Abstract
The Salmonella enterica serovar Typhi ompS2 gene codes for a 362-amino-acid outer membrane protein that contains motifs common to the porin superfamily. It is expressed at very low levels compared to the major OmpC and OmpF porins, as observed for S. enterica serovar Typhi OmpS1, Escherichia coli OmpN, and Klebsiella pneumoniae OmpK37 quiescent porins. A region of 316 bp, between nucleotides −413 and −97 upstream of the transcriptional start point, is involved in negative regulation, as its removal resulted in a 10-fold increase in ompS2 expression in an S. enterica serovar Typhi wild-type strain. This enhancement in expression was not observed in isogenic mutant strains, which had specific deletions of the regulatory ompB (ompR envZ) operon. Furthermore, ompS2 expression was substantially reduced in the presence of the OmpR D55A mutant, altered in the major phosphorylation site. Upon random mutagenesis, a mutant where the transposon had inserted into the upstream regulatory region of the gene coding for the LeuO regulator, showed an increased level of ompS2 expression. Augmented expression of ompS2 was also obtained upon addition of cloned leuO to the wild-type strain, but not in an ompR isogenic derivative, consistent with the notion that the transposon insertion had increased the cellular levels of LeuO and with the observed dependence on OmpR. Moreover, LeuO and OmpR bound in close proximity, but independently, to the 5′ upstream regulatory region. Thus, the OmpR and LeuO regulators positively regulate ompS2.
Salmonella enterica serovar Typhi is the causal agent of typhoid fever in humans, affecting a wide sector of the world population (37). Typhoid fever is a systemic infection, characterized by the presence of bacteria in the liver, spleen, and bone marrow, as S. enterica serovar Typhi invades and survives within macrophages (6). Specific humoral and cellular immune responses are mounted against Salmonella outer membrane proteins (OMPs) (2, 53, 51). Salmonella enterica serovar Typhimurium and S. enterica serovar Typhi OMPs have proven to be relevant immunogens for protection of mice challenged with virulent strains (23, 35). S. enterica serovar Typhimurium double ompC and ompF porin mutants showed attenuated virulence (7), and S. enterica serovar Typhimurium porins have been observed to trigger signal transduction in host cells (17). Hence, studies on the molecular features and regulation of Salmonella OMPs and porins should aid in further understanding their role during bacterium-host interactions.
S. enterica serovar Typhi synthesizes three highly abundant OMPs upon growth in standard laboratory media: the OmpC and OmpF porins and the OmpA structural protein (42). Expression of the ompC and ompF genes in S. enterica serovar Typhi is under the control of EnvZ and OmpR, a two-component signal transduction system encoded by the ompB (ompR envZ) locus. Interestingly, a shift in osmolarity only affects OmpF expression, and OmpC levels remain constant (43, 31). In contrast, the relative levels of OmpC and OmpF expression in E. coli are modulated in a reciprocal manner by changes in osmolarity, which also affect the level of the phosphorylated form of the response activator OmpR in the cell (reviewed in references 29 and 40).
In addition to its role in porin regulation, the Salmonella ompB operon has been implicated in the regulation of virulence. Mutants in ompR are attenuated in vivo, are defective in capsular Vi synthesis and in cytotoxicity towards macrophages, do not replicate within cultured macrophages, and show diminished expression from the ssrA/B genes in Salmonella pathogenicity island 2. ompR and envZ mutants are defective in the formation of Salmonella-induced filaments (13, 38, 28, 27, 33).
Adding to the porin repertoire of Salmonella, S. enterica serovar Typhi contains the ompS1 gene that codes for an OMP with all of the molecular features of the OmpC/OmpF family, which is, nevertheless, expressed at very low levels compared to the major OmpC and OmpF porins. In the absence of negative upstream regulatory sequences, ompS1 shows a hierarchical regulation, where expression occurs from a P1 promoter in the presence of OmpR or from an OmpR-independent P2 promoter that becomes activated in the absence of P1 activation (36). Here we report the isolation and initial characterization of ompS2 from S. enterica serovar Typhi, which shows sequence similarity to ompS1. Regulation of ompS2 was also negatively controlled by cis-acting sequences located upstream of the promoter, and its expression was positively regulated by the OmpR regulator as well. However, in addition, the LeuO regulator was found to be a positive regulator, and both OmpR and LeuO bound to the upstream regulatory region.
LeuO has been implicated in the bacterial response to stress: thus, it might be important for survival in natural environments outside the laboratory (14, 30). Moreover, overexpression of LeuO has been found to affect the expression of several genes influenced by mutations in hns, the gene for the H-NS nucleoid protein. Thus, LeuO relieved silencing of the bgl operon, the operon involved in the utilization of certain β-glucosides (52); suppressed the effect of an hns mutation on cadC, which codes for the activator of the acid-inducible lysine decarboxylase (50); and also suppressed the negative effect of the AT4 region, the 72-bp gene silencer region located between the leuO gene and the leuABCD leucine biosynthetic operon (8, 9). Besides, LeuO has a negative effect on the expression of the small regulatory DsrA RNA involved in RpoS translation (25).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in medium A (containing [per liter] 7 g of nutrient broth, 1 g of yeast extract, 2 g of glycerol, 3.7 g of K2HPO4, and 1.3 g of KH2PO4). To achieve high osmolarity, NaCl was added to a final concentration of 0.3 M. Cells were collected for protein, β-galactosidase, and RNA determinations at the mid-logarithmic (optical density at 600 nm [OD600], 0.4) phase of growth at 37°C. This was determined to be the point of optimum levels upon analysis of the whole growth curve (data not shown). When needed, 12 μg of tetracycline/ml or 30 μg of kanamycin/ml was used. Cells were collected by centrifugation and washed twice with 1× Z buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4 [pH 7]). After centrifugation, the bacterial pellet was resuspended in 1 ml of Z buffer.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Serovar Typhi strains | ||
| IMSS-1 | Mexican clinical isolate; 9,12, d:-, Vi | 42 |
| 81 | Serovar Typhi ΔompB::km isogenic IMSS-1 derivative | 31 |
| IMSS-40 | Serovar Typhi ΔompR::km isogenic IMSS-1 derivative | This study |
| IMSSTN103 | Serovar Typhi isogenic IMSS-1 derivative containing a Tn10::Cm insertion 160 bp upstream of the first ATG and 60 bp downstream of the promoter for the leuO gene | This study |
| Plasmids | ||
| pFM413S2 | pBR322-derived plasmid, containing the whole structural ompS2 gene plus its 5′ regulatory region, up to 413 bp upstream of the transcriptional start point | This study |
| pFM97S2 | pBR322-derivative, containing the ompS2 gene plus the regulatory region up to position −97 | This study |
| pFM413 to 34 series | pMC1871-derived plasmids containing translational fusions of the ompS2 5′ regulatory region, up to 413 to 34 bp upstream of the transcriptional start point, to the lacZ reporter gene | This study |
| pMPM-A3 | p15A-derived vector (Apr), with a multiple cloning site | 32 |
| pFM800TL | pMPMA-3-derived plasmid containing the serovar Typhi leuO gene together with Tn10 inserted in the upstream region | This study |
| pFMTrcleuO | pFMTrc12 (p15A1 lacI trcp Apr)-derived plasmid containing serovar Typhi leuO | This study |
| pKD46 | Red recombinase expression plasmid (Apr) | 10 |
| pKD4 | Template plasmid for gene inactivation with Kmr | 10 |
DNA techniques.
DNA manipulations were performed according to standard protocols (47). Oligonucleotides used for amplification by the PCR were purchased from BioSynthesis or provided by the Oligonucleotide Synthesis Facility at our institute. PCRs were done by using AmpliTaq (Perkin Elmer) according to the manufacturer's instructions. Restriction and modification enzymes were used according to the manufacturer's instructions (Amersham, Inc., Boehringer Mannheim, New England Biolabs, or Gibco BRL).
Southern blot hybridization.
Hybridization of 32P-labeled DNA probes (108 cpm/μg) was done overnight in 0.1 M NaH2PO4-Na2HPO4 (pH 7) at 65°C. Washes were performed at room temperature: first in 2× SSC (standard saline citrate; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) for 30 min and then in 1× SSC-0.1% SDS for 15 min. For higher stringency, washes were done at 65°C.
