Abstract
Background
Ovarian cancer is a lethal disease comprised of distinct histopathological types. There are few established biomarkers of ovarian cancer prognosis, in part because subtype-specific associations may have been obscured in studies combining all subtypes. We examined whether progesterone receptor (PR) and estrogen receptor (ER) protein expression were associated with subtype-specific survival in the international Ovarian Tumor Tissue Analysis (OTTA) consortium.
Methods
PR and ER were assessed by central immunohistochemical analysis of tissue microarrays for 2933 women with invasive epithelial ovarian cancer from 12 study sites. Negative, weak, and strong expression were defined as positive staining in <1%, 1–50%, and ≥50% of tumor cell nuclei, respectively. Hazard ratios (HRs) for ovarian cancer death were estimated using Cox regression stratified by site and adjusted for age, stage, and grade.
Results
PR expression was associated with improved survival for endometrioid (EC; p<0·0001) and high-grade serous carcinoma (HGSC; p=0·0006), and ER expression was associated with improved EC survival (p<0·0001); no significant associations were found for mucinous, clear cell, or low-grade serous carcinoma. EC patients with hormone receptor (PR and/or ER) positive (weak or strong) versus negative tumors had significantly reduced risk of dying from their disease, independent of clinical factors (HR, 0·33; 95% CI, 0·21–0·51; p<0·0001). HGSC patients with strong versus weak or negative tumor PR expression had significantly reduced risk of dying from their disease, independent of clinical factors (HR, 0·71; 95% CI, 0·55–0·91; p=0·0061).
Interpretation
PR and ER are prognostic biomarkers for endometrioid and high-grade serous ovarian cancers. Clinical trials, stratified by subtype and biomarker status, are needed to determine whether hormone receptor status predicts response to endocrine therapy, and can guide personalized treatment for ovarian cancer.
Funding
Carraressi Foundation, US National Institutes of Health, National Health and Medical Research Council of Australia, UK National Institute for Health Research, and others.
INTRODUCTION
Ovarian cancer causes over 140,000 deaths annually worldwide, and is the most lethal gynecologic malignancy in developed countries.1 Invasive epithelial ovarian cancer consists of five major histopathological types that are phenotypically, molecularly and etiologically distinct: high-grade serous carcinoma (HGSC), low-grade serous carcinoma (LGSC), mucinous carcinoma (MC), endometrioid carcinoma (EC), and clear cell carcinoma (CCC).2 It is now recognized that the association of tumor biomarker expression with survival varies substantially across subtypes, and can be obscured in analyses of all ovarian cancers combined.3 However, the infrequency of histopathological types other than HGSC have precluded robust subtype-specific analyses and hindered efforts to identify biomarkers of ovarian cancer survival to date.
Progesterone receptor (PR) and estrogen receptor (ER) mediate the effects of female steroid hormones on ovarian cancer cell proliferation and apoptosis.4 Immunohistochemical (IHC) assessment of ER/PR status is routinely performed for the clinical management of breast cancer.5 However, the utility of ER/PR for guiding ovarian cancer prognosis or therapy is uncertain. Previous studies have reported that PR6–8 or ER6, 9 protein expression was associated with improved ovarian cancer survival, independent of clinical prognostic factors, but these associations have not been consistently replicated.3, 10–13 These conflicting data are difficult to interpret for several reasons. First, most studies combined all disease subtypes,6–8, 10–12 which can obscure subtype-specific associations. Second, the few studies that focused on serous carcinoma9, 13 and other subtypes3 had limited sample sizes and statistical power. Finally, different methods of IHC analysis and biomarker scoring were employed, which could contribute to the heterogeneous results across studies.14
We formed the international Ovarian Tumor Tissue Analysis (OTTA) consortium to overcome the main obstacles that heretofore have prevented the development of clinically useful prognostic biomarkers for ovarian cancer. Here, we examine the association of tumor PR and ER protein expression with disease-specific survival by performing central IHC assessment and subtype-specific analyses of 2933 women with invasive epithelial ovarian cancer from 12 sites. This study is over five times larger than any previous study and the first pooled analysis of centrally collected IHC data, enabling robust assessment of subtype-specific associations of ER/PR status with ovarian cancer outcomes for the first time.
METHODS
Study participants and immunohistochemistry analysis
Twelve studies participating in the OTTA consortium contributed to this work, including sites located in Australia, the United States, Denmark, the United Kingdom, Poland, and Canada (Table 1). Study participants (N=2933) were diagnosed with invasive serous, mucinous, endometrioid, or clear cell carcinomas of the ovary; mixed and other histological types were excluded. Inclusion criteria were the availability of tissue microarrays (TMAs) for immunohistochemistry (IHC) analysis, clinical follow-up data, age at diagnosis, tumor grade and stage. Clinical data were obtained from medical records, cancer registries, death certificates, and pathology reports. Histological classifications were locally reviewed for consistency with World Health Organization guidelines15 by study physicians with expertise in gynecologic pathology, and was based upon slide review for approximately 70% of all patients (Table 1). Seven sites (AOC, HAW, MAL, MAY+MAC, NOT, TOR, VAN) provided information on the extent of residual disease. Four sites (AOC, MAL, MAY+MAC, STA) provided BRCA1 and BRCA2 germline mutation data.16 The data from each site were centrally harmonized and reviewed for consistency and quality.
