Abstract
Background and Purpose
The Heart of Glass (HEG) receptor binds KRIT1 and functions with KRIT1, CCM2 and PDCD10 in a common signaling pathway required for heart and vascular development. Mutations in KRIT1, CCM2 and PDCD10 also underlie human cerebral cavernous malformation (CCM), and postnatal loss of these genes in the mouse endothelium results in rapid CCM formation. Here we test the role of HEG in CCM formation in mice and humans.
Methods
We constitutively or conditionally deleted Heg and/or Ccm2 genes in genetically modified mice. Mouse embryos, brain and retina tissues were analyzed to assess CCM lesion formation.
Results
CCMs form in postnatal mice with Ccm2−/− but not Heg−/− or Heg−/−;Ccm2+/−endothelial cells. Consistent with these findings, human patients with CCM who lack exonic mutations in KRIT1, CCM2 or PDCD10 do not have mutations in HEG.
Conclusion
These findings suggest that the HEG-CCM signaling functions during cardiovascular development and growth, while CCMs arise due to loss of HEG-independent CCM signaling in the endothelium of the central nervous system after birth.
Introduction
Cerebral cavernous malformations (CCMs) are common vascular malformations that arise primarily in the central nervous system (CNS)1. CCMs are typically diagnosed in middle age, and constitute an important cause of stroke and neurologic deficit in younger individuals. One third of CCMs are familial, and positional cloning studies have identified loss of function mutations in three genes, KRIT1, CCM2 and PDCD10, as causal for this disease (reviewed in1). KRIT1, CCM2 and PDCD10 encode intracellular adaptor proteins that have been shown to form a single biochemical complex that is bound by the transmembrane receptor Heart of Glass (HEG)2, but the role of HEG in CCM disease has not been defined.
Fish and mouse genetic studies have demonstrated that HEG, KRIT1, CCM2 and PDCD10 function together in endothelial cells during formation of the heart and vasculature2–5. In addition, inducible endothelial deletion of Krit1, Ccm2 or Pdcd10 in neonatal mice results in the formation of retinal and hindbrain CCMs that accurately reproduce the human disease6, 7. In the present study we use genetically modified mice and studies of human patients with familial CCM to rigorously test the role of HEG in CCM formation.
Materials and Methods
Mice
Mutant Heg and Ccm2 mouse alleles and Cre transgenic mice have been described previously2, 8–10. The University of Pennsylvania IACUC approved all animal protocols.
Endothelial cell isolation and qPCR
Lung endothelial cells were isolated using anti-PECAM beads and qPCR performed after cDNA synthesis using SYBR Green (Applied Biosystems).
Evans blue dye extravasation assay
3mg/g Evans blue dye was administrated via tail vein injection 16 hours prior to sacrifice and pulmonary vascular perfusion with saline.
Human studies
Twenty-one unrelated patients and six healthy controls were used. The study was approved by the local ethics committee.
Sequencing and QMPSF analysis
HEG1 sequencing was performed after coding exon amplification using primers indicated in Supp. Table I. HEG genomic rearrangements were assessed using the Quantitative Multiplex PCR of Short Fluorescent fragments (QPMSF) method as described.
