Abstract
Protein scaffolds play an important role in signal transduction, functioning to facilitate protein interactions and localize key pathway components to specific signaling sites. Connector enhancer of KSR-2 (CNK2) is a neuronally-expressed scaffold recently implicated in non-syndromic, X-linked mental retardation (MRX) [1–3]. MRX patients have deficits in cognitive function and their neurons often exhibit dendritic spine abnormalities [4], suggesting a role for CNK2 in synaptic signaling and/or spine formation. To gain insight regarding how CNK2 might contribute to these processes, we used mass spectrometry to identify proteins that interact with the endogenous CNK2 scaffold. Here, we report that the major binding partner of CNK2 is Vilse/ARHGAP39 and that CNK2 complexes are enriched for proteins involved in Rac/Cdc42 signaling, including Rac1 itself, α-/β-PIX, GIT1/2, PAK3/4, and members of the cytohesin family. Binding between CNK2 and Vilse was found to be constitutive, mediated by the WW-domains of Vilse and a proline motif in CNK2. Through mutant analysis, protein depletion and rescue experiments, we identify CNK2 as a spatial modulator of Rac cycling during spine morphogenesis and find that the interaction with Vilse is critical for maintaining RacGDP/GTP levels at a balance required for spine formation.
Results and Discussion
The CNK2 Scaffold Interacts with Components Involved in Rho Family GTPase Signaling
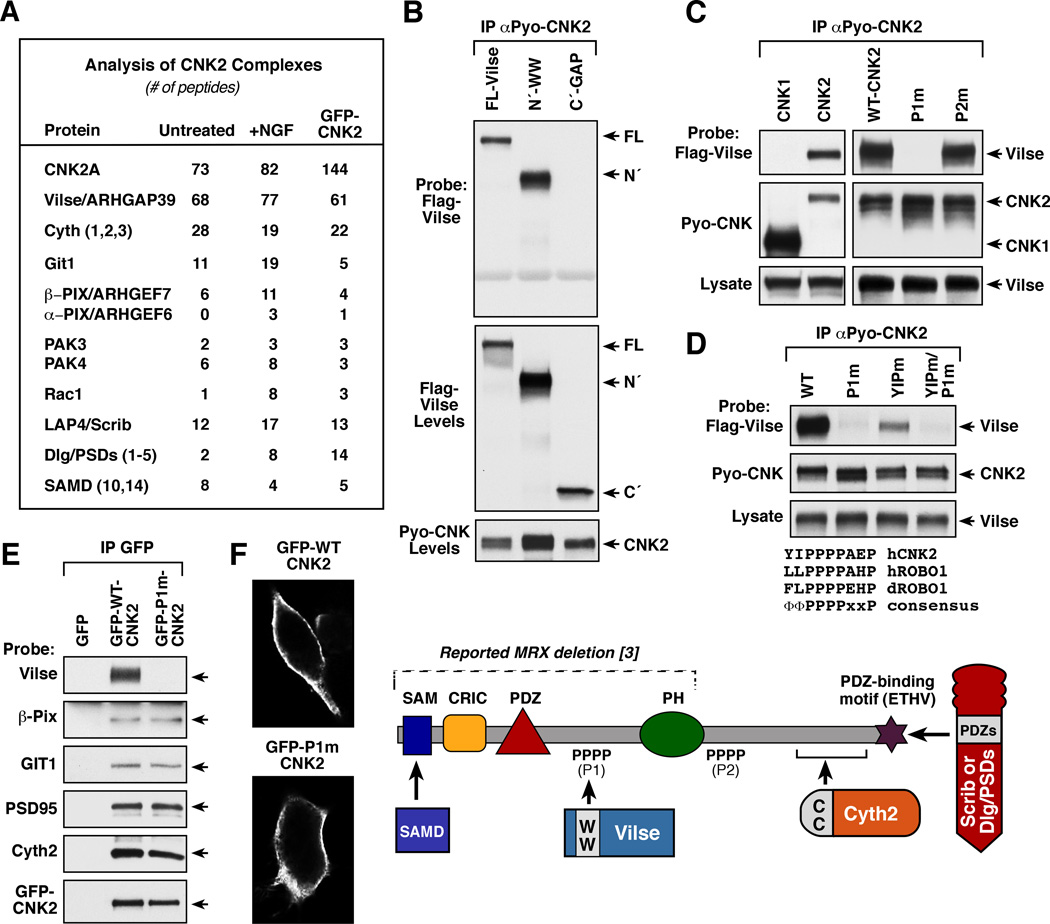
To gain insight regarding CNK2 function in neuronal signaling, we used mass spectrometry to identify proteins that interact with the endogenous CNK2 scaffold. CNK2 complexes were isolated from NG108 cells before and after 18 hrs of NGF treatment. The complexes were separated by SDS-PAGE, following which proteins were extracted from the gel matrix and analyzed by ion trap mass spectrometry. To control for CNK2-binding specificity, proteomic analysis was also performed on exogenous GFP-tagged CNK2 complexes isolated from NG108 cells (Figure 1A and Table S1). As confirmation of this approach, peptides from previously known CNK2-interacting proteins were detected, including PSD95/DLG5, and members of the SAMD, LAP, and cytohesin families [2, 5–7] (Figure 1A, S1). Of the previously unknown CNK2-binding partners, many were components involved in Rho family GTPase signaling. These include Vilse/ARHGAP39, which functions primarily as a Rac GTPase activating protein (GAP) [8, 9], the Rac/Cdc42 guanine nucleotide exchange factors (GEFs) α-/β-PIX, the Rac/Cdc42 effector kinases PAK3/4, as well as Rac1 itself. Interestingly, loss-of function mutations in two of these binding partners, α-Pix and PAK3, have also been reported in patients with MRX [10, 11]. The CNK2 complexes also contained GIT1/2, which contribute to Rac signaling through their interaction with α-/β-PIX [12]. Strikingly, of the proteins detected in the CNK2 complexes, the RacGAP Vilse was the predominant binding partner, with an almost equal stochiometry in the number of peptides detected for endogenous CNK2 and Vilse. Endogenous binding of CNK2 to Vilse, PSD95, Cytohesin-2, β-Pix, GIT1, and Scribble was further confirmed by immunoblot analysis (Figure S1A).