Table 1.
Description of twelve participating studies.
| Study site N=2933 |
HGSC N=1742 |
LGSC N=110 |
MC N=207 |
EC N=484 |
CCC N=390 |
Country (years) | Ascertainment of cases and clinical data | Pathology data and review |
|---|---|---|---|---|---|---|---|---|
|
AOC36 N=208 |
169 | 9 | — | 30 | — | Australia (2002–2006) | Treatment centers throughout Australia; cancer registries serving Queensland, South and West Australia; regular follow-up visits | Pathology reports and diagnostic slides reviewed by panel of gynecologic pathologists |
|
HAW37, 38 N=108 |
56 | 1 | 11 | 25 | 15 | US (1993–2008) | Hawaii Tumor Registry and medical records | Pathology reports and histological slides reviewed by study pathologist |
|
MAL39, 40 N=140 |
53 | 9 | 42 | 3 | 33 | Denmark (1994–1999) | Gynecological departments in Copenhagen, Frederiksberg and 7 surrounding counties | Review of pathology reports for all cases and histological slides for 30% by gynecologic pathologist |
|
MAY+MAC41 N=484 |
331 | 17 | 17 | 86 | 33 | US (2000–2009) | Mayo Clinic medical records and State death certificates | Pathology reports and histologic slides reviewed by Mayo Clinic gynecologic pathologists |
|
NOT42 N=232 |
127 | 11 | 21 | 48 | 25 | UK (1991–2008) | Hospital records and Trent cancer registry | Pathology reports reviewed by gynecologic pathologist |
|
POL43 N=81 |
50 | 4 | 8 | 13 | 6 | Poland (2000–2003) | Hospital records and cancer registries serving Warsaw and Lodz | Histological slides reviewed by study pathologist |
|
RPX44 N=281 |
224 | 12 | 10 | 14 | 21 | US (2000–2011) | Roswell Park Cancer Institute cancer registry and medical records | Pathology reports reviewed by Roswell Park Cancer Institute pathologists |
|
SEA45 N=179 |
96 | 3 | 13 | 40 | 27 | UK (1998–2008) | East Anglia and West Midlands Cancer Registry | Pathology reports reviewed by central pathologist |
|
STA46 N=276 |
172 | 19 | 20 | 35 | 30 | US (1997–2001) | Greater Bay Area Cancer Registry | Pathology reports and histological slides reviewed by study pathologist |
|
TOC47 N=101 |
— | — | 20 | 43 | 38 | Canada (1995–2003) | Ontario Cancer Registry | Pathology reports and histological slides reviewed by study pathologist |
|
UKO48 N=93 |
49 | 8 | 8 | 12 | 16 | UK (2006–2010) | 10 major Gynecologic Oncology NHS centers in England, Wales and Northern Ireland; cancer registries; NHS Information Centre for Health and Social Care (England and Wales) and Central Services Agency (Northern Ireland) | Central review of pathology reports by gynecologic oncologist |
|
VAN49, 50 N=750 |
415 | 17 | 37 | 135 | 146 | Canada (1984–2000) | Ovarian Cancer Registry serving British Columbia, and the Cheryl Brown Outcomes unit | Central review of pathology reports and histological slides by University of British Columbia pathologists |
HGSC=high-grade serous carcinoma; LGSC=low-grade serous carcinoma; MC=mucinous carcinoma; EC=endometrioid carcinoma; CCC=clear cell carcinoma.
IHC analyses were performed by a central laboratory (Genetic Pathology Evaluation Centre, Vancouver, BC, Canada). Each site submitted TMA sections on glass slides, and >75% of tumors were represented by at least two cores. We used the Ventana Discovery XT platform and antigen retrieval solution CC1. Staining was performed using rabbit monoclonal antibodies for ERα (Thermo clone SP1, 1:25 dilution) and PR (Roche clone 1E2, ready to use), which are routinely used for clinical testing of ER and PR in breast cancer.17, 18 The Universal Secondary Antibody (Ventana) and DAB MAP kit were used to detect and amplify the signal. Slides were visualized at 200× magnification and both biomarkers were scored using a three-tier system: negative, weak, and strong, defined a priori as positive staining in <1%, 1–50%, and ≥50% of tumor cell nuclei, respectively (Supplementary Figure 1).7, 17 Two pathologists (MK, TL) blindly scored each biomarker on a subset of 276 cases and the inter-observer agreement was 91% for ER and 92% for PR; one pathologist (MK) then scored all the remaining cases. Study protocols were approved by the respective Institutional Review Board for each site.