Results
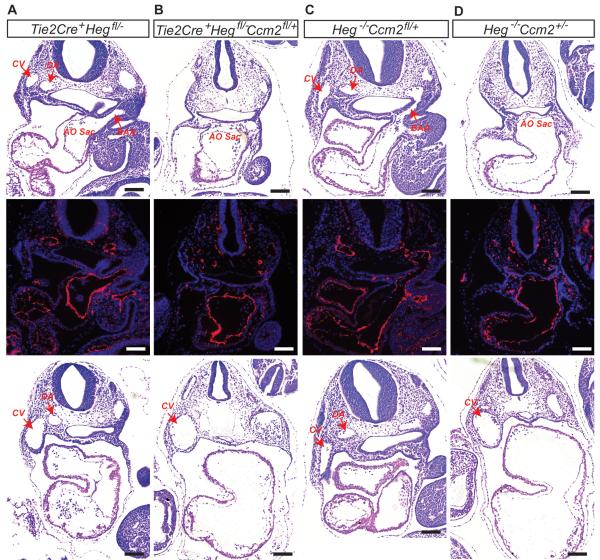
We have previously found that Heg−/−;Ccm2+/− die at E10 with vascular defects identical to those detected in Krit1−/− and Ccm2−/− embryos2. To determine if HEG and CCM proteins function together within endothelial cells we generated mice lacking both Heg alleles and a single Ccm2 allele exclusively in the endothelium. At E9.5 Tie2-Cre; Hegfl/− and Heg−/−;Ccm2fl/+ embryos exhibited normal blood circulation and patent branchial arch arteries (BAAs), but Tie2-Cre;Hegfl/−;Ccm2fl/+ littermates lacked patent BAAs, a phenotype identical to that of Heg−/−;Ccm2+/− embryos (Fig. 1A–D). Thus Heg and Ccm2 interact within endothelial cells during early cardiovascular development.
Figure 1. Heg and Ccm2 interact within endothelial cells during embryonic development.
H&E staining and Immunostaining with anti-Pecam antibody of transverse sections of E9.5 embryos reveal the presence of normally lumenized dorsal aortas (DA), cardinal veins (CV) and branchial arch arteries (BAA) in the Tie2Cre+;Heg1fl/−(A) and Heg−/−;Ccm2fl/+(C) embryos but not in Tie2Cre+;Heg1fl/−Ccm2fl/+(B) or Heg−/−;Ccm2+/−(D) embryos. H&E staining of transverse sections of caudal region of E9.5 embryos also revealed dilated cardinal veins and non-lumenized dorsal aortas in Tie2Cre+;Heg1fl/−Ccm2fl/+(B), and Heg−/−;Ccm2+/−(D) embryos. AO Sac, aortic sac; CV, cardinal vein; DA, dorsal aorta; BAA, Branchial arch ateries. Scale bars, 100μm.
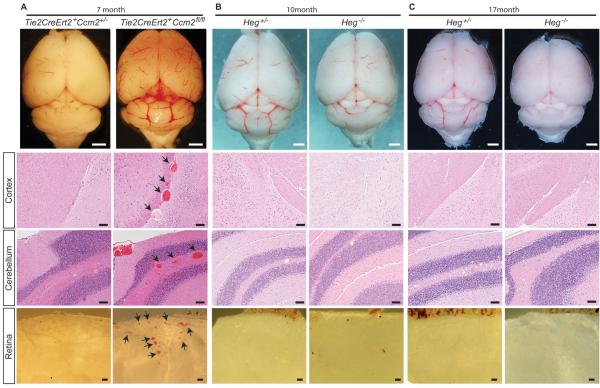
Unlike Ccm2−/− animals, Heg−/− mice survive past birth and approximately half live to advanced age2. To test the role of HEG in CCM formation we compared the brains of Heg−/− animals at ages 10 and 17 months with those of animals in which Ccm2 had been deleted postnatally in endothelial cells. All Tie2-CreERT2;Ccm2fl/fl mice exhibited CCM formation in the meninges, cerebellum and retina (Fig. 2A). In contrast, Heg−/− animals failed to exhibit CCM formation, even at 17 months of age (Fig 2B and C; N=34, Supp. Table II). Thus loss of HEG, in contrast to loss of CCM2, does not result in CCM formation in mice.
Figure 2. Loss of HEG does not result in CCM formation.
Postnatal deletion Ccm2 in Tie2CreErt2+; Ccm2fl/fl animals results in CCM formation in areas adjacent to the meninges, and in the cerebellum and retina (A). No CCM lesion were identified in the brains of Heg-deficient mice at age 10 months (B) or 17 months (C) age. Arrows indicate CCMs. White scale bars, 1mm; Black scale bars, 100μm.