S. enterica serovar Typhi genomic library construction and screening.
A genomic library was constructed by performing a complete SalI DNA digestion of S. enterica serovar Typhi IMSS-1 and cloning it into plasmid pUC18. Recombinant clones were pooled successively in groups of 10 and screened by Southern blot hybridization with radioactive ompS1; the pool containing the 2.3-kb band was further subdivided and probed until the desired individual plasmid, pM15, was isolated. This screening procedure avoided cross-hybridization with Escherichia coli host DNA, a strategy adopted previously with recombinant phage libraries (42).
Cloning of the ompS2 structural gene.
The 2.3-kb SalI insert from pM15, containing the entire structural ompS2 gene plus a 5′ regulatory region encompassing 413 bp upstream of the transcriptional start site, was cloned into pBR322 to generate pFM413S2. pFM97S2, carrying the entire ompS2 structural gene plus 97 bp upstream of the transcriptional start site, was constructed by PCR amplification with pFM413S2 as the template and the following oligonucleotides: 5′-ATATATGCGTTTCCGATAGTAAC-3′ (complementary to the omps2 regulatory region) and 5′-TCTGTGCCCAGGGATCCGTGCGTTCTGTTT-3′ (complementary to the 3′-terminal portion of the structural gene and generating a BamHI restriction site). The amplified fragment was cloned into pBR322, cut with BamHI and NruI.
Nucleotide and amino acid sequence analysis.
The entire 2.3-kb SalI insert from pM15 was sequenced by the dideoxy chain termination method (48) and deposited in the EMBL database. The GAP program (version 10) of the Computer Genetics Group (Madison, Wis.) was utilized for nucleotide and amino acid sequence alignments. Porin sequences were retrieved from the GenBank database. The S. enterica serovar Typhimurium and Salmonella enterica serovar Paratyphi ompS2 sequences were retrieved from the database of the Washington University School of Medicine Genome Sequencing Center.
Construction of translational ompS1-lacZ reporter fusions.
PCR was used for generating plasmids pFM413 and pFM266 from pFM413S2. Thus, oligonucleotides 5′-GTCGACAGATTTTACCTGCC-3′ and 5′-ATTACGTCTTTTTCGCTCAA-3′ were used, respectively, for amplifying from the 5′ upstream region; and oligonucleotide 5′-TACTTTTCTTTTCATTTTTT-3′ was used for the 3′ downstream portion. The regulatory regions were then cloned into the SmaI site of the pMC1871 fusion vector (Pharmacia) (49). Construction of the shorter pFM188, 145, 124, 97, 92, 85, 51, and 34 derivatives was done by digesting pFM266 with EcoRI, doing a time course digestion with Bal31, blunt-ending with Klenow polymerase, digesting with SauI, and ligating into pMC1871 cut with SauI and SmaI. Fusions were sequenced to determine their 5′ end points.
Primer extension analysis.
Ten micrograms of total RNA, isolated by using a commercial kit (RNeasy; Qiagen), from a sample collected after the culture reached an optical density at 600 nm (OD600) of 0.4 was linearized at 93°C for 3 min and then slowly cooled down to 42°C to favor annealing with a 32P-labeled oligonucleotide complementary to the 5′ terminus of the lacZ gene (5′-GCCAGGGTTTTCCCAGTCACGACG-3′). The oligonucleotide primer was extended with reverse transcriptase, and the extended product was collected with a Microcon-10 microconcentrator (Amicon) and analyzed by electrophoresis in 8% polyacrylamide-urea gels by following established procedures (3, 36). Samples for electrophoresis were quantified by OD260.
Microplate protein and β-galactosidase determinations.
Protein concentration determined by the Lowry method and β-galactosidase activity were adapted as a microtiter plate assay (31, 36).
OMP purification and electrophoresis.
OMPs were prepared as a Triton X-100 insoluble fraction and scaled down to a mini-prep level as described previously (44). They were separated by SDS-12% polyacrylamide gel electrophoresis (12% PAGE) and visualized by staining with Coomassie brilliant blue. Relative abundance was determined by densitometric scanning (Alpha Imager UV5.04; Alpha Innotech Corp.).
Disruption of S. enterica serovar Typhi ompR.
The procedure for the one-step inactivation of bacterial chromosomal genes by Datsenko and Wanner (10) with the pKD46 Red recombinase plasmid was used. For the nonpolar deletion of ompR, the specific H1 and H2 homologue extension sequences used were ompR5′-P1 (5′-GAGAATTATAAGATTCTGGTGGTTGATGACGATATGCGTCTGTGTAGGCTGGAGCTGCTTC-3′) and ompR3′-P2 (5′-GACGTAGCCCAGGCCCCATACGGTCTGAATATAACGCGGCATATGAATATCCTCCTTAGTTCC-3′), respectively, joined to the P1 and P2 priming sequences specific for the Kmr cassette gene from pKD4.
S. enterica serovar Typhi transposon mutagenesis.
Transposon Tn10, contained in plasmid pBSL181 (1), was used to perform random mutagenesis on S. enterica serovar Typhi IMSS-1 carrying lacZ fusion plasmid pFM413. An exponential bacterial culture (OD550 = 0.8) of 500 ml was concentrated to 5 ml for electroporation in a Bio-Rad electroporator (E. coli Pulser) by following the manufacturer's instructions. Three hundred microliters of the concentrated culture was electroporated with 8 μg of pBSL181. The cells were then placed in 1 ml of medium S.O.C. (Invitrogen) plus 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce the transposase for 1 h at 37°C. Selection was in NCE medium (3.94 g of KH2PO4, 6.46 g of K2HPO4 · 3H2O, 43.75 g of NaNH4HPO4 · 4H2O) plus 0.2% lactose, 0.1% tryptophan, 0.3% cysteine HCl H2O, 80 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/ml, 30 μg of chloramphenicol/ml, and 12 μg of tetracycline/ml. Three transformants, which showed a dark blue phenotype indicative of an increase in pFM413 lacZ expression, were obtained from the screening of a total of 30,000 transformants. S. enterica serovar Typhi lacks β-galactosidase activity; thus, the screening relied not only on color development but also on enhanced growth in minimal medium.
OmpR purification and phosphorylation.
OmpR was overexpressed and purified from a strain carrying a plasmid coding for a His6-OmpR fusion protein (36) by using a QIAexpressionist commercial kit (Qiagen) under 8 M urea-denaturing conditions. E. coli M15(pREP4), harboring the pQR254 plasmid, i.e., the pQE32 vector plasmid carrying the S. enterica serovar Typhi ompR structural gene fused to 6 histidine codons at the 5′ terminus, was grown to the mid-logarithmic phase. IPTG was added to a final concentration of 1 mM, and the bacteria were incubated for 4 to 5 h at 37°C and 250 rpm. Cells were then pelleted by centrifugation, resuspended in urea buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl [pH 8.0]), and disrupted by sonication. The suspension was centrifuged, and the supernatant was filtered through a HiTrap chelating column (Δkta prime automated liquid chromatography system; Amersham Pharmacia Biotech). The column was washed with 5 volumes of urea buffer (pH 8.0) and then with a gradient of urea buffer (pH 8.0 to 4.5). Fractions containing purified His6-OmpR protein were selected after SDS-PAGE and then loaded into a Slyde-A-Lyzer 10K cassette (Pierce), for extensive dialysis against OmpR storage buffer (20 mM HEPES [pH 7.9], 100 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.1 mM dithiothreitol [DTT], 20% glycerol), and quantified by the method of Lowry, as described above. OmpR phosphorylation was achieved by incubation in a fresh solution of 10 mM acetyl phosphate (Sigma) for 3 h at room temperature.
PCR and cloning of leuO.
PCR amplification of leuO together with the upstream inserted Tn10 element was performed by using the following forward and reverse, respectively, primers: 5′-TAGGGGAATGGCCTGTTAAAGGT-3′ and 5′-GATCGCTTATCGATCAAGGCGTAA-3′. The amplified product was cloned into vector pMPM-A3 (Table 1) (32) and digested with EcoRV, and protruding dT tails were added with Taq polymerase (47). Amplification of the leuO wild type for cloning into plasmid pFMTrc12, digested with NcoI and SmaI, was performed by using the forward primer 5′-GAGTTAACCATGGCAGAGGTCAAAAC-3′, which includes the NcoI site. The reverse primer was the same as above.