Figure 1. Identification of Vilse/ARHGAP39 as the Major Binding Partner of CNK2.
(A) Mass spectrometry analysis of endogenous CNK2 complexes and exogenous GFP-CNK2 complexes isolated from NG108 cells. (B–D) Pyo-CNK proteins were immunoprecipitated from lysates of 293 cells coexpressing the indicated Pyo-CNK and Flag-Vilse proteins. The immune complexes were then probed for the presence of Flag-Vilse by immunoblot analysis. Vilse proteins examined include full-length (FL), N´-WW (residues 1–698), and C´ (residues 699–1083). CNK proteins examined include CNK1, WT- and CNK2 mutants P1m, P2m, YIPm, and YIPm/P1m. Sequence alignment of the human CNK2 P1 motif with the human and Drosophila Robo1 CC2 proline motifs is shown in (D). (E) GFP immunoprecipitates were prepared from NG108 cells expressing GFP, GFP-WT-, or GFP-P1m-CNK2 proteins, and the immune complexes were probed for the binding of the indicated endogeneous proteins. (F) Localization of GFP-WT- and P1m-CNK2 proteins was visualized by confocal microscopy. A model depicting CNK2 and known protein interactions is also shown.
A Proline Motif on the CNK2 Scaffold Mediates Vilse Binding
To further analyze the significance of the CNK2/Vilse interaction, we sought to identify CNK2 residues required for Vilse binding. When truncation mutants of Vilse were examined for their ability to interact with CNK2 in coimmunoprecipitation assays, a protein encoding the N-terminal region of Vilse, which contains two WW domains, associated with CNK2, as did the full-length protein; however, a protein encoding the Vilse C-terminal region did not (Figure 1B). WW domains are known to interact with short proline-rich motifs [13], and CNK2 contains two such motifs, one at amino acid positions 354–357 (PPPP, P1) and one encompassing residues 703–706 (PPPP, P2). These motifs are not present in the CNK1 family member, and as expected, Vilse failed to co-immunoprecipitate with CNK1 (Figure 1C). The CNK2 P1 motif was further identified as the Vilse interaction site in that mutation of proline residues in the P1 motif (P1m), but not the P2 motif (P2m), disrupted Vilse binding (Figure 1C).
Vilse has been previously reported to interact with the axon guidance receptor Robo1 in a manner requiring the WW domains of Vilse and the CC2 proline-rich region of Robo1 [8]. Comparison of the sequences surrounding the CNK2 P1 and Robo1 CC2 motifs revealed that both contain hydrophobic amino acids in the −1 and −2 positions relative to the core PPPP sequence and a proline residue in the +3 position (Figure 1D). Interestingly, when the CNK2 residues in the −1, −2, and +3 positions were mutated to alanine (YIPm) in the absence of the core P1 proline mutations, binding between Vilse and CNK2 was significantly reduced (Figure 1D). These findings support the model that residues flanking the proline motif may provide additional specificity for WW domain interactions, and indicate that ΦΦPPPPxxP might represent a conserved binding motif for the WW domains of Vilse. Comparative mass spectrometry analysis of GFP-WT- and P1m-CNK2 scaffold complexes further revealed that Vilse was the only CNK2 binding partner not present in P1m-CNK2 complexes (Table S1). GFP-P1m-CNK2 was fully competent to bind all other CNK2-interactors, including PSD95, β-Pix, GIT1, and Cytohesin-2 (Table S1, Figure 1E), and it exhibited a similar plasma membrane localization as did GFP-WT-CNK2 ([14, 15]; Figure 1F).
