Abstract
Salmonella enterica serovar Typhi has a 134-kb island of DNA identified as salmonella pathogenicity island 7 (SPI7), inserted between pheU and ′pheU (truncated), two genes for tRNAPhe. SPI7 has genes for Vi exopolysaccharide, for type IVB pili, for putative conjugal transfer, and for sopE bacteriophage. Pulsed-field gel electrophoresis following digestion with the endonuclease I-CeuI, using DNA from a set of 120 wild-type strains of serovar Typhi assembled from several sources, identified eight strains in which the I-CeuI G fragment, which contains SPI7, had a large deletion. In addition, agglutination tests with Vi antiserum and phage typing with Vi phages show that all eight strains are Vi negative. We therefore tested these strains for deletion of SPI7 by multiplex PCR, by microarray analysis, and by sequencing of PCR amplicons. Data show that seven of the eight strains are precise deletions of SPI7: a primer pair flanking SPI7 results in a PCR amplicon containing a single pheU gene; microarrays show that all SPI7 genes are deleted. Two of the strains produce amplicons which have A derived from pheU at bp 27, while five have C derived from ′pheU at this position; thus, the position of the crossover which results in the deletion can be inferred. The deletion in the eighth strain, TYT1669, removes 175 kb with junction points in genes STY4465 and STY4664; the left junction of SPI7 and adjacent genes, as well as part of SPI7 including the viaB operon for Vi exopolysaccharide, was removed, while the right junction of SPI7 was retained. We propose that these deletions occurred during storage following isolation.
Of the more than 2,300 closely related Salmonella serovars recognized, Salmonella enterica serovar Typhi (referred to hereafter as serovar Typhi) is the only one which grows exclusively in humans and causes typhoid enteric fever. The closely related serovar S. enterica serovar Typhimurium is a host-generalist able to grow on a wide range of animals, and is causal agent of gastroenteritis rather than enteric fevers. In most enteric bacteria there is a strong conservation of chromosomal gene order, but serovar Typhi is an exception, because its chromosome shows major rearrangements in gene order in wild-type strains due to homologous recombination between the rrn operons (23). These rearrangements were detected by pulsed-field gel electrophoresis (PFGE) analysis involving complete and partial digestion of genomic DNA with intron-encoded endonuclease I-CeuI which cuts only in the rrn genes (20). PFGE analysis also revealed the presence of insertions in serovar Typhi, one predicted to be about 120 kb, carrying the viaB locus for Vi exopolysaccharide synthesis which is missing from most other Salmonella species (21, 23). Further analysis showed that this island contains genes for the type IV pili (37, 38).
This island is now shown in strain CT18 to be a 134-kb pathogenicity island, identified as SPI7, which is inserted between two genes which code for tRNAPhe, pheU and ′pheU (truncated) (29, 31) (Fig. 1A). Distribution of the island seems to be limited in S. enterica, with only serovar Typhi, serovar Dublin, and serovar Paratyphi C harboring it at the same pheU site; in Citrobacter freundii it can be inserted in the glyU gene for tRNA (31). The island is a mosaic structure and has been characterized into six distinct regions: the ′pheU to ssb region with genes for conjugal DNA transfer; pil genes for type IVB pili; genes which might determine DNA conjugational transfer; sopE bacteriophage genes; viaB operon for Vi exopolysaccharide; and integrases and pheU gene (31). The presence of genes for type IVB pili and for DNA transfer, suggests that SPI7 might have been obtained by Salmonella by horizontal transfer, perhaps as a conjugative transposon (31).
FIG. 1.
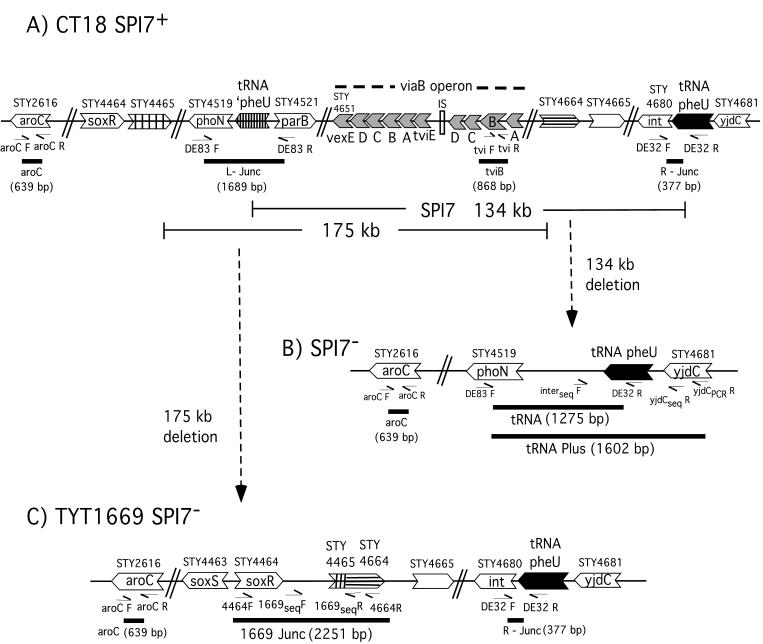
The genes in the region of SPI7 on the chromosome of serovar Typhi and their deletions. See the work of Pickard et al. (31) for a detailed map and annotations of gene function. (A) Vi-positive SPI7+ strain. The genes shown include the left junction, viaB operon, the right junction of SPI7 and genes to the “left” side of SPI7. The island is inserted between an intact 75-bp pheU gene (black arrow) for tRNAPheU and truncated ′pheU (striped arrow). Genes STY4465 and STY4664 are emphasized as they include the junction points of the deletions detected in TYT1669. The primers used for multiplex PCR and target sites are shown; the predicted amplicons (aroC and L-Junc, etc.) are shown as thick horizontal lines with their predicted sizes in base pairs. The map is not to scale. (B) Structure predicted if SPI7 is deleted by recombination between ′pheU and pheU. The predicted amplicons tRNA and tRNA Plus are illustrated as thick lines. (C) Schematic drawing of serovar Typhi TYT1669 which shows a partial deletion of SPI7 and regions on the ”left“ side of SPI7. Based on microarray data, primers were designed for both multiplex PCR and sequencing. The primers 4464F and 4664R result in the amplicon 1669Junc (2,251 bp); nested primers 1669seqF and -R were used to determine sequence of this amplicon and reveal the fusion of the STY4465 and STY4664.
The viaB locus in serovar Typhi in SPI7 consists of 10 genes: tviBCDE for Vi polysaccharide biosynthesis and vexABCDE for export, as well as tviA, which is activated by unlinked regulators rcsB-rcsC (2, 17) and ompR-envZ, which are themselves controlled by pH and osmolarity levels (30). Recently microarray work by Boyd et al. (4) and PCR analysis by Pickard et al. (31) have revealed that the 134-kb island is deleted from a serovar Typhi strain, SARB 64, one of the strains in the SARB set of strains constructed by Boyd et al. (3). The stability of SPI7 is of great interest as the Vi exopolysaccharide is currently used as a component for typhoid vaccines.