Purification of LeuO.
The leuO structural gene was cloned into pTrc99A, digested with NcoI and SmaI, with the same forward primer described above for cloning into plasmid pFMTrc12. The reverse primer 5′-TCATTACCCGGGTTAGTGGTGGTGGTGGTGGTGTCGCTTACAAACAGAGACTAATAA-3′included a SmaI site, the 6 histidine codons, and a portion the 3′ end of the leuO structural gene. LeuO-His6 was purified essentially as described above for His6-OmpR, except that it was under nondenaturing conditions. After growth and induction with IPTG, cells were pelleted by centrifugation, resuspended in 50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, and 5 mM imidazole, and disrupted by sonication. The suspension was then centrifuged, and the supernatant was filtered through a HiTrap chelating column (Δkta prime automated liquid chromatography system; Amersham Pharmacia Biotech). The column was washed with 5 volumes of the resuspension buffer with 20 mM imidazole and then with a gradient of 20 to 500 mM imidazole. Fractions containing LeuO-His6 purified protein were selected after SDS-PAGE, loaded into a Slyde-A-Lyzer 10K cassette (Pierce) for extensive dialysis against storage buffer (20 mM HEPES [pH 7.9], 100 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, 20% glycerol), and quantified by the method of Lowry, as described above.
DNA footprinting with LeuO and OmpR.
Plasmid pFM413 was used as a template for the PCR amplification of the ompS2 5′ regulatory region. A 32P-labeled oligonucleotide complementary to the 5′ coding sequence of ompS2 (5′-TACTTTTCTTTTCATTTTTT-3′) and an oligonucleotide complementary to bp −253 to −228 (5′-TCGCTCAAGAAAATTAAATTAA AAAA-3′) were used. Each assay contained 5 × 104 cpm (0.16 pmol) of template labeled with [γ-32P]ATP and polynucleotide kinase and was incubated at room temperature in 30 μl of 20 mM HEPES (pH 7.9), 100 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, 20% glycerol, and 1.0 μg of poly(dI-dC) (Boehringer)/ml. The DNA segments were separated by electrophoresis in 8% polyacrylamide-8 M urea gels.
Nucleotide sequence accession number.
The sequence of the SalI insert from pM15 was deposited in the EMBL database and given accession no. X89756.
RESULTS
Cloning and characterization of the ompS2 gene.
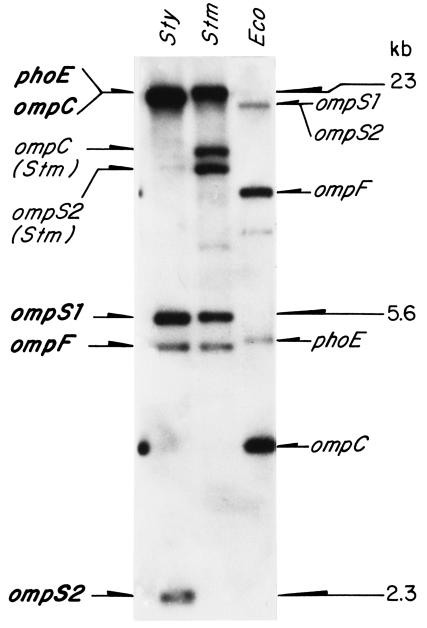
Hybridization of either the S. enterica serovar Typhi ompC (42), ompS1 (16), ompF (EMBL accession no. X89757; formerly ompS3), or phoE (EMBL accession no. X74595) 32P-labeled structural genes to Southern blots of genomic S. enterica serovar Typhi DNA, cut with the SalI restriction endonuclease, rendered a 2.3-kb band aside from the known ompS1 5.6-kb band (16). This 2.3-kb band was conspicuous, as it did not correspond to other known porin genes: it was particularly evident upon hybridization with S. enterica serovar Typhi ompS1 (Fig. 1).
FIG. 1.
Southern blot hybridization of genomic DNA from S. enterica serovar Typhi (Sty), S. enterica serovar Typhimurium (Stm), and E. coli (Eco) digested with SalI restriction endonuclease, with the S. enterica serovar Typhi ompS1 gene (37) labeled with 32P. The identities of the hybridizing bands for Salmonella genes are shown on the left, including a novel conspicuous band that corresponds to ompS2, and those for E. coli genes are shown on the right.
Thus, the 2.3-kb SalI fragment containing the S. enterica serovar Typhi ompS2 gene was isolated from a recombinant S. enterica serovar Typhi genomic library constructed in pUC18, as described in Materials and Methods. Analysis of the nucleotide sequence of the 2.3-kb fragment allowed the identification of an open reading frame (ORF) coding for a 362-amino-acid mature protein (40,043 Da) plus a putative 21-amino-acid leader peptide. Homology searches revealed that the nucleotide sequence of this ORF shares the following identities with other porin genes: Klebsiella pneumoniae kpn37, 77%; E. coli ompN, 76%; S. enterica serovar Typhi ompS1, 72%; E. coli ompC, 72%; S. enterica serovar Typhi ompC, 71%; S. enterica serovar Typhi ompF, 66%; E. coli ompF, 66%. This gene, thus named ompS2, shares 98 and 90% identity with its counterparts in S. enterica serovar Typhimurium and S. enterica serovar Paratyphi A, respectively. Furthermore, Southern blot hybridization with S. enterica serovar Typhi ompS2 as a probe rendered a strong hybridization signal with a 2.3-kb SalI fragment not only with S. enterica serovar Typhimurium and S. enterica serovar Paratyphi A but also with various other salmonellae DNA (data not shown), indicative of its high conservation. Hence, the hybridizing bands labeled ompS1 and ompS2 actually correspond to a truncated ORF (36) and to the ompN gene, respectively, in E. coli (Fig. 1).
Analysis of the deduced amino acid sequence showed that the OmpS2 protein maintains the pattern of alternate variable (loop) and conserved (β-strand) regions characteristic of the porin superfamily (12, 24), with the conservation of the Arg-37, Arg-75, Asp-106, and Arg-126 that have been implicated in determining pore permeability (4, 34). The similarities and identities of the OmpS2 sequence with the amino acid sequences from other porins are as follows: K. pneumoniae Kpn37, 87 and 83%; E. coli OmpN, 84 and 84%; S. enterica serovar Typhi OmpS1, 76 and 72%; S. enterica serovar Typhi OmpC, 76 and 71%; E. coli OmpC, 73 and 69%; S. enterica serovar Typhi OmpF, 68 and 64%; E. coli OmpF, 68 and 64%.
The highest identity of S. enterica serovar Typhi ompS2 outside of the salmonellae was with the K. pneumoniae kpn37 and E. coli ompN genes and the respective proteins (12, 41). Nevertheless, ompS2 in S. enterica serovar Typhi is located downstream of the rstA gene and upstream of an ORF, for which no homology was found to a characterized gene. On the other hand, ompN in E. coli is not located at an equivalent site: it maps at centisome 30.9 between the ynaF and ydbK loci, while the rstA-rstB operon is located at centisome 36.21 (Entrez Genome, National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?db=Genome]).
ompS2 expression.
The ompS2 regulatory region (Fig. 2) shares only 34 to 38% identity with those from other porin genes, including kpn37 and ompN, its putative counterparts in K. pneumoniae and E. coli, respectively (12, 41). In contrast, for instance, the regulatory regions of the ompC and ompF homologues in S. enterica serovar Typhi and E. coli share 75 and 77% identity, respectively.
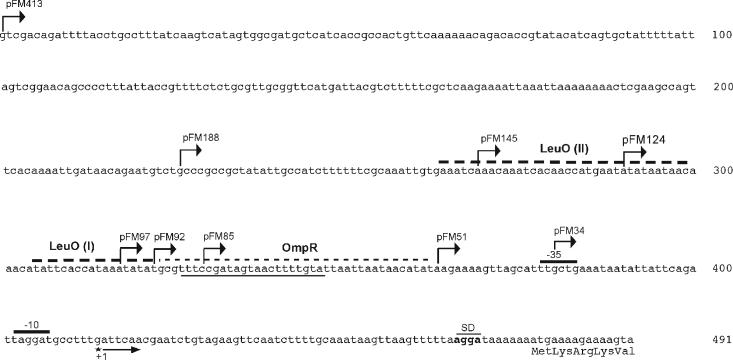
FIG. 2.