Functional Analysis of the CNK2/Vilse Interaction in NG108 Cells
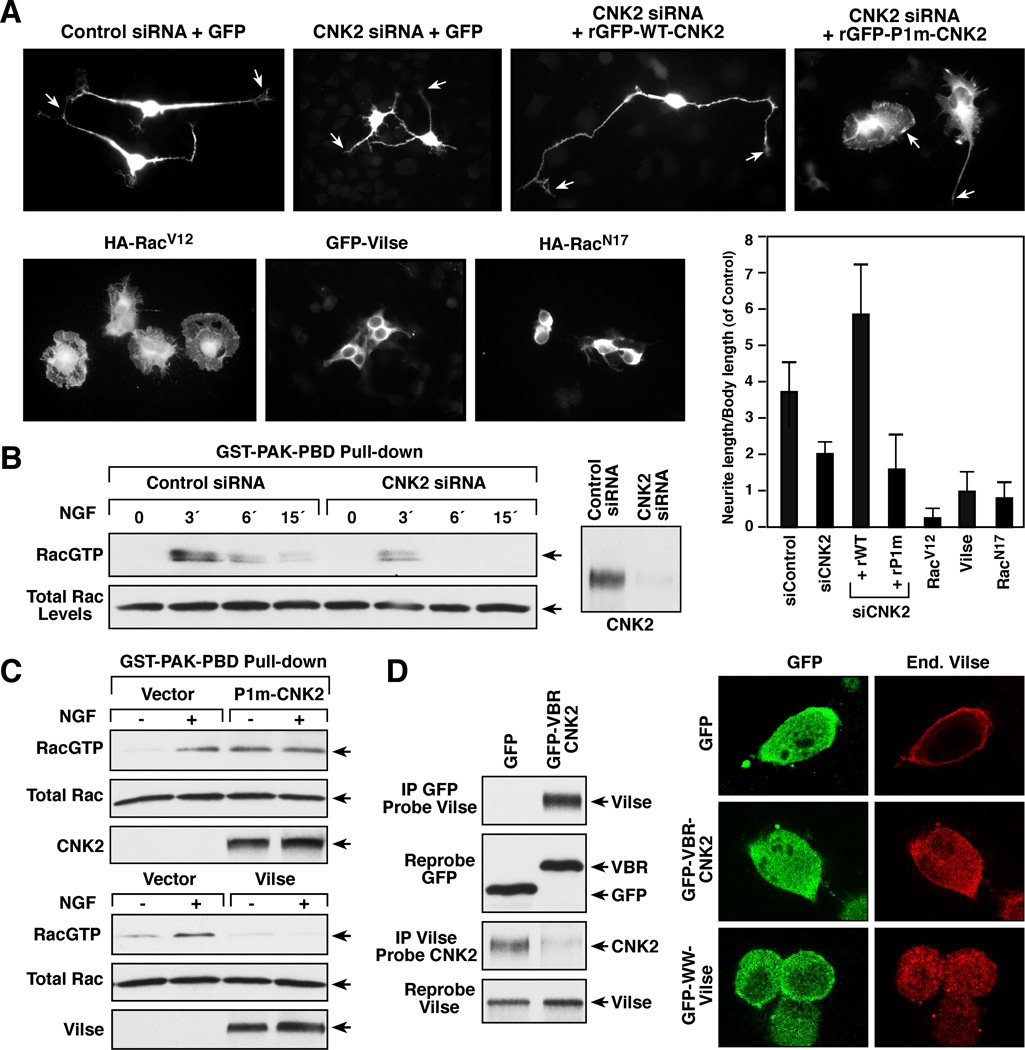
To investigate the functional importance of the CNK2/Vilse interaction, our initial experiments were conducted in NG108 cells, due to the ease of their biochemical analysis. For these studies, we first monitored NGF-induced neurite outgrowth and RacGTP loading, given that 1) the periodic cycling of Rac between its GDP and GTP-bound states is required for NGF-induced neurite outgrowth [16, 17], 2) CNK2 interacts with numerous proteins, including Vilse, that modulate Rac cycling, and 3) CNK2 has been reported to contribute a function to NGF-induced neurite outgrowth that is distinct from any effects on ERK cascade signaling [18]. As shown in Figure 2A and B, depletion of CNK2 protein levels using a small interfering RNA (siRNA) caused a significant reduction in the levels of RacGTP induced by NGF treatment and resulted in reduced neurite outgrowth. When siRNA-resistant CNK2 proteins were re-expressed in the CNK2-depleted cells, WT-CNK2 restored neurite outgrowth and even increased neurite length was observed (Figure 2A). In contrast, cells expressing the P1m-CNK2 mutant defective in Vilse-binding had shorter neurites and exhibited spreading of the cell body, a phenotype similar to that observed in cells expressing constitutively active RacV12 ([16]; Figure 2A) and which correlated with elevated basal RacGTP levels (Figure 2C), suggesting that the CNK2/Vilse interaction might impact RacGDP/GTP cycling. As expected, Vilse functioned as a RacGAP in NG108 cells in that its overexpression blocked NGF-induced RacGTP loading (Figure 2C). Moreover, overexpression of Vilse inhibited neurite outgrowth and resulted in a retracted cell body shape similar to that observed in cells expressing dominant negative RacN17 (Figure 2A).