Previous analysis carried out by our research group on a set of 120 wild-type strains of serovar Typhi by endonuclease I-CeuI digestion followed by PFGE revealed that some of these strains had reductions in the size of fragment I-CeuI G which contains the viaB operon; some of these strains were also nontypeable by Vi phage typing indicating they were Vi negative (unpublished data). In this study we used PFGE analysis with I-CeuI digestion to confirm the DNA deletion in these strains of serovar Typhi. Multiplex PCR and nonredundant microarray analysis were used to determine the exact locations of these deletions which had been predicted by PFGE. Following this, the junction points in the deletion strains were determined by nucleotide sequencing.
MATERIALS AND METHODS
Bacterial strains.
The serovar Typhi strains which were used in the study and their sources are listed in Table 1. The strains are maintained in Luria-Bertani medium (Bacto Tryptone, 10 g/liter; Bacto Yeast extract, 5 g/liter; NaCl, 5 g/liter) with 20% glycerol at −70°C in the collection of the Salmonella Genetic Stock Centre (www.ucalgary.ca/∼kesander).
TABLE 1.
Serovar Typhi strain information and phenotypic and genotypic characteristics related to SP17
| Strain no. | SGSC no. | Origin | Source | Yr of isolation | Phage type | Altered fragment (PFGE) | MP-PCR amplicons (bp) | Microarray |
|---|---|---|---|---|---|---|---|---|
| Ty2a | 2408 | 1919 | E1 | I-CeuI G, unaltered | tviB (868), aroC (639), R-junction (377), L-junction (1,689) | SP17 intact | ||
| T202b | 3189 | Thailand | T. Pang | 1990 | UT (Vi-neg) | I-CeuI G-130 kb | tRNA (1,275), aroC (639) | SP17 deleted |
| T104b | 3188 | Thailand | T. Pang | 1990 | UT (Vi-neg) | I-CeuI G-130 kb | tRNA (1,275), aroC (639) | SP17 deleted |
| SARB64 IP.E.88.353c | 2521 | Dakar, Senegal | R. K. Selander | 1988 | UT (Vi-neg) | I-CeuI G-130 kb | tRNA (1,275), aroC (639) | SP17 deleted |
| R1962d | 2693 | Alberta | Provincial laboratory | 1994 | UT (Vi-neg) | I-CeuI G-130 kb | tRNA (1,275), aroC (639) | SP17 deleted |
| 415Tye | 3213 | Indonesia | B. Stocker | 1982 | UT (Vi-neg) | I-CeuI G-130 kb | tRNA (1,275), aroC (639) | SP17 deleted |
| 421Tye | 3219 | Indonesia | B. Stocker | 1984 | UT (Vi-neg) | I-CeuI G-130 kb | tRNA (1,275), aroC (639) | SP17 deleted |
| 26.003f | 3126 | Alberta | LCDC | 1993 | UT (Vi-neg) | I-CeuI G-130 kb | tRNA (1,275), aroC (639) | SP17 deleted |
| TYT1669g | 3488 | Chile | G. Mora | UT (Vi-neg) | I-CeuI G-170 kb | aroC (639), R-junction (377), 1669 Junc (2,251) | Part of SP17 deleted |
Ty2 was originally isolated in the USSR in 1919 and provided via the Walter Reed Army Institute Hospital, Bethesda, Md., and via D. Hone, Baltimore, Md. (14).
Strains T202 and T104 were obtained from T. Pang in Malaysia, who obtained them from Thailand.
Strain IP.E.88.353 was obtained by the Institut Pasteur, Paris, France, from Dakar, Senegal; it is strain SARB64 of the Salmonella Reference Collection B (3).
Strain R1962 was provided by the Provincial laboratory, Calgary, Alberta, Canada.
Strains 415Ty and 421Ty were obtained from M. Hori in Japan via Le Minor, Paris, France, and B. Stocker, Stanford, Calif. Both strains were originally isolated in Indonesia.
Strain 26.003 was isolated in Alberta, Canada, and provided by the Laboratory Center for Disease Control (LCDC), Ottawa, Ontario, Canada, by D. Woodward.
Strain TYT1669 was provided by G. C. Mora from Chile via S. Maloy.
Chemicals and growth media.
Endonuclease I-CeuI was from New England Biochemicals. Taq polymerase and deoxynucleotides were from Amersham. Most other chemicals, including agarose, were from Sigma Chemicals. All strains were grown in Luria-Bertani medium at 37°C.
Phage typing and agglutination with antiserum to Vi exopolysaccharide.
Bacteriophage typing was performed at the Laboratory Center for Disease Control, Ottawa, Ontario, Canada, using the Vi-specific phages of Typhi according to the methods of Khakhria and Lior (18) and Anderson and Williamson (1). Slide agglutination with antiserum for Vi exopolysaccharide (Becton Dickinson and Co.) was carried out according to manufacturer's instructions.
Endonuclease digestion and PFGE methods.
Preparation of intact genomic DNA, I-CeuI endonuclease cleavage of DNA in agarose blocks, and separation of the DNA fragments by PFGE were done as described previously (19, 24, 25). PFGE was performed with the Bio-Rad CHEF Mapper or Bio-Rad CHEF DRII electrophoresis system (19).
Multiplex PCR analysis.
Chromosomal DNA used in multiplex PCR was isolated using the Wizard genomic DNA purification kit (Promega) according to the manufacturer's instructions.
Primers were designed according to the CT18 sequence (29) (GenBank accession no. NC_003198) using Primer3program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi/) and synthesized by the University Core DNA Services (Health Sciences Center, University of Calgary) (Table 2, Fig. 1A and C). The primers were designed to detect conservation of deletion and insertion points at tRNAPheU involving SPI7 and other genes related to the absence or presence of SPI7. The following five primer pairs were initially tested as individual pairs, but were subsequently mixed together in the multiplex PCR system: aroC-F and -R (amplicon produced is aroC gene); DE83-F and -R (left junction); tviF and -R (tviB gene); DE32F and -R (right junction) (all these are illustrated in Fig. 1 A as heavy horizontal lines); and 4464F and 4664R (amplicon produced is 1669Junc) (see Fig. 1C). The 639-bp aroC product acts as a positive control for the presence of Salmonella DNA.
TABLE 2.