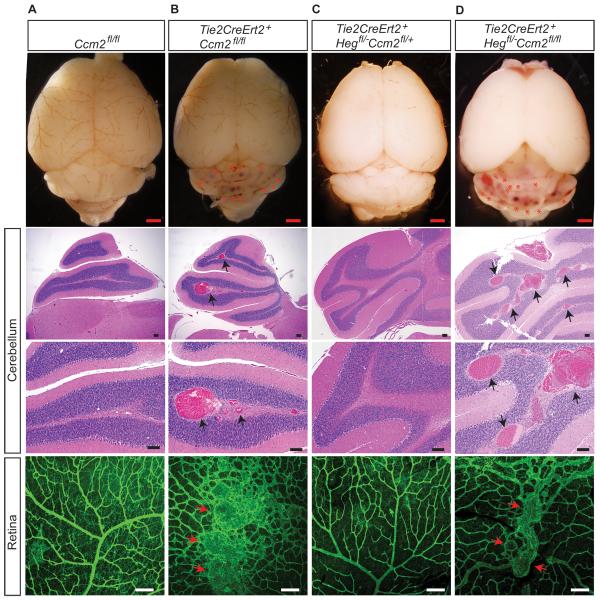
To further address the role of HEG in CCM formation we conditionally deleted one Ccm2 allele in Tie2-Cre;Heg−/−;Ccm2fl/+ animals, creating postnatal animals with the same genotype as embryos that exhibit vascular defects identical to those of endothelial Ccm2−/− animals. Although endothelial deletion of Ccm2 immediately after birth conferred rapid CCM formation by P17 (Fig. 3A–B and Supp. Table II; N= 4), deletion of one allele of Ccm2 in Heg−/− animals failed to confer CCM formation in either the retina or hindbrain (Fig. 3C and Supp. Table II; N=16). The number of visible hindbrain CCM lesions in endothelial Ccm2−/− animals did not differ from that in endothelial Heg−/−;Ccm2−/− animals (mean lesion number of 17 and16 respectively; N=3 for both groups) (Fig. 3D).
Figure 3. Heg and Ccm2 do not interact in endothelial cells during CCM formation.
(A–D) Whole-mount brain imaging, H&E staining and whole-mount isolectin staining reveals numerous CCM lesions in the cerebellum and retina of P18 Tie2CreErt2+;Ccm2fl/fl (B) and Tie2CreErt2+;Hegfl/−;Ccm2fl/fl (D) mice, but not in Tie2CreErt2+; Hegfl/−;Ccm2fl/+(C) mice following postnatal deletion of Heg and Ccm2 in the endothelium. Arrows indicate CCMs. Red scale bars, 1mm; Black and white scale bars, 100μm.
To determine if HEG1 might be a human CCM disease gene, we analyzed this gene in 21 unrelated patients with CCMs identified by cerebral MRI and/or pathological examination, and in whom no point mutation or copy number anomaly was detected in KRIT1, CCM2 or PDCD10. Eighteen cases were sporadic with multiple lesions and 3 were familial (at least one relative with CCM lesions) (Supp.Table III). None of these CCM patients exhibited germline mutations or large deletions in HEG1. Sixteen exonic polymorphisms were detected. All were present in SNP databases (Supp. Table IV) and 14 have a frequency >5%.
A role for HEG-CCM signaling in endothelial barrier function has been demonstrated in vitro and in vivo, and proposed to participate in CCM disease pathogenesis11, 12. Unlike CCM2-deficient lung endothelial cells, lung endothelial cells harvested from Heg−/− mice expressed normal levels of the endothelial cell junction genes Claudin5 and VE-cadherin by qPCR (Supp. Fig. I). In addition, while endothelial loss of Ccm2 conferred a 60% increase in Evans blue extravasation in the lungs of Cdh5-CreERT2; Ccm2fl/fl mice, no difference was observed in Heg−/− mice (Supp. Fig. I). The role of endothelial barrier function in CCM pathogenesis remains speculative, but these studies suggest that HEG is not required in the CCM signaling pathway that supports vascular integrity.
Discussion
How loss of CCM signaling causes CCM formation and why CCMs form so specifically in the CNS remain unanswered questions. Our studies reveal roles for HEG during embryonic CCM signaling, but not in the postnatal pathway that underlies CCM pathogenesis.