Figure 2. Functional Analysis of the CNK2/Vilse Interaction in NG108 cells.
(A) NG108 cells transfected with the indicated siRNAs or cDNA constructs were treated with NGF and monitored for neurite outgrowth 72 hrs after transfection. Neurite length was quantified for 3 independent experiments and error bars indicate SD. (B) NG108 cells transfected with control or CNK2 siRNAs were treated with NGF for the indicated times prior to lysis. Lysates were incubated with glutathione beads containing GST-PAK-PBD, and binding of endogenous GTP-bound Rac to GST-PAK-PBD was determined by immunoblot analysis. Total Rac levels are also shown. (C) Cells expressing vector control, GFP-P1m-CNK2, or Flag-Vilse were examined for RacGTP levels as in (B). (D) Cells expressing GFP or GFP-VBR-CNK2 were examined for the localization of endogenous Vilse and for complex formation between endogenous Vilse and CNK2.
To further assess the effects of CNK2/Vilse binding, GST- and GFP-fusion proteins containing the isolated Vilse-binding region of CNK2 (VBR-CNK2) were generated. In in vitro GAP activity assays, we found that when GST-VBR-CNK2 was allowed to bind purified Vilse, no significant change in the GAP activity of Vilse was observed (Figure S2A). However, when GFP-VBR-CNK2 was expressed in NG108 cells, the endogenous interaction between CNK2 and Vilse was disrupted, and endogenous Vilse, which normally localized with CNK2 at the plasma membrane, no longer exhibited membrane staining (Figure 2D, S2C). In a similar manner, when a GFP protein containing the isolated WW-domains of Vilse (GFP-WW-Vilse) was used to disrupt the CNK2/Vilse interaction, Vilse showed little to no membrane localization (Figure 2D, S2B). Taken together these findings suggest that physical binding of Vilse to CNK2 does not directly modulate Vilse activity, but rather functions to regulate Vilse localization.
CNK2 is Required for Spine Morphogenesis in Hippocampal Neurons
Deletions in the human CNK2 gene are associated with non-syndromic, X-linked mental retardation (MRX) [3]; V. Kalscheur personal communication] and given that patients with MRX display abnormalities in the number and shape of their dendritic spines [4, 19], CNK2 may have a biological function in spine formation. Dendritic spines are small actin-rich protrusions on neurons that receive the majority of excitatory synaptic inputs in the brain [20]. During development, spine formation begins with the extension of highly, dynamic filopodia-like protrusions from the dendritic shafts [21]. Upon axonal contact, the protrusions subsequently stabilize and mature into spines that are stubby and mushroom-shaped [22]. Spine morphogenesis requires dynamic changes in the actin cytoskeleton, and Rac is a key regulator of actin structures during spine formation [20]. Moreover, dysregulated Rac signaling can cause spine defects, with expression of constitutively active RacV12 inducing many long, thin protrusions that fail to mature into spines and expression of dominant-negative RacN17 preventing both protrusion and spine formation [23, 24]. As previously reported [25], CNK2 was detected in the dendritic spines of primary hippocampal rat neurons cultured for 18 days in vitro (18 DIV), and it colocalized with the excitatory synaptic marker PSD95 (Figure 3A). Endogenous Vilse also colocalized with PSD95 (Figure 3A) and could be co-immunoprecipitated with CNK2 from lysates of 18 DIV neurons (Figure 3A), confirming the endogenous interaction of these proteins in hippocampal neurons.