Primers for multiplex PCR amplification, to investigate the presence or absence of SP17 from serovar Typhi strains, and for sequencing of the amplicons
| Name of amplicon | Amplified DNA region(s) | Nucleotide position in CT18 genome | Primer | Primer sequence | Product size (bp) |
|---|---|---|---|---|---|
| L-Junc | phoN to parB | 4408374-4408399 | DE83-Fa | TCATCTTCAGGACGGCAGGTAGAATG | 1,689 |
| 4410044-4410686 | DE83-R | GATCGTTGGAAATGATGACG | |||
| tviB | tviB | 4522951-4522971 | tviF | GGAACCCTCAACGTTAATTCC | 868 |
| 4523797-4523816 | tviR | AATCTCGTCAGGTTGTTGGC | |||
| aroC | aroC | 2449777-2449797 | aroC-Fa | GGCACCAGTATTGGCCTGCT | 639 |
| 2450396-2450415 | aroC-Ra | CATATGCGCCACAATGTGTTG | |||
| R-Junc | int to tRNA pheU | 4542762-4542781 | DE32F | GCCAGACGATTTTCCTTACC | 377 |
| 4543116-4543139 | DE32Ra | GCTCAGTCGGTAGAGCAGGGGATT | |||
| tRNA | phoN to pheU (if SP17 is deleted) | 4408374-4408399 | DE83-F | TCATCTTCAGGACGGCAGGTAGAATG | 1,275 |
| 4543116-4543139 | DE32R | GCTCAGTCGGTAGAGCAGGGGATT | |||
| tRNA Plus | phoN to yjdC | 4408374-4408399 | DE83-F | TCATCTTCAGGACGGCAGGTAGAATG | 1,602 |
| 4543452-4543470 | yjdcPCRR | GCGGCACATGATTTTACCC | |||
| 1669 Junc | soxR to STY4664 (in TYT1669) | 4349581-4349598 | 4464F | CCGCCCTGCACTTCTATG | 2,251 |
| 4576965-4526984 | 4664R | ACCCGATTTATCCGCAGAAG |
Primer sequence provided by Claire Kidgell (personal communication).
PCR was performed in a volume of 25 μl containing 1× PCR buffer, 2.5 mM MgCl2, 0.1 mM deoxynucleoside triphosphates, 0.1 μM (each) primers, and 1 U of TaqDNA polymerase. The template was 40 ng of genomic DNA. Amplification was performed in an Eppendorf Gradient Thermal Cycler programmed as follows: initial denaturation at 95°C for 3 min and then 28 cycles of denaturation (30 s, 95°C), annealing (1 min, 54°C), and elongation (5 min, 72°C). The amplified product was electrophoresed at 100V on a 1.6% agarose gel (type II medium electrophoresis grade) in a 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 10 mM EDTA [pH 8]). Following electrophoresis the gel was stained in ethidium bromide (1 μg/ml) and photographed under UV light.
Microarray analysis.
Construction of the serovar Typhimurium and serovar Typhi microarray (which consists of PCR products representing 471 Typhi-specific genes and gene fragments which were added to genes or gene fragments representing 4,442 Typhimurium genes), preparation of the labeled genomic DNA probes, hybridization, data acquisition and data analysis have been previously described (4, 32, 33). Overall, coverage of the serovar Typhi genome is 94.5% (the microarray contains 4348 genes). This nonredundant microarray was probed with labeled genomic DNA from nine serovar Typhi strains of interest (Table 1; see Fig. 4).
FIG.4.
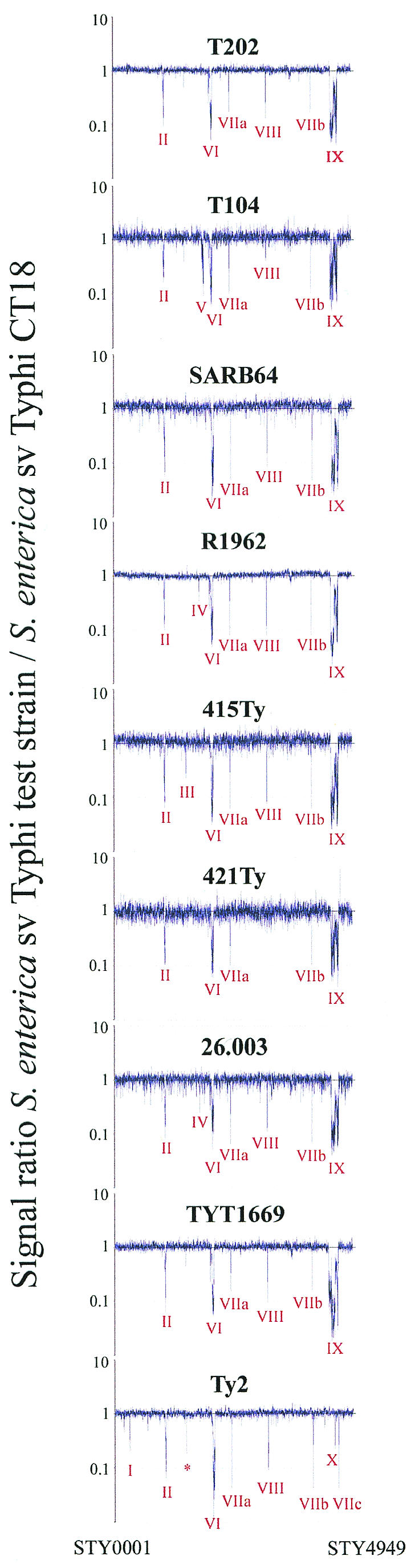
Comparative genome hybridization of nine S. enterica serovar Typhi strains versus serovar Typhi CT18. Genes are plotted in order of their position on the Typhi CT18 genome. The 10 regions of more than one gene deleted from other Typhi strains are indicated. The narV gene partially deleted from strains Ty2, T202, T104 and TYT1669 is marked by an asterisk.
DNA sequencing of the deleted SPI7 junctions.
PCR amplicons were purified by Wizard PCR preps DNA purification system (Promega). DNA sequencing of these amplicons was done with nested primers by the chain-terminating method of Sanger et al. (35) at the University Core DNA and Protein Services, DNA Sequencing Laboratory, Health Sciences, University of Calgary. Computer analysis was done by using BLAST programs and database services produced by National Center for Biotechnology Information, Bethesda, Md. (www.ncbi.nlm.nih.gov).
RESULTS
I-CeuI digestion and PFGE analysis.