One interpretation of these studies is that there exist multiple upstream inputs to the CCM signaling pathway in endothelial cells, e.g. HEG during cardiovascular growth and another to prevent CCM formation and perhaps maintain vascular barrier function elsewhere. Definitive proof of distinct upstream activators of CCM signaling will require the molecular identification of such proteins and genetic studies linking their function to CCM formation. Alternatively, it remains possible that HEG couples to CCM signaling in the CNS endothelium but that its loss does not disable the pathway to the extent required for lesion formation. The lack of CCMs in postnatal Tie2-CreERT2;Heg−/−; Ccm2fl/+ animals that carry an endothelial deficiency state equivalent to that which causes embryonic phenotypes identical to those conferred by complete KRIT1 or CCM2 deficiency suggests that these studies have a reasonable sensitivity to detect a role for HEG in CCM formation. In either case, our studies indicate that HEG cannot be the sole upstream activator of CCM signaling in the CNS endothelium; thus the remarkable specificity of this disease for that organ is likely to reflect the function of other activator(s) that remain to be identified.
Supplementary Material
Acknowledgments
Source of Funding These studies were supported by NIH grants R01HL094326 and R01HL102128 (MLK) and AHA grant 11SDG7430025 (XZ).
Footnotes
Disclosures: none.
References
- 1.Riant F, Bergametti F, Ayrignac X, Boulday G, Tournier-Lasserve E. Recent insights into cerebral cavernous malformations: The molecular genetics of ccm. FEBS J. 2010;277:1070–1075. doi: 10.1111/j.1742-4658.2009.07535.x. [DOI] [PubMed] [Google Scholar]
- 2.Kleaveland B, Zheng X, Liu JJ, Blum Y, Tung JJ, Zou Z, et al. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med. 2009;15:169–176. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead KJ, Chan AC, Navankasattusas S, Koh W, London NR, Ling J, et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via rho gtpases. Nat Med. 2009;15:177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng X, Xu C, Di Lorenzo A, Kleaveland B, Zou Z, Seiler C, et al. Ccm3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J Clin Invest. 2010;110:2795–2804. doi: 10.1172/JCI39679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mably JD, Chuang LP, Serluca FC, Mohideen MA, Chen JN, Fishman MC. Santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;123:3129–3146. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- 6.Chan AC, Drakos SG, Ruiz OE, Smith AC, Gibson CC, Ling J, et al. Mutations in 2 distinct genetic pathways result in cerebral cavernous malformations in mice. J Clin Invest. 2011;111:1871–1881. doi: 10.1172/JCI44393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulday G, Rudini N, Maddaluno L, Blecon A, Arnould M, Gaudric A, et al. Developmental timing of ccm2 loss influences cerebral cavernous malformations in mice. J Exp Med. 2011;208:1835–1847. doi: 10.1084/jem.20110571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng X, Xu C, Smith AO, Stratman AN, Zou Z, Kleaveland B, et al. Dynamic regulation of the cerebral cavernous malformation pathway controls vascular stability and growth. Dev Cell. 2011;23:342–355. doi: 10.1016/j.devcel.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korhonen H, Fisslthaler B, Moers A, Wirth A, Habermehl D, Wieland T, et al. Anaphylactic shock depends on endothelial gq/g11. J Exp Med. 2009;206:411–420. doi: 10.1084/jem.20082150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, et al. Ephrin-b2 controls vegf-induced angiogenesis and lymphangiogenesis. Nature. 2010;465:483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 11.Glading A, Han J, Stockton RA, Ginsberg MH. Krit-1/ccm1 is a rap1 effector that regulates endothelial cell cell junctions. J Cell Biol. 2007;179:247–254. doi: 10.1083/jcb.200705175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stockton RA, Shenkar R, Awad IA, Ginsberg MH. Cerebral cavernous malformations proteins inhibit rho kinase to stabilize vascular integrity. J Exp Med. 2010;207:881–96. doi: 10.1084/jem.20091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.