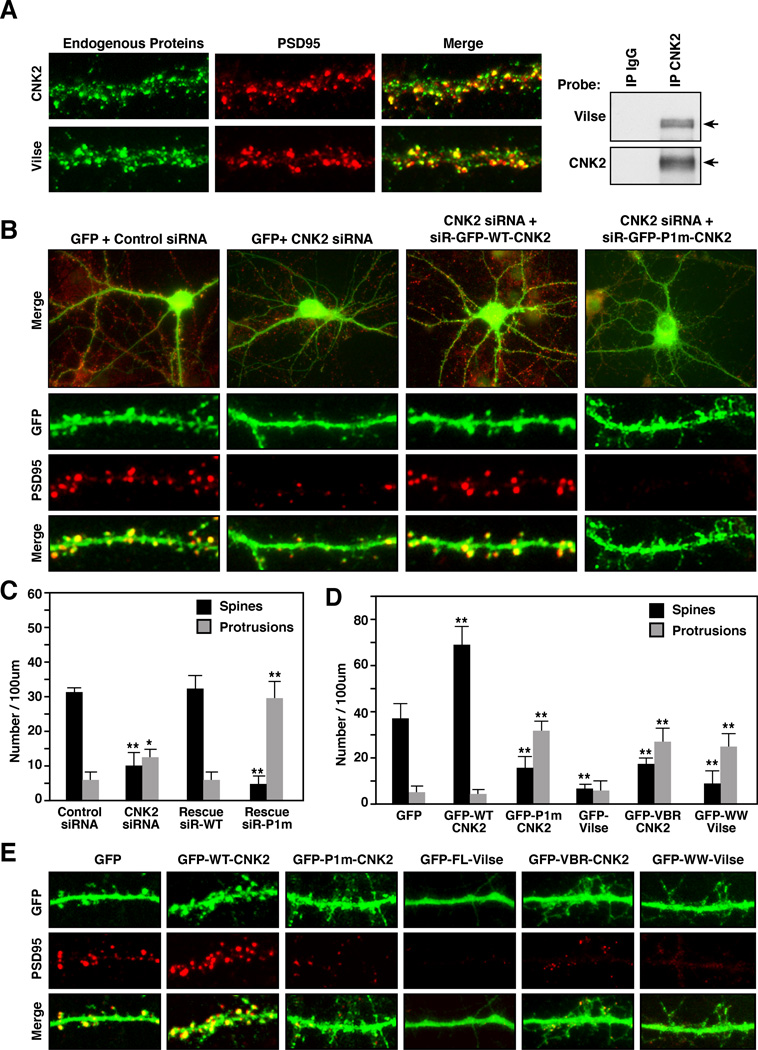
Figure 3. CNK2 is Required for Dendritic Spine Morphogenesis.
(A) Primary rat hippocampal neurons were cultured for 18 DIV and then fixed and stained for PSD95 and endogenous CNK2 or Vilse. Endogenous CNK2 was immunoprecipitated from lysates of 18 DIV neurons, and the immune complexes were probed for Vilse binding. (B,E) Neurons were transfected with the indicated siRNAs and/or cDNA constructs at 7 DIV. Morphology of neurons fixed and stained at 18 DIV is shown. (C,D) Quantification of dendritic spines and protrusions in the transfected neurons is shown. Error bars indicate SD and **p<0.001.
To determine whether depleting CNK2 protein levels would alter spine morphogenesis, control or CNK2 siRNAs were co-transfected with a GFP-encoding construct into neurons at 7 DIV, following which the neurons were fixed and immunostained for GFP and the synaptic marker PSD95 at 18 DIV. As shown in Figure 3B, mature mushroom-shaped spines were clearly visible on the dendrites of control siRNA transfected cells, and the majority of the spines were associated with clusters of PSD95 staining. In neurons transfected with CNK2 siRNAs, the number of mature spines was dramatically reduced, whereas the number of dendritic protrusions was increased (Figure 3B,C). These effects were specifically attributable to the loss of CNK2 in that formation of mature spines was restored when a siRNA-resistant GFP-WT-CNK2 was coexpressed with the CNK2 siRNA (Figure 3B,C). In contrast, coexpression of the siRNA-resistant GFP-P1m-CNK2 mutant, which lacks Vilse binding, did not restore spine formation, but rather caused a more dramatic increase in the number of dendritic protrusions (Figure 3B,C), a phenotype again similar to that observed in cells overexpressing constitutively active RacV12.
Next, we examined the effect of upregulating CNK2 protein levels. In comparison to GFP-expressing neurons, those overexpressing WT-CNK2 had a significant increase in the number of mature spines and PSD clusters (Figure 3D,E). In contrast, neurons overexpressing the P1m-CNK2 mutant had an increased number of dendritic protrusions and fewer mature spines (Figure 3D,E). These defects appeared to be due to the specific loss in Vilse binding in that a similar increase in dendritic protrusions was observed in neurons overexpressing either GFP-VBR-CNK2 or GFP-WW-Vilse to disrupt the CNK2/Vilse interaction (Figure 3D,E). Overexpression of Vilse also had an effect on spine formation, with neurons expressing GFP-Vilse having smooth dendritc shafts and few if any spines or dendrites (Figure 3D,E), a phenotye similar to that reported for neurons expressing RacN17 [23]. Spine maturation could be restored in the Vilse-expressing neurons when Vilse was coexpressed with RacV12, and the spine defects observed in P1m-CNK2 neurons were suppressed when P1m-CNK2 was coexpressed with either RacN17 or Vilse, suggesting that these morphogenic defects were due to effects on Rac GDP/GTP cycling (Figure 4A).