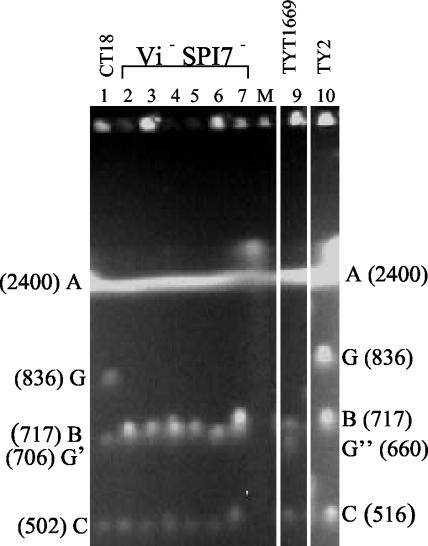
DNA from about 120 independent wild-type serovar Typhi strains, which had been assembled from numerous sources, was digested with I-CeuI and the fragments were separated by PFGE. The sizes of the seven I-CeuI fragments in CT18 and in Ty2 (Fig. 2, lanes 1 and 10) are as follows: I-CeuI A (2,400 kb), B (717 kb), C (516 kb), D (134 kb), E (149 kb), F (42 kb), and G (836 kb); these sizes are slightly corrected, using nucleotide sequence for CT18 (29), from the PFGE sizes previously reported (22) (the small fragments D, E, and F are not shown in this figure). The endonuclease I-CeuI digests at a 19-bp site (26) which is present in all seven rrl genes for 23S-rRNA, but at no other site; thus, the seven fragments produced from closely related strains are expected to be identical unless there have been insertions, deletions, or other rearrangements. As expected, in most of the 120 strains the sizes of the seven fragments were indistinguishable from the sizes for CT18 and Ty2, but in a few cases specific fragments show differences. An example of these differences is in the six strains in lanes 2 to 7; I-CeuI fragment sizes are indistinguishable from CT18 and Ty2 except that I-CeuI-G is reduced in size (now called I-CeuI-G′) so that B and G′ both appear as one major spot (Table 1; Fig. 2, lanes 2 to 7) indicating that the size of G has been reduced by about 130 kb. A seventh strain, 26.003, which shows the same pattern, is not shown here. In addition, in strain TYT1669, I-CeuI G was reduced in size to about 660 kb (I-CeuI G′′), suggesting a deletion of about 170 kb (Table 1; Fig. 2, lane 9). In practically all the other serovar Typhi strains the seven fragment sizes, including that of I-CeuI G, are indistinguishable from those of Ty2 and CT18.
FIG. 2.
Complete digestion of DNA of strains of serovar Typhi with endonuclease I-CeuI, separation by PFGE and staining with ethidium bromide. Fragment designations are given on the right for SPI7 positive serovar Typhi Ty2 (lane 10) and CT18 (lane 1), which are used as controls, and on the left for the SPI7 negative serovar Typhi strains (lanes 2 to 7, and lane 9). The I-CeuI D (134 kb), E (149 kb) and F (42 kb) fragments are not shown, as these fragments were identical sizes for all the strains studied. Lane 2, strain T202; lane 3, strain T104; lane 4, strain SARB64; lane 5, strain R1962; lane 6, strain 415Ty; lane 7, strain 421Ty; lane 9, strain TYT1669. One of the strains used in the study, strain 26.003, was not shown in the figure but it had the same banding pattern as the other 6 strains in lanes 2 to 7. The sizes of the fragments are shown in kilobases (kb). The λ concatemer marker is shown in lane 8 (M) though not very visible. All the lanes shown are from the same gel run. Note that I-CeuI B(717 kb) and I-CeuI G′ (ca. 706) are difficult to separate.
We had previously shown that I-CeuI-G of serovar Typhi Ty2 contains a large insertion of additional DNA (initially thought to be about 120 kb), including the viaB operon for synthesis of the Vi exopolysaccharide (22); these genes are not present in other serovars such as Typhimurium. This insertion has been recently identified by sequence as SPI7, containing the genes for the Vi exopolysaccharide, and shown to be 134 kb (29, 31). The above-described set of 120 wild-type strains of Typhi had been tested for phage type by R. Khakhria (personal communication) with adapted Vi II phages and with unadapted Vi I+IV phages, all of which use the Vi antigen for adsorption to the cell, and a phage type was determined for most strains; however, all eight strains which have their I-CeuI G fragment reduced in size (Fig. 2) were not lysed by any of these phages (they are therefore labeled nontypeable) and are concluded to be Vi negative (Table 1). The wild-type strain Ty2 was found to be phage type E1, which shows that it is Vi positive; similarly, a phage type was determined for most of the other 120 strains (data not shown). We confirmed that strain Ty2 is Vi positive and that the eight serovar Typhi strains labeled nontypeable are Vi negative by testing for slide agglutination with antiserum to Vi exopolysaccharide. The coincidental loss of the Vi exopolysaccharide and of a block of DNA similar in size to SPI7 led us to test all these strains to see if SPI7 has been deleted, using multiplex PCR, microarray analysis, and nucleotide sequencing.
Multiplex PCR for detection and characterization of SPI7.
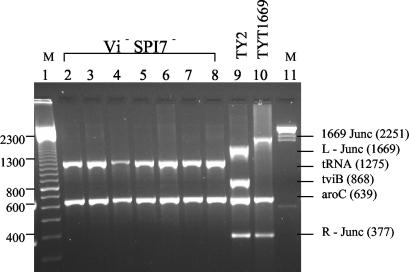
Our multiplex PCR system used five primer pairs; the amplicons resulting from these primer pairs are aroC, L-Junc, tviB, and R-Junc (Fig. 1A) and 1669Junc (Fig. 1C). With template DNA from the Vi-positive strain Ty2, the four amplicons which were expected, according to the genes in Fig. 1A, are produced (L-Junc, aroC, tviB, and R-Junc) (Fig. 3, lane 9); this confirms that the strain is SPI7+. Template DNAs from the seven Vi-negative strains in lanes 2 to 8 all have the aroC amplicon, as expected, but lack the L-Junc, R-Junc, and tviB amplicons; however, they produce the tRNA amplicon (1,275 bp) which is the size expected from the primers DE83F and DE32R if the entire SPI7 island between tRNA genes ′pheU and pheU has been deleted and the chromosome has been fused (Fig. 1B).
FIG. 3.
Multiplex PCR of serovar Typhi. Lanes 2 to 8 represent SPI7-negative strains that clearly show the absence of tviB gene (868 bp), left (1,689 bp) and right (377 bp) junctions of SPI7. The 1,275-bp amplicon seen in these SPI7-negative strains indicates the deletion between the phoN and tRNAPheU genes. Lane 9 is serovar Typhi Ty2 used as a SPI7-positive control strain, hence the amplification of the right and left junction of SPI7 and the tviB gene. Lane 10 depicts serovar Typhi TYT1669 that harbors a partial SPI7 deletion. TYT1669 has the right junction of SPI7, but a deletion between STY4664 (last gene present of the island) and STY4464 (first gene present after the left flank deletion). The 2,251-bp product is an amplification between STY4464 and STY4664 due to the 175-kb deletion between the 2 genes. The 639-bp aroC gene was produced in all the strains and acts as a control for Salmonella DNA. Lane 1, 100-bp marker (Amersham); lane 11, λ HindIII marker (Amersham).
With DNA of strain TYT1669 (Vi negative) as template, the R-Junc and aroC amplicons were produced, but tviB, tRNA, and L-Junc are not produced (Table 1; Fig. 1C; Fig. 3, lane 10). Based on microarray results (explained below) primers were designed to determine the deleted regions in TYT1669. Primer 4464F in STY4464 (which is the first gene determined by microarray to be present just before the left end deletion) and primer 4664R in STY4664 (the first gene detected by microarray to be flanking the right end of the deletion) produced a product of 2,251 bp, indicating that the deletion was indeed between the two genes as predicted by microarray analysis (Table 3; Fig. 1C; Fig. 3, lane 10). These data suggest that the segment between STY4464 and STY4664 has been deleted; this was tested (as described below) by nucleotide sequencing.
TABLE 3.