Nucleotide sequence of the S. enterica serovar Typhi ompS2 5′ upstream regulatory region, featuring the first five codons of the structural gene, the putative Shine-Dalgarno ribosome-binding sequences (SD), the −10 and −35 promoter sequences, and the single transcription initiation at a G residue obtained by primer extension analysis. Also shown are the boundaries for various pFM translational fusions, where the indicated segments of the regulatory region plus five codons were fused to the lacZ reporter gene. A sequence with similarity to the consensus OmpR-binding site (thick underline), the site for the binding of OmpR (thin dashed line), and two boxes for the binding of LeuO (I and II) (thick dashed lines) are also represented.
A 491-bp DNA segment, encompassing 413 bp upstream of the first transcribed nucleotide, plus 63 transcribed and nontranslated nucleotides (leader mRNA) and the first 5 codons of the structural gene, was used to generate a translational fusion to the lacZ reporter gene, thus constituting plasmid pFM413 (Fig. 2; Table 1). This region corresponds to the 5′ upstream sequence present in plasmid pFM413S2, which in addition, contains the whole structural ompS2 gene (Table 1). Similarly, a series of plasmids containing shorter portions of the regulatory region were generated, as described in Materials and Methods (Fig. 2; Table 1).
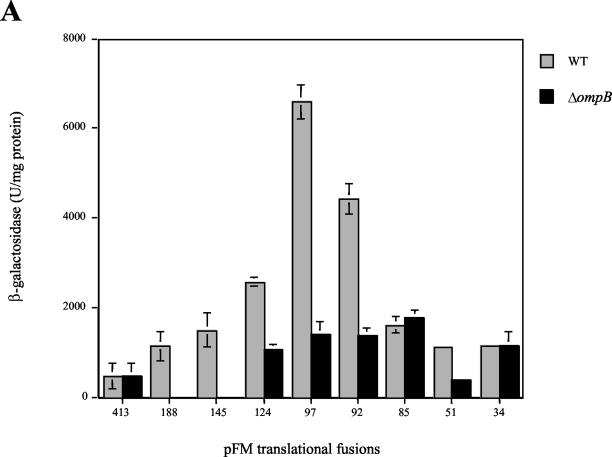
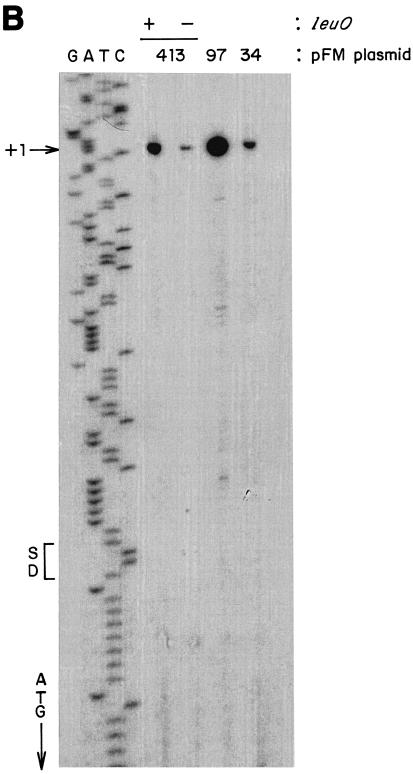
As can be seen in Fig. 3A, expression in wild-type S. enterica serovar Typhi increased more than 10-fold upon removal of 317 bp, between positions −413 and −97, as shown by the relative activities directed by the fusion plasmids pFM413 and pFM97. Hence, this region contains negative-control elements that maintain low levels of ompS2 expression. On the other hand, the absence of 12 residues in pFM85 resulted in a significant decrease in expression (about fourfold) in comparison with pFM97 (Fig. 3). Thus, a relevant region for positive control of expression is located downstream of position −97. Figure 3B depicts the mapping of the ompS2 promoter by primer extension from reporter plasmids. Accordingly, expression is the lowest from pFM413 and increases substantially upon removal of the 317 bp upstream of −97, in pFM97, or by the addition of the plasmid carrying the cloned leuO gene, ptrcleuO (see below). No other promoter was detected downstream of the major transcriptional start (+1). The relative increase in expression in pFM34 is from the only promoter and is likely due to the formation of a −35-like box upon fusion of the plasmid sequences and the ompS2 regulatory sequences, i.e., TTCCC/GCTGAAA (consensus residues are underlined).
FIG. 3.
(A) Activity of the ompS2 gene depends on the length of the regulatory region and on the ompB operon. β-Galactosidase-specific activities rendered by pFM fusion plasmids, containing various lengths of the 5′ regulatory region as indicated by their numeral denomination (Fig. 2), harbored in S. enterica serovar Typhi are shown. The gray bars depict activities in the wild-type (WT) strain; black bars show activity in the isogenic S. enterica serovar Typhi 81 (ΔompB::Km) strain. (B) Mapping by primer extension of the first (+1) transcribed G residue of the ompS2 gene. The initiation of transcription is shown from pFM413, in the absence (−) and in the presence (+) of the leuO gene (cloned in ptrcleuO); from pFM97, lacking 317 bp between −413 and −97; and from pFM34. The Shine-Dalgarno (SD) sequence, the first methionine of the ORF, and the GATC sequence ladder of the complementary strand are shown.
Dependence of ompS2 expression on OmpR.
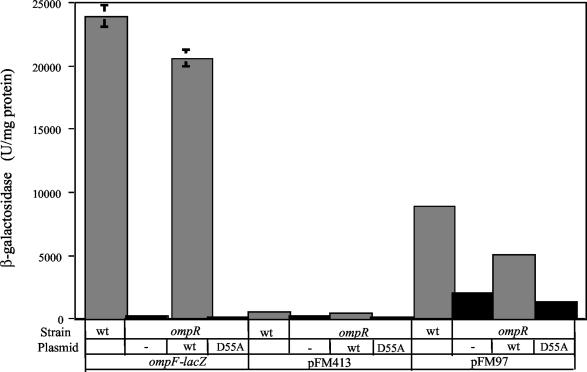
As observed in Fig. 3, most of the active ompS2-lacZ fusions (particularly pFM97) in the wild-type strain showed lower levels of expression in the isogenic S. enterica serovar Typhi 81 mutant strain (ΔompB), similar to those for pFM85. Furthermore, ompS2 full expression from the most-active pFM97 construct was dependent on the presence of wild-type OmpR (Fig. 4). This OmpR-dependent activation required the main OmpR phosphorylation site, since a significant lowering in reporter gene activity was observed in the presence of the OmpR D55A variant (11). The fact that there is a residual expression both in the absence of OmpR and in the presence of the OmpR D55A derivative reflects the promoter strength in the absence of OmpR and of the upstream negative regulatory region.
FIG. 4.
Effect of the OmpR D55A mutant on ompS2 activity. Activities are shown for an S. enterica serovar Typhi ompF-lacZ fusion, used as a control, and of pFM413 and pFM97 in either the IMSS-1 wild type (wt) or in the isogenic S. enterica serovar Typhi IMSS-40 ompR derivative (ΔompR::Km), not complemented (−) or complemented with either a plasmid harboring the wild-type (wt) OmpR gene or the D55A mutant gene, mutated in the codon for the main phosphorylation site (11).
OmpS2 is an OMP.