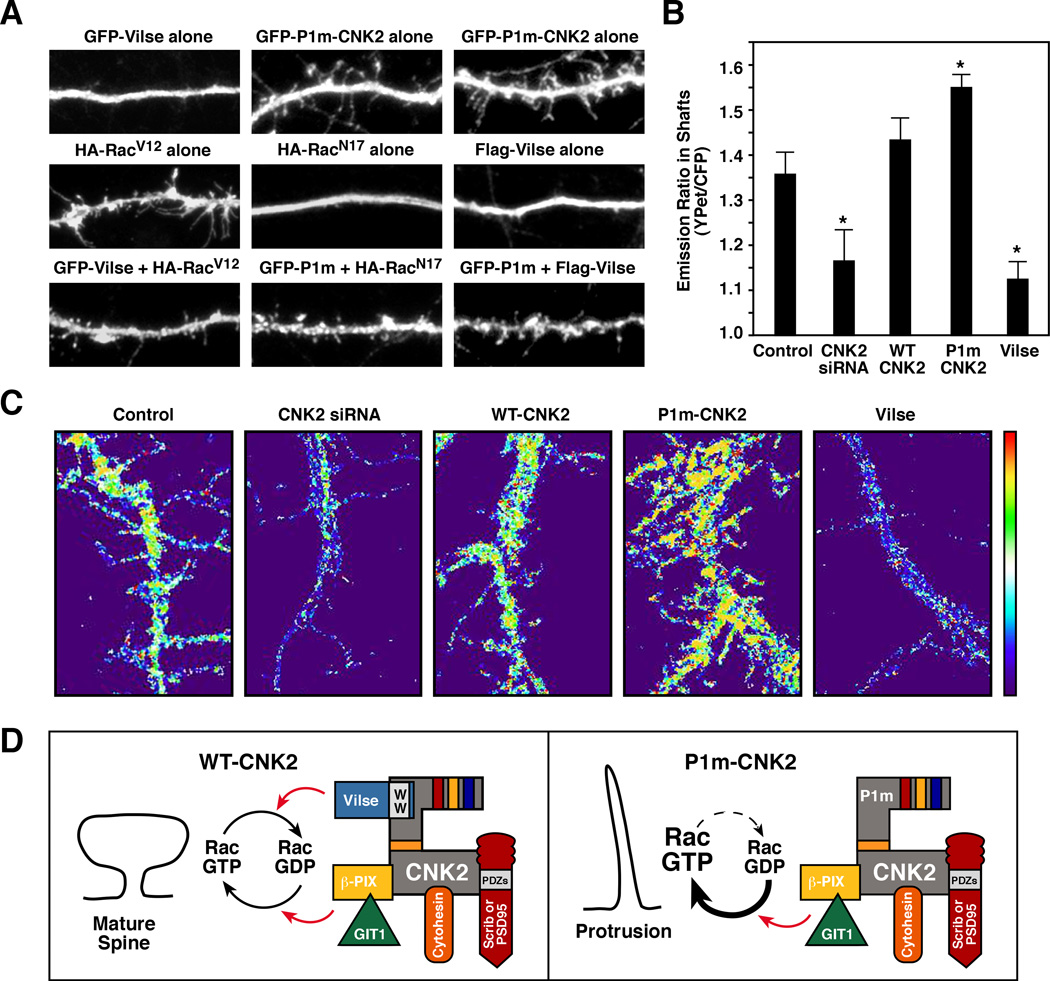
Figure 4. CNK2 Modulates RacGDP/GTP Cycling During Spine Morphogenesis.
(A) Neurons transfected with the indicated constructs at 7 DIV were fixed and stained at 14–18 DIV. RacV12 was able to reverse Vilse overexpression defects, and both Vilse and RacN17 were able to suppress P1m-CNK2 defects. (B, C) Neurons were transfected with the RaichuEV-Rac1 FRET reporter and CNK2 siRNA or Pyo-WT-CNK2, Pyo-P1m-CNK2, Flag-Vilse constructs at 8 DIV and FRET analyses were performed 48–60 hrs later. Quantification of emission ratios (B) and ration images of FRET efficiency (C) in a representative dendritic shaft is shown. Error bars indicate SEM; *p < 0.05. (D) Model depicting CNK2 function as a spatial modulator of RacGDP/GTP cycling.
CNK2 Modulates Rac Activity During Spine Morphogenesis
The above findings suggest a model whereby CNK2 tethers regulators that promote Rac cycling and functions to modulate RacGDP/GTP levels at localized sites during spine morphogenesis. Therefore, to monitor Rac activity in live neurons, we used a fluorescent resonance energy transfer (FRET) biosensor RaichuEV-Rac1 that contains YPet, the PAK PBD, an EV linker (to reduce background FRET), Rac1, and CFP [26]. Upon GTP-loading, Rac1 interacts with the neighboring PAK-PBD domain, thus bringing YPet in close proximity to CFP and inducing FRET. To determine whether changes in CNK2 levels affect Rac cycling, we expressed the RaichuEV-Rac1 probe in neurons that were depleted of CNK2 or that overexpressed WT-CNK2, P1m-CNK2 or Vilse (Figure 4B,C and Figure S3). In comparison to control neurons, a decrease in FRET efficiency was observed in the dendrites of CNK2-depleted neurons, correlating with the decrease in spine number. Consistent with its RacGAP activity, FRET efficiency was also reduced in neurons overexpressing Vilse. In contrast, a small but reproducible increase in FRET efficiency was seen in cells expressing WT-CNK2, and a more significant increase was observed in neurons expressing P1m-CNK2, with increased FRET detected throughout the dendritic protrusions and in the shafts. Thus, when the CNK2 scaffold cannot bind Vilse, RacGTP levels are elevated, correlating with the increase in dendritic protrusions.