S. enterica serovar Typhi CT18 genomic regions of two or more genes which are missing in Vi-negative serovar Typhi isolates, and in strain Ty2, determined by microarray analysis
| Region | STY gene no. | No. of genes | Major product(s) | Missing regiond
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T202 | T104 | SARB 64 | R1962 | TY415 | TY421 | 26.003 | TYT1669 | Ty2 | ||||
| I | 0311-0312 | 2 | x | |||||||||
| II | 1048-1073 | 26 | Phage | x | x | x | x | x | x | x | x | x |
| III | 1505-1508 | 3 | GlgX | x | ||||||||
| IV | 1779-1779a | 2 | Rfc | x | x | |||||||
| V | 1849-1891 | 42 | PagCD, EnvE, MsgA, CspH | x | ||||||||
| VI | 2038-2077a | 42 | Phage | x | x | x | x | x | x | x | x | x |
| VII | ||||||||||||
| a | 2419-2420 | 2 | IS1 | x | x | x | x | x | x | x | x | x |
| b | 4124-4125 | 2 | IS1 | x | x | x | x | x | x | x | x | x |
| c | 4657-4658 | 2 | IS1 | x | x | x | x | x | x | x | x | x |
| VIII | 3188-3193 | 5 | Integrase | xb | xb | x | x | x | x | x | x | |
| IX | 4521-4680 | 149 | SP17 | x | x | x | x | x | x | x | xc | |
| X | 4580-4582 | 2 | x | |||||||||
Inconsistent absent or divergent gene status also from STY2007 to STY2015.
Some genes of the region are called divergent, or present.
Region is shifted to STY4465 to STY4663.
x, region not present in the genome.
About six other wild-type strains of Typhi from the set of 120 strains were nontypeable by phage typing (and are therefore Vi negative) but have I-CeuI fragment sizes which were indistinguishable from those of CT18 and Ty2. We postulated that they are SPI7+ but that they are Vi negative due to mutations in the viaB operon or in other genes (such as rcsBC or ompR-envZ which are required for Vi exopolysaccharide expression). We proved that they are all SPI7+, because in multiplex PCR tests they produce the same four amplicons as for Ty2 (Fig. 3, lane 9) but do not produce the 1,275-bp tRNA amplicon as illustrated in Fig. 3 (data not shown); we have not further tested the nature of their mutations.
Detection of deleted genes by microarray analysis.
The microarray data (Table 3; Fig. 4 ) indicates different DNA regions (I to X) that are present or absent in the genome of serovar Typhi strains relative to the CT18 genome which was used to construct the Typhi-specific components of the microarray. For example, region I which is Typhi-specific and comprises two genes that encode hypothetical proteins (STY0311 to STY0312) is absent from serovar Typhi Ty2 but present in all other eight strains studied, whereas regions II (comprising 26 genes) and region VI (STY2038 to STY2077) are Typhi strain CT18-specific bacteriophages that are absent or divergent from all other Typhi (Table 3; Fig. 4).
Region V contains a set of 42 genes that code for outer membrane invasion protein (pagC), putative outer membrane virulence protein (pagD), putative cold shock protein (cspH), putative virulence protein (msgA), putative lipoprotein (envE), and many other hypothetical proteins, which are deleted from strain T104 only (Table 3; Fig. 4). Unlike the other regions (4), deletion of region V was not previously observed.
Region IX, which is the main focus of this study, contains 149 genes including STY4521 to STY4682 which constitute the gene content of SPI7 (31). All seven strains which are precise deletions of SPI7 according to PCR data (Fig. 3) are missing all these genes in region IX (Fig. 4); Boyd et al. (4) previously reported the same conclusion for strain SARB64.
In strain TYT1669, genes STY4465 to STY4663 are deleted according to microarray data (Fig. 4; Table 3). Thus, the genes from STY4465 to STY4519 (phoN) (which are outside SPI7) and from STY4521 to STY4663 (including viaB genes) have been deleted—thus removing the left junction. However, the last 17 genes on the “right” side of the island (STY4664 to STY4680) have been retained, thus maintaining the right junction.
As expected, the wild-type strain Ty2, which has the Vi antigen, contained all the genes in region IX (SPI7); it differs from CT18 in some of the genes in regions I to X, as described earlier from microarray data (4) and sequencing data (7).
DNA sequencing of the excision regions in the SPI7 deleted strains.
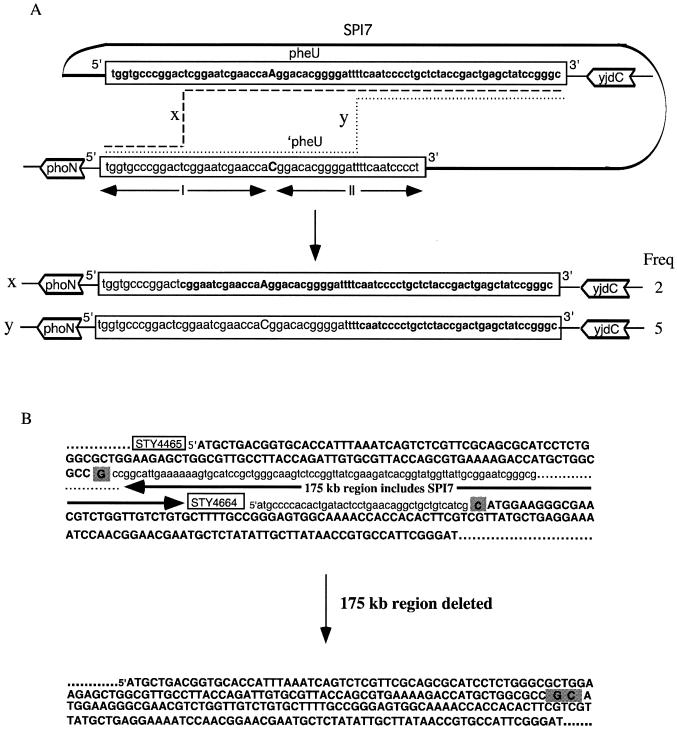
The truncated tRNA pheU gene (51 bp) at the left junction of SPI7 (labeled ′pheU in Fig. 5A) and the complete tRNA pheU gene (75 bp) at the right junction of SPI7 initially reported by Parkhill et al. (29) and Pickard et al. (31) are illustrated with their adjacent genes, phoN and yjdC (Fig. 5A). The 51 bp at the 5′ ends of ′pheU and pheU are identical except for an A/C difference at bp 27; in addition the 24 bp from the 3′ end of ′pheU are missing. (The tRNA genes are shown in this figure as DNA, not as the tRNA transcript). Using the primers DE83F and yjdCPCRR, a 1,602-bp amplicon containing the pheU gene and adjacent regions (labeled tRNA Plus) (Fig. 1B) was produced from each of the seven serovar Typhi SPI7− strains; using nested primers (interseqF and yjdCseqR), the nucleotide sequence of the amplicon was determined. In all seven strains the complete pheU gene was present, with the phoN-containing segment at the 5′ end and the yjdC-containing segment at the 3′ end (about 200 bp was determined on each side of the pheU gene in all amplicons, and no nucleotide divergence from that of CT18 was detected in any case). In two of the SPI7− strains (415Ty and 421Ty) an A at position 27 in pheU indicated that a recombination between the direct repeats occurred between ′pheU and pheU to the left of position 27 in the segment marked I; this is indicated as crossover X, though its exact location cannot be determined. In the remaining five SPI7− strains with precise deletions, a C at position 27 in pheU indicated recombination at crossover Y in the segment marked II (Fig. 5A).