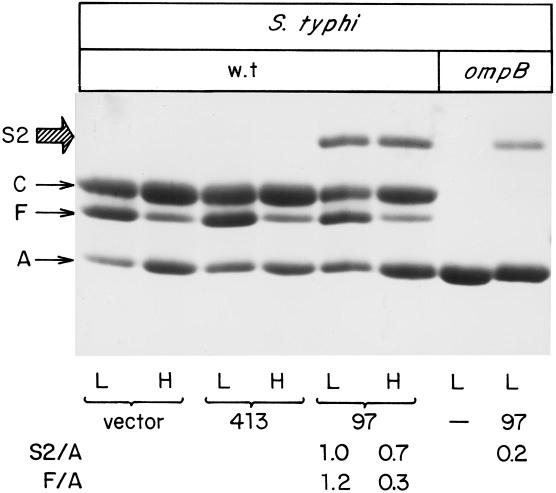
Figure 5 illustrates the presence of OmpS2 in the outer membrane upon expression from plasmid pFM97S2 (Table 1), which contains the ompS2 structural gene expressed under a regulatory region encompassing 97 bp upstream of the transcriptional start point. This region directed the highest ompS2-lacZ activity (Fig. 3). In contrast, no expression was observed from plasmid pFM413S2 (Fig. 5), where the structural gene was under the control of 413 bp upstream of the transcriptional start, also in accord with the translational fusion studies (Fig. 3). The amount of OmpS2 in the outer membrane was determined by densitometric scanning relative to the corresponding OmpA protein (S2/A). Similarly, the amount of OmpF was determined in some of the lanes where the osmolarity was changed (F/A) (Fig. 5, bottom). No effect by osmolarity was observed on OmpS2 expression from pFM97S2, compared with the expected lowering of expression at high osmolarity for OmpF (43). The same result was obtained when expression from the pFM97 ompS2-lacZ fusion was assessed at low and high osmolarity (data not shown).
FIG. 5.
Expression of the OmpS2 protein. PAGE profile of OMP preparations from wild-type (wt) S. enterica serovar Typhi, grown at either low (L) or high (H) osmolarity, and harboring either the pBR322 vector (vector), pFM413S2 (413; containing the structural ompS2 gene under the control of a 413-bp regulatory region), or pFM97S2 (97; ompS2 under a 97-bp regulatory region), S. enterica serovar Typhi ΔompB::Km, containing either no plasmid (−) or pFM97S2 (97), at low osmolarity. The major OMPs OmpC, OmpF, and OmpA are indicated as C, F, and A, respectively. The relative abundance of the OmpS2 (S2) or OmpF (F) protein with respect to OmpA (A) is shown for some lanes and was obtained by densitometric analysis.
In accordance with the general pattern of the lacZ fusion (Fig. 3) and transcriptional data obtained by primer extension (data not shown), expression of the OmpS2 protein from pFM97S2 was lowered in an S. enterica serovar Typhi ΔompB background (Fig. 5).
LeuO positively regulates ompS2.
In a search for additional proteins involved in the regulation of ompS2, random mutagenesis of wild-type S. enterica serovar Typhi IMSS-1 with transposon Tn10 rendered 3 transformants, from a total of 30,000, with increased expression of reporter plasmid pFM413. In all three, the transposon inserted upstream of the leuO gene, which codes for the LeuO regulator, a member of the LysR family of regulatory proteins (21).
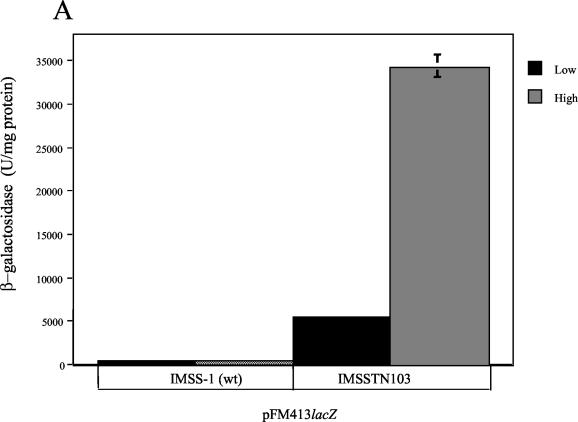
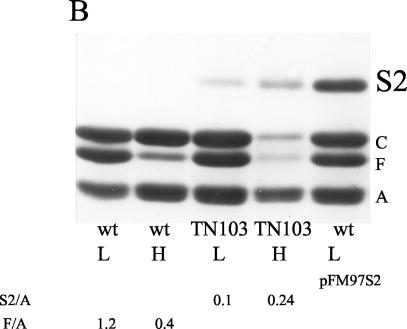
In one mutant, IMSSTN103, the site for Tn10 insertion was 160 bp upstream of the first ATG and 60 bp downstream of the promoter, as determined by nucleotide sequence analysis (data not shown). The β-galactosidase activity was quantified for the pFM413 reporter plasmid (Fig. 6A), and the expression of the OmpS2 protein from the chromosomal gene was observed upon electrophoresis of OMP preparations (Fig. 6B). Interestingly, expression was higher at high osmolarity in both instances. The amount of OmpS2 in the outer membrane was quantified by densitometric scanning, relative to the corresponding OmpA protein (S2/A). As a control, the amount of OmpF (F/A) was lower at high osmolarity in the absence of cloned leuO (Fig. 6B), as expected (43). With respect to the lowering of OmpC and OmpF in the outer membrane in IMSSTN103 at high osmolarity (Fig. 6B), it appears to be due to a posttranslational effect, as the expression of ompC-lacZ and ompF-lacZ translational fusions in IMSSTN103 was not affected compared to the wild type (data not shown).
FIG. 6.
LeuO participates in the regulation of ompS2. (A) Activity of ompS2 from pFM413, both in S. enterica serovar Typhi IMSS-1 (wild type [wt]) and in the IMSSTN103 insertion mutant (Tn10::leuO), at low (black bars) and high (gray bars) osmolarity. (B) PAGE profile of OMP preparations from S. enterica serovar Typhi IMSS-1 (wt) and from the IMSSTN103 insertion mutant (TN103) at low (L) and high (H) osmolarity, depicting the expression of OmpS2 (S2) from the chromosomal gene. The expression of OmpS2 from pFM97S2 was included as a control marker. The major OMPs OmpC, OmpF, and OmpA are indicated as C, F, and A, respectively. The relative abundance of the OmpS2 (S2) or OmpF (F) protein with respect to OmpA (A) is shown for some lanes and was obtained by densitometric analysis.
It is worth noting that it has been previously observed that the insertion of a Tn10 derivative, 26 or 19 bp upstream of the leuO ORF, caused a negative effect on the expression of the rpoS gene or a positive effect on the expression of the bgl operon, respectively. Hence, it was postulated that leuO expression increased due to the activity of the Pout promoter from the IS10 element, and therefore, the LeuO regulator exerted an effect on either rpoS or bgl, respectively (25, 52).
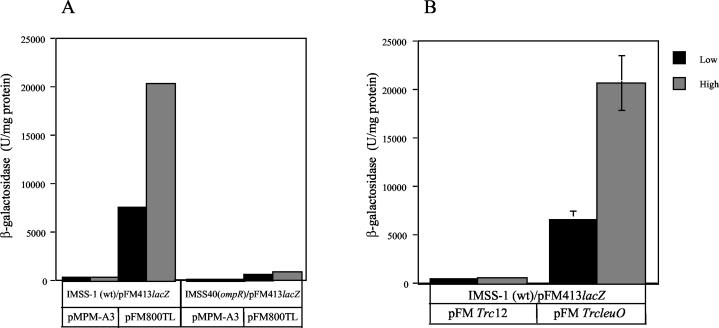
To test the idea that ompS2 expression was due to an increase in expression of leuO in IMSSTN103, the S. enterica serovar Typhi leuO gene together with the upstream Tn10 insertion was amplified by PCR and cloned to render pFM800TL (Table 1). Transformation of wild-type S. enterica serovar Typhi IMSS-1 with pFM800TL, harboring pFM413, resulted in a substantial increase in ompS2-lacZ reporter activity, being higher at high osmolarity (Fig. 7A), similar to what was observed in the IMSSTN103 insertion mutant (Fig. 6). This effect did not occur in the isogenic ompR derivative IMSS-40 (Fig. 7A). Thus, besides supporting the notion that the increase in ompS2 expression was the result of augmented levels of LeuO, the dependence on the OmpR transcriptional regulator was further substantiated.
FIG. 7.