Conclusions
CNK2 is a multi-domain scaffold protein expressed primarily in brain tissues. Although deletions in the human CNK2 gene have been reported in MRX patients, an explanation for why loss of the CNK2 scaffold would contribute to MRX has been unclear. By taking a proteomics approach to identify proteins that interact with the CNK2 scaffold and, in turn, may contribute to its biological function, we find that CNK2 interacts with numerous Rac/Cdc42 pathway components and functions in the spatial regulation of RacGDP/GTP cycling during spine morphogenesis (Figure 4D). CNK2 localizes to the dendrites of hippocampal neurons, and by interacting with regulators of Rac cycling, helps to maintain RacGTP/GDP levels at a concentration conducive for spine formation (Figure 4D). Thus, when the interaction between CNK2 and a regulator such as Vilse is disrupted, the localized balance in RacGTP/GDP levels is perturbed, resulting in spine defects. In particular, when CNK2/Vilse interaction is lost, the modulators that promote RacGTP loading go unchecked and RacGTP levels become elevated (Figure 4D). These findings define a molecular mechanism for CNK2 function in spine morphogenesis and reveal a new role for the Vilse RacGAP in neuronal signaling.
Experimental Procedures
Mass Spectrometry Analysis
Untreated NG108 cells or those treated for 18 hr with 50 ng/ml NGF, or expressing GFP-CNK2A proteins were lysed with low salt lysis buffer (30 mM Tris-HCl at pH 8.0, 75 mM NaCl, 10% glycerol, 1% Triton X-100, 0.15 U/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride [PMSF], 20 uM leupeptin, and 5 mM sodium orthovanadate). Endogenous CNK2 or GFP-CNK2A complexes were isolated using covalently coupled anti-CNK2 or anti-GFP sepharose beads, respectively, and analyzed by mass spectrometry as described in Lim et al. [7].
Immunocytochemistry and FRET Imaging
Transfected E18 rat hippocampal neurons and NG108 cells were fixed and stained as described in Supplemental Experimental Methods. Cell morphology and localization of proteins were visualized by confocal or epifluorescence microscopy. To determine spine and protrusion density, 10–20 neurons from three separate cultures were examined and multiple dendrites were counted from each neuron. Spines were identified as mushroom-shaped protrusions with associated PSD95 staining. For FRET imaging, neurons were co-transfected with RaichuEV-Rac1 and CNK2 siRNA or various plasmids at 8 DIV and FRET analysis was performed 48–60 hours after transfection using a Zeiss LSM 710 confocal microscope.
Supplementary Material
Highlights.
The CNK2 scaffold interacts with components of Rac/Cdc42 pathway signaling.
The primary binding partner of endogenous CNK2 is Vilse/ARHGAP39.
CNK2 is required for proper dendritic spine morphorgenesis in hippocampal neurons.
CNK2 is a spatial modulator of RacGDP/GTP cycling during spine morphogenesis.
Acknowledgements
We thank Elizabeth Terrell and Suzanne Specht for technical support, and Dr. Stephen Lockett and Alla Brafman at Leidos-FNL for assistance with FRET imaging and analysis. This project was funded by federal funds from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes three figures, one table and Supplemental Experimental Procedures.