FIG. 5.
Nucleotide sequence analysis of deletions in serovar Typhi. (A) The sequence of an SPI7+ strain is shown at the top, in the gene order phoN, ′pheU, (SPI7), pheU, yjdC; this is based on published sequence of the genome. Under this it the sequence of the tRNA Plus amplicon (Fig. 1B) produced from seven strains with precise deletions of SPI7. Two of the strains have A in the pheU gene at bp 27 (postulated to be due to crossover X in region I), and five have C due to crossover Y in region II. (B) The sequence of the N-terminal part of gene STY4465 and of the N-terminal part of gene STY4664 is shown. This sequence was determined from amplicon 1669 Junc (Fig. 1C). The sequences which are deleted from the strain TYT1669 are shown in lowercase type, inferred from published genomic sequences; the sequences which are included in the junction are in boldface uppercase type; the G and C at the junction points are shown in shading. The deletion in the strain is inferred to be 175 kb; the sequence of the N-terminal portion of the resulting gene is shown at the bottom of the figure.
Microarray data (Fig. 4) predicted that all genes between STY4464 and STY4664 were deleted from strain TYT1669. Primers 4464F and 4664R (Fig. 1C) resulted in a PCR amplicon of about 2.2 kb, labeled 1669Junc (Fig. 3), confirming that there was a large deletion. Nested primers (1669seqF and 1669seqR) were used to sequence the 1669Junc amplicon; these sequences show that the junction point is within the gene STY4465 and the gene STY4664 (Fig. 1C; Fig. 5B). The total size of the deletion is 175 kb. According to the annotated sequence, the N-terminal portion of gene STY4465 is expected to be translated to the junction point shown by shaded GC (Fig. 5B). This results in a frameshift as it enters STY4664; a stop codon is predicted about 40 amino acids after the junction point so this protein is presumably nonfunctional. STY4465 is annotated as a putative glutathione S-transferase, and STY4664 is annotated as a putative DNA helicase (29) but there is no indication that the loss of these genes has functional significance.
DISCUSSION
Seven serovar Typhi strains are proposed to have precise deletions of the entire 134 kb of SPI7, based on the following data: PFGE data shows that all have a reduction in size of the I-CeuI G fragment, which is known to contain SPI7, by about 130 kb (Fig. 2); based on their nontypeability with Vi typing phages, and their lack of agglutination with antiserum for the Vi antigen, all strains must be Vi negative; template DNA of these strains yields multiplex PCR products (tRNA in Fig. 3) from a primer pair outside the left and right junctions of SPI7 but not from most other primer pairs (Fig. 3); microarrays reveal that all the genes within SPI7 are missing (Fig. 4); nucleotide sequencing of the amplicon called tRNA (Fig. 1B) reveals that it is a junction fragment containing the genes phoP, pheU, and yjdC (Fig. 5A). All these strains are proposed to have originated by recombination between the direct repeats at the boundaries of SPI7, ′pheU and pheU (Fig. 5A).
The strain TYT1669 is also nontypeable by Vi phages and nonagglutinable with Vi antiserum, and therefore Vi negative, but the size of its I-CeuI G fragment indicates a larger deletion, estimated as 170 kb. Multiplex PCR indicates that it lacks the left junction of SPI7 and the tviB gene but has the right junction. Microarray analysis, showing that all the genes between STY4464 to STY4664 were missing, facilitated the design of a primer pair which spans the postulated breakpoints of the deletion; a PCR amplicon of the predicted size (about 2.2 kb) was produced. Nucleotide sequencing of this amplicon revealed that a segment of 175 kb was deleted, with breakpoints in genes STY4465 and STY4664. No sequence motifs have been detected at the junction points, so this mutational event, which has been detected only in this one strain, appears to be an illegitimate recombination. The genes from STY4465 to STY4519 (phoN) which are deleted in TYT1669 are part of the core genome of Salmonella; we conclude this because the genes STM4267 to STM4319 (phoN) from serovar Typhimurium give the highest BlastP hit to this block of genes from Typhi. Obviously this group of about 54 genes does not contain essential functions because they can be deleted with no obvious effect on phenotype when cells are grown on Luria-Bertani broth or agar. However, they contain a number of named genes such as proP (betaine-proline transporter) and melAB (melibiose utilization), so changes in phenotype might be detectable. The 12-kb DNA segment to the left of ′pheU (including STY4503 to STY4519 (phoN) was originally proposed to be horizontally transferred to Salmonella, since it is missing from Escherichia coli K-12 and O157:H7 (9, 32)
The eight strains of serovar Typhi which are Vi negative either due to precise deletion of SPI7 (seven strains) or to a probable illegitimate recombination leading to deletion of part of the island and the segment on the left side (one strain, TYT1669) are each from independent wild-type strains of serovar Typhi with different isolation and storage histories. The normal serotype of serovar Typhi is 9,12,[Vi]:d:-, i.e., 9, 12 for LPS antigen, Vi positive, flagellar antigen d in phase 1 and negative in phase 2. Part of the history of each strain is in the footnotes to Table 1. We believe that these serotypes were all determined at initial isolation, including the test for Vi, and in some cases we have confirmed that these strains were initially tested for the Vi exopolysaccharide by agglutination tests as part of the serology which identified the strains as serovar Typhi. This suggests that the deletions have occurred in the strains during storage in laboratory cultures. However, in spite of this evidence that genomic rearrangements have occurred in culture, data from complete genome sequencing of two independent wild-type strains of serovar Typhi, CT18 (29) and Ty2 (7), and microarray analysis of these strains and others (4) indicates that in general the genome is quite stable during storage. For example, Ty2 was isolated in 1919 in the USSR and maintained initially in a variety of ways as it was passed between laboratories, and then it was maintained recently by low-temperature freezing; CT18 was isolated in 1993 in Vietnam and then immediately frozen in glycerol at −70°C, and it has been maintained in this fashion since then (29). A comparison of sequence of Ty2 with CT18 reveals that genome content and organization is very similar, with over 4,000 open reading frames (ORFs) and pseudogenes identical in both and 282 ORFs which differ by a single point mutation (7). Both have similar IS elements except that CT18 has three copies of IS1, while Ty2 has no IS1. In total there are 29 ORFs unique to Ty2, whereas 84 are unique to CT18; many of these are associated with putative prophages. CT18 has 204 pseudogenes, nine of which are intact in Ty2; Ty2 has 206 pseudogenes, all of which are intact in CT18 (7). Analysis by microarrays confirms many of these conclusions (4). Surprisingly, we found that cells of strain Ty2, which has been in culture for many years, are still Vi positive when tested by slide agglutination tests, whereas CT18, a much more recent isolate, is Vi negative; we find that both strains are SPI7+. Pickard et al. (31) also noted that CT18 is Vi negative and proposed that this may be because an IS1 element is inserted between tviE and tviD genes which may disrupt an essential Shine-Dalgarno ribosome binding site; the strain was Vi positive on initial isolation but has become Vi negative by transposition of the IS1. Since Ty2 does not have any IS1 elements this specific rearrangement cannot occur.