Induction of ompS2 expression in the presence of cloned leuO. β-Galactosidase-specific activity from pFM413 is shown. (A) S. enterica serovar Typhi IMSS-1 (wild type [wt]) or the isogenic IMSS-40 ΔompR derivative, transformed with either the plasmid vector (pMPM-A3) or pFM800TL, harboring the cloned leuO gene with the Tn10 insertion in the upstream region, at low (black) and high (gray) osmolarity. (B) S. enterica serovar Typhi IMSS-1, transformed with the pFMTrc12 vector plasmid or with the pFMTrcleuO plasmid, carrying leuO under the control of the trc promoter, at low (black) and high (gray) osmolarity.
To observe the effect of osmolarity on ompS2 expression in the absence of the transposon upstream of leuO, the structural leuO wild-type gene was amplified by PCR and cloned into vector plasmid pFMTrc12 under the control of the trc promoter to render pFMTrcleuO (see Materials and Methods). Again, the activity of the pFM413 reporter gene was increased in the wild type upon transformation with pFMTrcleuO, and the level of expression was higher at high than at low osmolarity (Fig. 7B), in a similar manner as observed when leuO was cloned together with the upstream inserted transposon (Fig. 7A) or in the IMSSTN103 insertion mutant (Fig. 6).
Binding of LeuO and OmpR-P to the ompS2 5′ regulatory region.
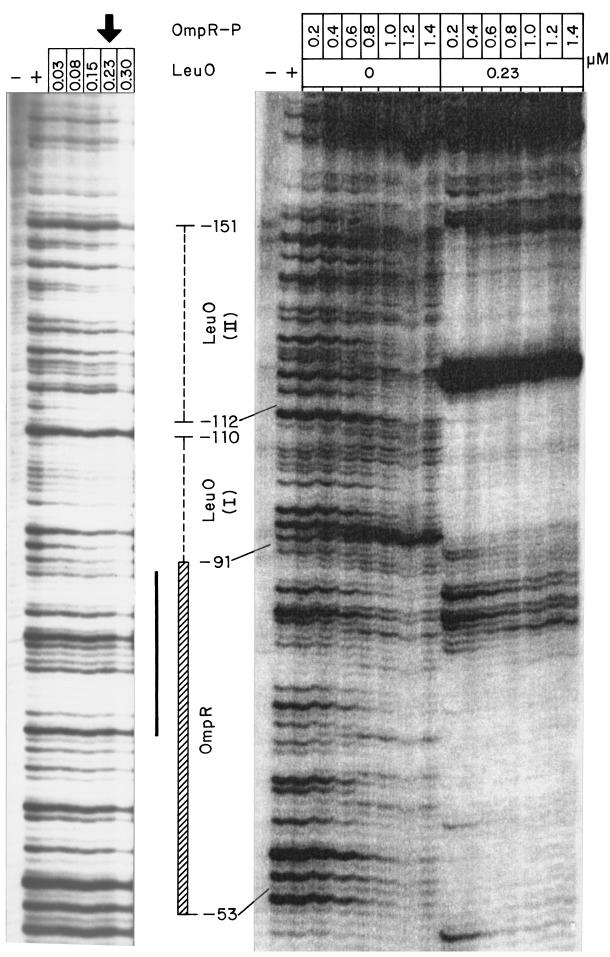
The experiments described above indicated that both the LeuO and the OmpR regulators were involved in the positive regulation of ompS2 expression. Thus, to determine whether LeuO and OmpR directly interacted with the 5′ upstream regulatory region of ompS2, DNA footprinting experiments were performed. As can be seen in Fig. 8, protection with LeuO was obtained at 0.03 μM and higher, upstream of position −91 at two boxes, named LeuO (I) and LeuO (II) (also shown in Fig. 2). In the presence of OmpR-P, LeuO also bound to both boxes I and II, and there was an enhanced sensitivity to DNase digestion at −118, indicative of a change in conformation upon binding of both regulators (Fig. 8). OmpR-P bound at 0.8 μM and above, at and around a 20-bp sequence with similarity to the OmpR-binding F1 and C1 type boxes (20) (Fig. 8; also Fig. 2). Interestingly, OmpR-P bound at 0.2 μM and above, in the presence of LeuO.
FIG. 8.
DNA footprinting at the 5′ regulatory region of ompS2 with LeuO and OmpR-P. On the left panel, the DNase-minus/protein-minus lane is shown with a minus sign (−) and the DNase-plus/protein-minus lane is depicted with a plus sign (+). The following lanes contained 0.03, 0.08, 0.15, 0.23 (vertical arrow), and 0.30 μM LeuO, respectively. The right panel also contains a DNase-minus/protein-minus lane (−) and a DNase-plus/protein-minus lane (+), followed by seven lanes with increasing concentrations of OmpR-P (0.2, 0.4, 0.6, 0.8, 1.0, 1.2, and 1.4 μM). The next lanes contain the same increasing concentrations of OmpR-P but in the presence of 0.23 μM LeuO. Binding boxes LeuO (I) and LeuO (II) are shown, as is the binding region for OmpR. The solid black bar represents the 20-bp sequence with similarity to the OmpR-binding F1 and C1 type boxes.
DISCUSSION
The recent interest in quiescent porins, such as ompS1 in S. enterica serovar Typhi (16, 36), nmpC and ompN in E. coli (41), and ompK37 in K. pneumoniae (12), has opened many questions regarding their role in bacterial physiology. It is of interest that the S. enterica serovar Typhi ompS2 gene shows a high identity with K. pneumoniae ompK37 (12) and with E. coli ompN (41) and that all three genes contain complete ORFs that are strongly down-regulated, although there is much less conservation in their regulatory regions. In contrast, the counterpart of S. enterica serovar Typhi ompS1 is truncated in E. coli, an observation that opens thoughts on the evolutionary circumstances that caused its loss of function (36). The porin properties of E. coli OmpN have been found to resemble those of E. coli OmpC (41); hence, even though some variation in size and in anion or cation selectivity has been observed among the pores formed by E. coli and S. enterica serovar Typhimurium, no clear functional difference has yet been established between them, and it is not readily apparent why a bacterium synthesizes a variety of structurally related porins (5, 18, 26).
Studies with various translational lacZ fusions, containing different portions of the ompS2 5′ regulatory region (Table 1; Fig. 2 and 3), allowed the identification of both a negative-control region located between positions −413 and −97 and a positive-control region located downstream from position −97. It is worth noting the similarities with the ompS1 gene, which also presents a negative and positive mode of regulation and where expression was increased upon removal of distal 5′ upstream portions and lowered when 5′-proximal sequences were removed (36). With respect to the region for negative control (from −413 to −97), it could be determining a necessary structure for negative regulation or it could be a region for the binding of an unknown negative effector or both.
Expression of S. enterica serovar Typhi ompS2 was lowered in isogenic ΔompB and ΔompR strains (Fig. 3 to 5 and 7), thus pointing towards a role of the EnvZ/OmpR two-component signal transduction system in ompS2 expression, similar to what has been observed for S. enterica serovar Typhi ompS1 (16, 36). Interestingly, overlapping the positive control region, between positions −88 and −69, there is a 20-bp box which is 50% identical to the high-affinity F1 and C1 type boxes found in the E. coli ompF and ompC regulatory regions: 5′-TTTCCGATAGTAACTTTTGT-3′ (Fig. 2 and 8) (19, 39, 22, 20). (In all cases, underlining shows the most conserved residues.) The box contains the GXXAC central motif of the F1, F2, C1, and C2 boxes plus other F1 and C1 consensus nucleotides (underlined).
Interestingly, OmpR-P protected this 20-bp box from DNase digestion, together with a downstream segment that contains the motif 5′-ACATAT-3′, which is also present in the 3′ portion of the F1 and C1 boxes and in the Salmonella ssrA A1 box, although in the latter two it is 5′-ACATCT-3′ and 5′-ACAATTT-3′, respectively (15, 20) (Fig. 2 and 8). Moreover, the 3′ portion of the ompS2 20-bp box contains the motif 5′-ACTTTTG-3′, which is present in the 5′ portion of the F1 and C1 boxes, although in the latter it is 5′-ACATTTTG-3′ (20) (Fig. 2). The role of these motifs and of their distribution towards the binding of OmpR-P is certainly a matter of further research. In this respect, in the footprinting experiments, OmpR-P bound to the ompS2 regulatory region at 0.8 μM (Fig. 8), similar to what was observed for the footprinting of OmpR-P on ssrA box A1, which has been considered just below the highest affinity F1 and C1 boxes (15).