REFERENCES
- 1.Therrien M, Wong AM, Rubin GM. CNK, a RAF-binding multidomain protein required for RAS signaling. Cell. 1998;95:343–353. doi: 10.1016/s0092-8674(00)81766-3. [DOI] [PubMed] [Google Scholar]
- 2.Yao I, Hata Y, Ide N, Hirao K, Deguchi M, Nishioka H, Mizoguchi A, Takai Y. MAGUIN, a novel neuronal membrane-associated guanylate kinase-interacting protein. J Biol Chem. 1999;274:11889–11896. doi: 10.1074/jbc.274.17.11889. [DOI] [PubMed] [Google Scholar]
- 3.Houge G, Rasmussen IH, Hovland R. Loss-of-Function CNKSR2 Mutation Is a Likely Cause of Non-Syndromic X-Linked Intellectual Disability. Mol Syndromol. 2012;2:60–63. doi: 10.1159/000335159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renieri A, Pescucci C, Longo I, Ariani F, Mari F, Meloni I. Non-syndromic X-linked mental retardation: from a molecular to a clinical point of view. J Cell Physiol. 2005;204:8–20. doi: 10.1002/jcp.20296. [DOI] [PubMed] [Google Scholar]
- 5.Ohtakara K, Nishizawa M, Izawa I, Hata Y, Matsushima S, Taki W, Inada H, Takai Y, Inagaki M. Densin-180, a synaptic protein, links to PSD-95 through its direct interaction with MAGUIN-1. Genes Cells. 2002;7:1149–1160. doi: 10.1046/j.1365-2443.2002.00589.x. [DOI] [PubMed] [Google Scholar]
- 6.Rajakulendran T, Sahmi M, Kurinov I, Tyers M, Therrien M, Sicheri F. CNK and HYP form a discrete dimer by their SAM domains to mediate RAF kinase signaling. Proc Natl Acad Sci U S A. 2008;105:2836–2841. doi: 10.1073/pnas.0709705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim J, Zhou M, Veenstra TD, Morrison DK. The CNK1 scaffold binds cytohesins and promotes insulin pathway signaling. Genes Dev. 2010;24:1496–1506. doi: 10.1101/gad.1904610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundstrom A, Gallio M, Englund C, Steneberg P, Hemphala J, Aspenstrom P, Keleman K, Falileeva L, Dickson BJ, Samakovlis C. Vilse, a conserved Rac/Cdc42 GAP mediating Robo repulsion in tracheal cells and axons. Genes Dev. 2004;18:2161–2171. doi: 10.1101/gad.310204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu H, Li M, Labrador JP, McEwen J, Lai EC, Goodman CS, Bashaw GJ. Cross GTPase-activating protein (CrossGAP)/Vilse links the Roundabout receptor to Rac to regulate midline repulsion. Proc Natl Acad Sci U S A. 2005;102:4613–4618. doi: 10.1073/pnas.0409325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- 11.Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, et al. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet. 2000;26:247–250. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
- 12.Premont RT, Perry SJ, Schmalzigaug R, Roseman JT, Xing Y, Claing A. The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell Signal. 2004;16:1001–1011. doi: 10.1016/j.cellsig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 14.Yao I, Ohtsuka T, Kawabe H, Matsuura Y, Takai Y, Hata Y. Association of membrane-associated guanylate kinase-interacting protein-1 with Raf-1. Biochem Biophys Res Commun. 2000;270:538–542. doi: 10.1006/bbrc.2000.2475. [DOI] [PubMed] [Google Scholar]
- 15.Lanigan TM, Liu A, Huang YZ, Mei L, Margolis B, Guan KL. Human homologue of Drosophila CNK interacts with Ras effector proteins Raf and Rlf. FASEB J. 2003;17:2048–2060. doi: 10.1096/fj.02-1096com. [DOI] [PubMed] [Google Scholar]
- 16.Aoki K, Nakamura T, Matsuda M. Spatio-temporal regulation of Rac1 and Cdc42 activity during nerve growth factor-induced neurite outgrowth in PC12 cells. J Biol Chem. 2004;279:713–719. doi: 10.1074/jbc.M306382200. [DOI] [PubMed] [Google Scholar]
- 17.Nikolic M. The role of Rho GTPases and associated kinases in regulating neurite outgrowth. Int J Biochem Cell Biol. 2002;34:731–745. doi: 10.1016/s1357-2725(01)00167-4. [DOI] [PubMed] [Google Scholar]
- 18.Bumeister R, Rosse C, Anselmo A, Camonis J, White MA. CNK2 couples NGF signal propagation to multiple regulatory cascades driving cell differentiation. Curr Biol. 2004;14:439–445. doi: 10.1016/j.cub.2004.02.037. [DOI] [PubMed] [Google Scholar]
- 19.Dierssen M, Ramakers GJ. Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes Brain Behav. 2006;5(Suppl 2):48–60. doi: 10.1111/j.1601-183X.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama AY, Luo L. Intracellular signaling pathways that regulate dendritic spine morphogenesis. Hippocampus. 2000;10:582–586. doi: 10.1002/1098-1063(2000)10:5<582::AID-HIPO8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 21.Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005;75:161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Matus A. Growth of dendritic spines: a continuing story. Curr Opin Neurobiol. 2005;15:67–72. doi: 10.1016/j.conb.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Webb DJ, Asmussen H, Horwitz AF. Synapse formation is regulated by the signaling adaptor GIT1. J Cell Biol. 2003;161:131–142. doi: 10.1083/jcb.200211002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iida J, Nishimura W, Yao I, Hata Y. Synaptic localization of membrane-associated guanylate kinase-interacting protein mediated by the pleckstrin homology domain. Eur J Neurosci. 2002;15:1493–1498. doi: 10.1046/j.1460-9568.2002.01987.x. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, Kamioka Y, Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.