Most pathogenicity islands are flanked by specific DNA regions (in most cases direct repeat DNA sequences); around 75% of the virulence genes that are associated with bacterial pathogenicity are flanked by tRNA genes (11, 16, 34). Pathogenicity islands that are inserted into tRNA genes are unstable due to the flanking direct repeats which not only promote integration into the bacterial genome but also excision out of it via the possible involvement of integrases and/or excisionases (5, 10). Studies in a range of bacteria show that many different tRNA genes serve as targets, and the overlap in the direct repeats at the ends of the island can vary from 9 to as much as 135 bp (16). In SPI7 the ′pheU gene at the left end of SPI7 is an almost-precise direct repeat of the pheU gene at the right end of SPI7 (except for C instead of A at bp 27), covering 51 bases at the 3′ end of the gene (when shown in the tRNA form). In the seven strains of serovar Typhi in which SPI7 was deleted due to recombination between the direct repeats ′pheU and pheU, thus producing a hybrid tRNA of normal (75 bp) length in all seven cases, two strains have the A of the normal pheU (which we have designated pheU-A) while five have the C of the truncated ′pheU (designated pheU-C). The normal pheU-A gene will make a tRNA with complementary U at bp 27 from the 3′ end, which pairs with A in the pseudouracil loop; the unusual pheU-C gene will make tRNA with complementary G which will mispair with A in the loop; it is not clear if this mispaired tRNA will be functional. This may not be functionally important, because there is another unlinked gene on the Typhi chromosome, pheV, which has tRNA sequence which is identical to pheU. A BlastN search of the NCBI database with the pheU-C sequence revealed many genomes of different genera with the normal pheU-A at position 27; there was no case detected of a complete pheU-C gene except for the ones detected in this study.
As mentioned earlier, strain TYT1669 has a larger deletion (175 kb) in which the deletion process does not involve the duplicated pheU regions, thus suggesting the possibility of illegitimate recombination. Multiplex PCR using primers 4464F and 4664R (Fig. 3), and sequencing data have shown that genes STY4465 and STY4664 are fused in strain TYT1669.
The minimum sequence length sufficient to initiate homologous recombination in vivo varies between the different bacterial species. Homologous recombination is RecA-mediated and can occur where homologous repeat segments are as small as 20 to 30 bp, but efficiency of the process drops sharply where the intervening sequence between the repeats is larger than 400 bp (28); where the intervening sequence is many kilobases as in SPI7, homologous recombination between the pheU genes, which have direct repeats of 51 bp, may be rare (28). Recombination between short sequences, often less than 20 bp, is considered to be site-specific recombination, involving site-specific elements such as phages and transposable elements, and requires site-specific recombinases such as integrases, resolvases, and DNA invertases that recognize two specific sites in the DNA and promote recombination between them (8, 28). Deletions of SPI7 may be due to site-specific recombination, analogous to the deletions of a 66-kb island from Shigella flexneri 2a which occurred at frequency of 10−5 per cell from the wild-type strain, but at much lower frequency from strains mutated in an int gene in the island (36); int functions may have a similar role in SPI7, for int genes have been annotated within the island, including gene STY4680 located next to pheU (30).
Illegitimate recombination on the other hand involves junctions of nonhomologous or very short homologous DNA sequences (often less than 3 bp) which are not recognized by site-specific enzymes. Since BlastN analysis did not reveal any sequence motifs in common between the genes STY4465 and STY4664, we presume that a rare illegitimate recombination resulted in the 175-kb deletion in strain TYT1669.
The element CTnscr94, postulated to be a conjugative transposon which carries genes for sucrose fermentation, transfers by conjugation from S. enterica serovar Senftenberg to E. coli K-12 and integrates in a RecA-independent fashion at two attachment sites on the chromosome, one of which was proven to be pheV while the other was postulated to be pheU (12). In the integration site identified within pheV, which is the structural gene for tRNAPhe, CTnscr94 was bracketed on one side by pheV and on the other side by a truncated gene phe′V containing 50 bp of pheV at the 3′ end. The sequence of the pheV in E. coli K-12 is identical to pheV and also to pheU in serovar Typhi, determined from BlastN searches of genomic sequences; similarly, the sequence of the 50-bp phe′V in the transconjugant is identical to the ′pheU in SPI7+ serovar Typhi strains (except that ′pheU in serovar Typhi has 51 bp conserved rather than 50, and has a C at bp 27 in ′pheU) (Fig. 5A). Thus, following transfer of CTnscr94 to E. coli K-12 by a mechanism postulated to be conjugal transposition, sequences almost identical to those found on both sides of SPI7 in serovar Typhi are found on both sides of the element. This observation strengthens the proposal, which was based on the presence of genes for DNA replication and transfer, that SPI7 resembles CTnscr94 in being a conjugative transposon (31).
SPI7 has genes for synthesis and shufflon regulation of type IVB pili; it has been postulated that serovar Typhi cells use these pili to enter human intestinal epithelial cells (37, 38). If the pili have a role in cell entry and thus in virulence, then loss of SPI7 would affect virulence due to loss of these pili as well as due to loss of Vi exopolysaccharide.
The Vi antigen of serovar Typhi is currently the basis for licensed injectable vaccine against typhoid fever. Hornick et al. (15) have pointed out that the infectivity of Vi-expressing strains of serovar Typhi is greater than that of strains which do not express the antigen. However, Vi is not essential for infection to be established in humans because spontaneous Vi-negative mutants of serovar Typhi can cause infection in humans (13, 15). Indeed, there have been reports of outbreaks caused by Vi negative serovar Typhi in India (27). Bueno et al. (6) have shown in an independent study that some Chilean clinical serovar Typhi isolates are Vi negative by agglutination and have deletions for SPI7. Therefore, if Vi-negative strains retain pathogenicity the production of Vi-negative cells by chromosomal mutations in regulatory genes or due to loss of SPI7 by deletion might result in strains which can escape immune protection resulting from use of Vi exopolysaccharide as a vaccine. Several methods, including PFGE, multiplex PCR, and microarray analysis were all used in this study for the detection of SPI7 deletions which are Vi negative. Multiplex PCR, used for targeting regions related to the island, rapidly detects SPI7 deletions, and could have future implications in monitoring serovar Typhi profiles where vaccination programs for typhoid are planned. In doing this, it must also be remembered that Vi-negative cells can also result from mutations in the regulatory genes for Vi exopolysaccharide production.