Moreover, the LeuO activator of the LysR family was identified (21) as a regulator for ompS2. Upon random mutagenesis, a mutant with increased expression of ompS2 was found where the Tn10 element had been inserted upstream of the leuO structural gene and downstream of its native promoter (Fig. 6). The increase in leuO expression upon the upstream insertion of a Tn10 element has been previously observed and substantiated in a similar manner as done here, upon analysis of collections of random mutants to establish regulators for the bgl operon and for the DsrA small regulatory RNA (25, 52). In this respect, ompS2 expression substantially increased in wild-type S. enterica serovar Typhi upon transformation with cloned leuO, either together with the upstream inserted Tn10 element (pFM800TL) (Fig. 7A) or under the control of the trc promoter (pFMTrcleuO) (Fig. 7B). This is consistent with the notion that an increase in the expression of LeuO causes an increase in ompS2 expression.
LeuO bound to the 5′ upstream regulatory region of ompS2 in two defined boxes, LeuO (I) and LeuO (II), at 0.03 μM and higher (Fig. 2 and 8). The lowest concentration is similar to the concentrations of LeuO used to protect the 25-bp AT7 box, which is part of the AT4 box, upstream of leuO and involved in LeuO binding, as defined in another footprinting study (9). Interestingly, Box II shares the motif 5′-AATCACA-3′ with the AT7 box and the 5′-AATCA-3′ motif with motif III, which is required for the LeuO-mediated negative effect on dsrA expression, as assessed with gene reporter fusions (46). Box I has a similar 5′-ATTCACCA-3′ motif to the one in box II (Fig. 2 and 8). Interestingly, as can be seen in Fig. 8, in the presence of LeuO there was an increased the affinity of OmpR-P for the regulatory region.
Our present model for ompS2 expression implies that LeuO relieves repression by disrupting either a structure in the region from −413 to −97 or by antagonizing the effect of an unknown repressor that binds to such region or both. Thus, OmpR-P would then be allowed to bind at and around the consensus OmpR-binding box, permitting ompS2 expression mostly at high osmolarity, where there is a higher abundance of OmpR-P (41). This is based on the following observations. The addition of an active cloned leuO gene allowed expression of the pFM413 construct at the level of the pFM97 construct, i.e., it had the same effect as the removal of the −413 to −97 region (Fig. 3). This was observed at low osmolarity, although expression increased at high osmolarity (Fig. 6 and 7). Furthermore, LeuO and OmpR-P bound in close proximity at the 5′ regulatory region (Fig. 2 and 8), and the activity rendered by pFM97 required phosphorylation at the major OmpR D55 residue (Fig. 4).
It is of interest that the effect of osmolarity was not observed in the expression of the OmpS2 protein under a −97 bp regulatory region (Fig. 5), yet it was evident upon expression from the chromosomal gene in the IMSSTN103 mutant (Fig. 6B) and from the full-length regulatory region reporter fusion (Fig. 6A and 7). This implies that OmpR-P is especially relevant in the presence of a full-length regulatory region and of the LeuO regulator. Hence, when the −413 to −97 region is absent, even relatively low concentrations of OmpR-P at low osmolarity would be sufficient for full expression. Thus, it is possible that osmolarity is affecting the structure of the −413 to −97 region, allowing for higher expression at high osmolarity. In this respect, no osmoregulation of leuO expression was observed by primer extension studies on pFMTrcleuO (data not shown), in support of a specific effect of osmolarity on ompS2 expression.
Hence, there are several similarities between ompS1 and ompS2 regarding their mode of expression. They both have an upstream negative regulatory region: in ompS1 it encompasses bp −310 to −88 upstream of the first transcribed nucleotide (36), and in ompS2 it encompasses bp −413 to −97. Also, they are both positively regulated by OmpR-P. On the other hand, a positive regulator such as LeuO has only been found for ompS2, although there is the distinct possibility that there is such a regulator for ompS1.
Certainly, the precise aspects of the mechanism of ompS2 activation and repression remain to be solved, together with the environmental cues that trigger ompS2 expression. In this respect, the stoichiometry of the OmpR-P and LeuO interactions with the ompS2 regulatory region, as well as the possible involvement of the H-NS nucleoid protein in repression should be assessed, together with the physiological conditions that affect the expression of both leuO and ompS2. Recently, a homologue of ompS2 has been implicated in the pathogenesis of Edwardsiella tarda in fish (45), suggestive of signals in the host that trigger its expression.
Acknowledgments
We thank M. L. Gutiérrez for the isolation of pM15, the original recombinant plasmid carrying the ompS2 gene, as the subject of her Master's degree thesis. We also thank Linda J. Kenney, Ricardo Oropeza, and Rob Edwards for helpful discussions and suggestions and Paul Gaytán and Eugenio Bustos for oligonucleotide synthesis.
This work was supported by grants to E.C. from the Universidad Nacional Autónoma de México (DGAPA IN229001), from the Consejo Nacional de Ciencia y Tecnología, México (CONACyT 37738-N), and by an International Research Scholar Award (75191-527102) from the Howard Hughes Medical Institute, Chevy Chase, Md. J.L.P. is funded by DGAPA, CONACyT, and HHMI.
REFERENCES
- 1.Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of Gram-negative bacteria. Gene 160:59-62. [DOI] [PubMed] [Google Scholar]
- 2.Aron, L., G. Faundez, C. González, E. Roessler, and F. Cabello. 1993. Lipopolysaccharide-independent radio-immunoprecipitation and identification of structural and in vivo induced immunogenic surface proteins of Salmonella typhi in typhoid fever. Vaccine 11:10-17. [DOI] [PubMed] [Google Scholar]
- 3.Barrios, H., H. M. Fischer, H. Hennecke, and E. Morett. 1995. Overlapping promoters for two different RNA polymerase holoenzymes control Bradyrhizobium japonicum nifA expression. J. Bacteriol. 177:1760-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, S. A., J. L. Occi, and B. A. Sampson. 1988. Mutations that alter the pore function of the OmpF porin of Escherichia coli K-12. J. Mol. Biol. 203:961-970. [DOI] [PubMed] [Google Scholar]
- 5.Benz, R., A. Schmid, and R. E. W. Hancock. 1985. Ion selectivity of gram-negative bacterial porins. J. Bacteriol. 162:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calva, E., J. L. Puente, and J. J. Calva. 1988. Research opportunities in typhoid fever: epidemiology and molecular biology. BioEssays 9:173-177. [DOI] [PubMed] [Google Scholar]
- 7.Chatfield, S. N., C. J. Dorman, C. Hayward, and G. Dougan. 1991. Role of ompR-dependent genes in Salmonella typhimurium virulence: mutants deficient in both OmpC and OmpF are attenuated in vivo. Infect. Immun. 59:449-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C.-C., M. Fang, A. Majumder, and H.-Y. Wu. 2001. A 72-base pair AT-rich DNA sequence element functions as a bacterial gene silencer. J. Biol. Chem. 276:9478-9485. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C.-C., M. Ghole, A. Majumder, Z. Wang, S. Chandana, and H.-Y. Wu. 2003. LeuO-mediated transcriptional derepression. J. Biol. Chem. 278:38094-38108. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgado, J., S. Forst, S. Harlocker, and M. Inouye. 1993. Identification of a phosphorylation site and functional analysis of conserved aspartic acid residues of OmpR, a transcriptional activator for ompF and ompC in Escherichia coli. Mol. Microbiol. 10:1037-1048. [DOI] [PubMed] [Google Scholar]
- 12.Doménech-Sánchez, A., S. Hernández-Allés, L. Martínez-Martínez, V. J. Benedí, and S. Albertí. 1999. Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in β-Lactam antibiotic resistance. J. Bacteriol. 181:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorman, C. J., S. Chatfield, C. F. Higgins, C. Hayward, and G. Dougan. 1989. Characterization of porin and ompR mutants of a virulent strain of Salmonella typhimurium: ompR mutants are attenuated in vivo. Infect. Immun. 57:2136-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang, M., A. Majumder, K.-J. Tsai, and H.-Y. Wu. 2000. ppGpp-dependent leuO expression in bacteria under stress. Biochem. Biophys. Res. Commun. 276:64-70. [DOI] [PubMed] [Google Scholar]
