-6. [DOI] [PubMed] [Google Scholar]
- 35.Kingham PJ, McLean WG, Walsh MT, et al. Effects of eosinophils on nerve cell morphology and development: the role of reactive oxygen species and p38 MAP kinase. Am J Physiol Lung Cell Mol Physiol. 2003;285:L915–L924. doi: 10.1152/ajplung.00094.2003. [DOI] [PubMed] [Google Scholar]
- 36.Walsh MT, Curran DR, Kingham PJ, et al. Effect of eosinophil adhesion on intracellular signaling in cholinergic nerve cells. Am J Respir Cell Mol Biol. 2004;30:333–341. doi: 10.1165/rcmb.2003-0188OC. [DOI] [PubMed] [Google Scholar]
- 37.Kingham PJ, McLean WG, Walsh MT, et al. Effects of eosinophils on nerve cell morphology and development: the role of reactive oxygen species and p38 MAP kinase. Am J Physiol Lung Cell Mol Physiol. 2003;285:L915–L924. doi: 10.1152/ajplung.00094.2003. [DOI] [PubMed] [Google Scholar]