Acknowledgments
We thank Rasik Khakhria, who used Vi-specific phages to determine the phage type of strains of serovar Typhi, and Claire Kidgell, who designed some of the PCR primers which we used. We also thank John Wain for discussions and for exchanging with us information on studies on deletion mutants of SPI7.
The work was supported by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) to K.E.S. and to S.-L.L. and by grants from the National Institute of Allergy and Infectious Diseases of the NIH (AI034829-14 and AI52237-02) to K.E.S. and M.M.
REFERENCES
- 1.Anderson, E. S., and R. E. Williamson. 1956. Bacteriophage typing of enteric pathogens and staphylococci and its use in epidemiology. J. Clin. Pathol. 9:94-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arricau, N., D. Hermant, H. Waxin, C. Ecobichon, P. S. Duffey, and M. Y. Popoff. 1998. RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol. Microbiol. 29:835-850. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139:1125-1132. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, E. F., S. Porwollik, F. Blackmer, and M. McClelland. 2003. Differences in gene content among Salmonella enterica serovar Typhi isolates. J. Clin. Microbiol. 41:3823-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchrieser, C., R. Brosch, S. Bach, A. Guiyoule, and E. Carniel. 1998. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 30:965-978. [DOI] [PubMed] [Google Scholar]
- 6.Bueno, S. M., C. A. Santiviago, A. A. Murillo, J. A. Fuentes, A. N. Trombert, P. Youderian, and G. C. Mora. 2004. Excision of the large pathogenicity island of Salmonella enterica serovar Typhi. J. Bacteriol. 186:3202-3213. [DOI] [PMC free article] [PubMed]
- 7.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrlich, S. D., H. Bierne, E. d'Alenson, D. Vilette, M. Petranovic, P. Noirot, and B. Michel. 1993. Mechanisms of illegitimate recombination. Gene 135:161-166. [DOI] [PubMed] [Google Scholar]
- 9.Groisman, E. A., M. H. Saier, and H. Ochman. 1992. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO J. 11:1309-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 123:1089-1097. [DOI] [PubMed] [Google Scholar]
- 11.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 12.Hochhut, B., K. Jahreis, J. W. Lengeler, and K. Schmid. 1997. CTnscr94, a conjugative transposon found in enterobacteria. J. Bacteriol. 179:2097-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hone, D. M., S. R. Attridge, B. Forrest, R. Morona, D. Daniels, J. T. LaBrooy, R. C. Bartholomeusz, D. J. Shearman, and J. A. Hackett. 1988. galE via (Vi antigen-negative) mutant of Salmonella typhi Ty2 retains virulence in humans. Infect. Immun. 56:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hone, D. M., A. M. Harris, S. Chatfield, G. Dougan, and M. M. Levine. 1991. Construction of genetically defined double aro mutants of Salmonella typhi. Vaccine 9:810-816. [DOI] [PubMed] [Google Scholar]
- 15.Hornick, R. B., S. E. Greisman, T. E. Woodward, H. L. Dupont, A. T. Dawkins, and M. J. Snyder. 1970. Typhoid fever: pathogenesis and immunologic control. N. Engl. J. Med. 283:686-691. [DOI] [PubMed] [Google Scholar]
- 16.Hou, Y. M. 1999. Transfer RNAs and pathogenicity islands. Trends Biochem. Sci. 24:295-298. [DOI] [PubMed] [Google Scholar]
- 17.Houng, H. S., K. F. Noon, J. T. Ou, and L. S. Baron. 1992. Expression of Vi antigen in Escherichia coli K-12: characterization of ViaB from Citrobacter freundii and identity of ViaA with RcsB. J. Bacteriol. 174:5910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khakhria, R., and H. Lior. 1981. Phage typing of Salmonella typhi in Canada (1967-1976). Can. J. Public Health 72:30-36. [PubMed] [Google Scholar]
- 19.Liu, G. R., A. Rahn, W.-Q. Liu, K. E. Sanderson, R. N. Johnston, and S.-L. Liu. 2002. The evolving genome of Salmonella enterica serovar Pullorum. J. Bacteriol. 184:2626-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, S. L., and K. E. Sanderson. 1995. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J. Bacteriol. 177:3355-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, S. L., and K. E. Sanderson. 1995. Rearrangements in the genome of the bacterium Salmonella typhi. Proc. Natl. Acad. Sci. USA 92:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, S. L., and K. E. Sanderson. 1995. Genomic cleavage map of Salmonella typhi Ty2. J. Bacteriol. 177:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, S. L., and K. E. Sanderson. 1996. Highly plastic chromosomal organization in Salmonella typhi. Proc. Natl. Acad. Sci. USA 93:10303-10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, S. L., and K. E. Sanderson. 1998. Homologous recombination between rrn operons rearranges the chromosome in host-specialized species of Salmonella. FEMS Microbiol. Lett. 164:275-281. [DOI] [PubMed] [Google Scholar]
- 25.Liu, S. L., and K. E. Sanderson. 1992. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J. Bacteriol. 174:1662-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall, P., and C. Lemieux. 1991. Cleavage pattern of the homing endonuclease encoded by the fifth intron in the chloroplast large subunit rRNA-encoding gene of Chlamydomonas eugametos. Gene 104:241-245. [DOI] [PubMed] [Google Scholar]
- 27.Mehta, G., and S. C. Arya. 2002. Capsular Vi polysaccharide antigen in Salmonella enterica serovar Typhi isolates. J. Clin. Microbiol. 40:1127-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michel, B. 1999. Illegitimate recombination in bacteria, p. 129-150. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. ASM Press, Washington, D.C.
- 29.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 30.Pickard, D., J. Li, M. Roberts, D. Maskell, D. Hone, M. Levine, G. Dougan, and S. Chatfield. 1994. Characterization of defined ompR mutants of Salmonella typhi: ompR is involved in the regulation of Vi polysaccharide expression. Infect. Immun. 62:3984-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickard, D., J. Wain, S. Baker, A. Line, S. Chohan, M. Fookes, A. Barron, P. A. Gaora, J. A. Chabalgoity, N. Thanky, C. Scholes, N. Thomson, M. Quail, J. Parkhill, and G. Dougan. 2003. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 185:5055-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porwollik, S., J. Frye, L. D. Florea, F. Blackmer, and M. A. McClelland. 2003. A non-redundant microarray of genes for two related bacteria. Nucleic Acids Res. 31:1869-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiter, W. D., P. Palm, and S. Yeats. 1989. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 17:1907-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner, S. A., S. N. Luck, H. Sakellaris, K. Rajakumar, and B. Adler. 2001. Nested deletions of the SRL pathogenicity island of Shigella flexneri 2a. J. Bacteriol. 183:5535-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, X. L., C. Morris, and J. Hackett. 1997. Molecular cloning, nucleotide sequence, and function of a site-specific recombinase encoded in the major ‘pathogenicity island’ of Salmonella typhi. Gene 202:139-146. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, X. L., I. S. Tsui, C. M. Yip, A. W. Fung, D. K. Wong, X. Dai, Y. Yang, J. Hackett, and C. Morris. 2000. Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 68:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]