- 15.Feng, X., R. Oropeza, and L. J. Kenney. 2003. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol. Microbiol. 48:1131-1143. [DOI] [PubMed] [Google Scholar]
- 16.Fernández-Mora, M., R. Oropeza, J. L. Puente, and E. Calva. 1995. Isolation and characterization of ompS1, a novel Salmonella typhi outer membrane protein-encoding gene. Gene 158:67-72. [DOI] [PubMed] [Google Scholar]
- 17.Galdiero, M., M. Vitiello, E. Sanzari, M. D'Isanto, A. Tortora, A. Longanella, and S. Galdiero. 2002. Porins from Salmonella enterica serovar Typhimurium activate the transcription factors activating protein 1 and NF-kappaB through the Raf-1-mitogen-activated protein kinase cascade. Infect. Immun. 70:558-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock, R. E. W. 1987. Role of porins in outer membrane permeability. J. Bacteriol. 169:929-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harlocker, S. L., L. Bergstrom, and M. Inouye. 1995. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 270:26849-26856. [DOI] [PubMed] [Google Scholar]
- 20.Head, C. G., A. Tardy, and L. J. Kenney. 1998. Relative binding affinities of OmpR and OmpR-phosphate in the ompF and ompC. J. Mol. Biol. 281:857-870. [DOI] [PubMed] [Google Scholar]
- 21.Henikoff, S., G. W. Haughn, J. M. Calvo, and J. C. Wallace. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. USA 85:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, K.-J., and M. M. Igo. 1996. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J. Mol. Biol. 262:615-628. [DOI] [PubMed] [Google Scholar]
- 23.Isibasi, A., V. Ortiz, M. Vargas, J. Paniagua, C. González, J. Moreno, and J. Kumate. 1988. Protection against Salmonella typhi infection in mice after immunization with outer membrane proteins isolated from Salmonella typhi 9, 12, d, Vi. Infect. Immun. 56:2953-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeanteur, D., J. H. Lakey, and F. Pattus. 1991. The bacterial porin superfamily: sequence alignment and structure prediction. Mol. Microbiol. 5:2153-2164. [DOI] [PubMed] [Google Scholar]
- 25.Klauck, E., J. Böhringer, and R. Hengge-Aronis. 1997. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25:559-569. [DOI] [PubMed] [Google Scholar]
- 26.Klebba, P. E., and S. M. Newton. 1998. Mechanisms of solute transport through outer membrane porins: burning down the house. Curr. Opin. Microbiol. 2:238-248. [DOI] [PubMed] [Google Scholar]
- 27.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindgren, S. W., I. Stojilijkovic, and F. Heffron. 1996. Macrophage killing is an essential virulence mechanism of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:4197-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, X., and T. Ferenci. 2001. An analysis of multifactorial influences on the transcriptional control of OmpF and OmpC porin expression under nutrient limitation. Microbiology 148:2981-2989. [DOI] [PubMed] [Google Scholar]
- 30.Majumder, A., M. Fang, K.-J. Tsai, C. Ueguchi, T. Mizuno, and H.-Y. Wu. 2001. LeuO expression in response to starvation for branched-chain amino acids. J. Biol. Chem. 276:19046-19051. [DOI] [PubMed] [Google Scholar]
- 31.Martínez-Flores, I., R. Cano, V. H. Bustamante, E. Calva, and J. L. Puente. 1999. The ompB operon partially determines differential expression of OmpC in Salmonella typhi and Escherichia coli. J. Bacteriol. 181:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer, M. P. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163:41-46. [DOI] [PubMed] [Google Scholar]
- 33.Mills, S. D., S. R. Ruschkowski, M. A. Stein, and B. B. Finlay. 1998. Trafficking of porin-deficient Salmonella typhimurium mutants inside HeLa cells: ompR and envZ mutants are defective for the formation of Salmonella-induced filaments. Infect. Immun. 66:1806-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misra, R., and S. A. Benson. 1988. Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J. Bacteriol. 170:3611-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muthukummar, S., and V. R. Muthukkaruppan. 1993. Mechanism of protective immunity induced by porin-lipopolysaccharide against murine salmonellosis. Infect. Immun. 61:3017-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oropeza, R., C. L. Sampieri, J. L. Puente, and E. Calva. 1999. Negative and positive regulation of the non-osmoregulated ompS1 porin gene in Salmonella typhi: a novel regulatory mechanism that involves OmpR. Mol. Microbiol. 32:243-252. [DOI] [PubMed] [Google Scholar]
- 37.Pang, T., Z. A. Bhutta, B. B. Finlay, and M. Altwegg. 1995. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol. 3:253-255. [DOI] [PubMed] [Google Scholar]
- 38.Pickard, D., J. Li, M. Roberts, D. Maskell, D. Hone, M. Levine, G. Dougan, and S. Chatfield. 1994. Characterization of defined ompR mutants of Salmonella typhi: ompR is involved in the regulation of Vi polysaccharide expression. Infect. Immun. 62:3984-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pratt, L. A., and T. J. Silhavy. 1995. Identification of base pairs important for OmpR-DNA interaction. Mol. Microbiol. 17:565-573. [DOI] [PubMed] [Google Scholar]
- 40.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 41.Prilipov, A., P. S. Phale, R. Koebnik, C. Widmer, and J. P. Rosenbusch. 1998. Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli BE. J. Bacteriol. 180:3388-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puente, J. L., V. Flores, M. Fernández, Y. Fuchs, and E. Calva. 1987. Isolation of an ompC-like outer membrane protein gene from Salmonella typhi. Gene 61:75-83. [DOI] [PubMed] [Google Scholar]
- 43.Puente, J. L., A. Verdugo-Rodríguez, and E. Calva. 1991. Expression of Salmonella typhi and Escherichia coli OmpC is influenced differently by medium osmolarity; dependence on Escherichia coli OmpR. Mol. Microbiol. 5:1205-1210. [DOI] [PubMed] [Google Scholar]
- 44.Puente, J. L., D. Juárez, M. Bobadilla, C. F. Arias, and E. Calva. 1995. The Salmonella ompC gene: structure and use as a carrier for heterologous sequences. Gene 156:1-9. [DOI] [PubMed] [Google Scholar]
- 45.Rao, P. S. S., T. M. Lim, and K. Y. Leung. 2003. Functional genomics approach to the identification of virulence genes involved in Edwardsiella tarda pathogenesis. Infect. Immun. 71:1343-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Repoila, F., and S. Gottesman. 2001. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 183:4012-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Sanger, F., A. R. Coulson, B. G. Barrell, A. J. M. Smith, and B. A. Roe. 1980. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J. Mol. Biol. 143:161-178. [DOI] [PubMed] [Google Scholar]
- 49.Shapira, S. T., J. Chou, F. V. Richaud, and M. J. Casadaban. 1983. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of β-galactosidase. Gene 25:71-82. [DOI] [PubMed] [Google Scholar]
- 50.Shi, X., and G. N. Bennet. 1995. Effects of multicopy LeuO on the expression of the acid-inducible lysine decarboxylase gene in Escherichia coli. J. Bacteriol. 177:810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh, M., H. Vohra, L. Kumar, and N. K. Ganguly. 1999. Induction of systemic and mucosal immune response in mice immunized with porins of Salmonella typhi. J. Med. Microbiol. 48:79-88. [DOI] [PubMed] [Google Scholar]
- 52.Ueguchi, C., T. Ohta, C. Seto, T. Suzuki, and T. Mizuno. 1998. The leuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J. Bacteriol. 180:190-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verdugo-Rodríguez, A., Y. López-Vidal, J. L. Puente, G. M. Ruiz-Palacios, and E. Calva. 1993. Early diagnosis of typhoid fever by an enzyme immunoassay using Salmonella typhi outer membrane protein preparations. Eur. J. Clin. Microbiol. Infect. Dis. 12:248-254. [DOI] [PubMed] [Google Scholar]