Statistical analysis
The primary endpoint was disease-specific survival within 10 years of diagnosis with ovarian cancer. We used left truncation to avoid potential survival bias from the inclusion of patients (<10%) enrolled >1 year after diagnosis. Women were considered to have died from disease if the underlying cause of death was ovarian cancer (N=1312) or unknown (N=197) because most deaths among women with ovarian cancer are due to disease. Women were censored at the earliest time of: last follow-up (N=875) or 10 years (N=389) if alive because the data were limited after 10 years; or death from other causes unrelated to their disease (N=160). To evaluate the influence of misclassification of the cause of death, we performed sensitivity analyses classifying deaths due to unknown causes as censored rather than disease-related, and also analyzed overall survival irrespective of the causes of death.
Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated for each disease subtype using Cox regression, stratified by site. We evaluated potential confounding by age at diagnosis, tumor stage, tumor grade, extent of residual disease after primary cytoreductive surgery, and pre-treatment CA125 levels. All models included age, fit as age (years) and age-squared to allow for nonlinearity. Stage at diagnosis was categorized as localized (FIGO IA, IB), regional (FIGO IC, II), or advanced (FIGO III, IV) using cancer registry and/or FIGO stage information from each site according to SEER guidelines (http://seer.cancer.gov/). Grade was assigned using cancer registry, FIGO, Silverberg, and/or two-tier serous carcinoma19 grade information from each site. Grade was categorized as well, moderately, or poorly differentiated in EC and MC models; moderately or poorly differentiated in HGSC models; and grade was not included in CCC models because it was not a significant predictor or confounder. We performed sub-analyses, adjusting for macroscopic residual disease (no, yes, unknown), in addition to age, stage, and grade for seven sites with this information. Pre-treatment CA125 levels did not predict ovarian cancer survival or confound the associations of interest, and was not included in final models. The Cox proportional hazards assumption was evaluated by testing for a nonzero slope of the scaled Schoenfeld residuals regressed on time. We evaluated potential heterogeneity among different study sites by performing likelihood ratio tests of the interactions with site. All statistical tests were two-sided and performed using Stata 11.
Role of the funding source
No funding agency or sponsor had any role in the study design; collection, analysis, or interpretation of the data; or writing of the report. WS, MK, and SJR had full access to all of the data and the final responsibility to submit for publication.
RESULTS
Clinical and pathologic characteristics of the 2933 women with ovarian cancer (1742 HGSC, 110 LGSC, 207 MC, 484 EC, 390 CCC) are summarized in Table 2. 1669 (56·9%) patients died within ten years of their diagnosis. Of the 1669 patients who died, the cause of death was ovarian cancer for 1312 (78·6%), unrelated to ovarian cancer for 160 (9·6%), and unknown for 197 (11·8%). Information regarding extent of residual disease following primary cytoreductive surgery was available for 2023 patients (1151 HGSC, 64 LGSC, 148 MC, 370 EC, 290 CCC).
Table 2.
Characteristics of women with invasive epithelial ovarian cancer, by subtype.
| Characteristic | HGSC N=1742 n (%) |
LGSC N=110 n (%) |
MC N=207 n (%) |
EC N=484 n (%) |
CCC N=390 n (%) |
|---|---|---|---|---|---|
|
| |||||
| Age at diagnosis, mean ± SD | 60·9 ± 11·4 | 56·2 ± 14·7 | 55·0 ± 14·1 | 56·3 ± 12·4 | 56·8 ± 11·8 |
| Years followeda, mean ± SD | 4·1 ± 2·8 | 5·2 ± 3·1 | 5·5 ± 3·6 | 5·8 ± 3·2 | 5·7 ± 3·4 |
| Overall survival, median (IQR), years | 3·5 (1·8-7·0) | 6·1 (3·0-10+) | 10+ (1·8-10+) | 10+ (4·8-10+) | 10+ (2·9-10+) |
| Vital statusa | |||||
| Alive | 496 (28·5) | 51 (46·4) | 130 (62·8) | 344 (71·1) | 243 (62·3) |
| Died | 1246 (71·5) | 59 (53·6) | 77 (37·2) | 140 (28·9) | 147 (37·7) |
| Cause of deatha, % deaths | |||||
| Ovarian cancer | 1014 (81·4) | 48 (81·4) | 52 (67·5) | 86 (61·4) | 112 (76·2) |
| Other causes | 97 (7·8) | 6 (10·2) | 12 (15·6) | 26 (18·6) | 19 (12·9) |
| Unknown | 135 (10·8) | 5 (8·5) | 13 (16·9) | 28 (20·0) | 16 (10·9) |
| Stage | |||||
| Localized (FIGO IA, IB) | 87 (5·0) | 20 (18·2) | 111 (53·6) | 159 (32·9) | 136 (34·9) |
| Regional (FIGO IC, II) | 305 (17·5) | 24 (21·8) | 57 (27·5) | 217 (44·8) | 176 (45·1) |
| Distant (FIGO III, IV) | 1350 (77·5) | 66 (60·0) | 39 (18·8) | 108 (22·3) | 78 (20·0) |
| Gradeb | |||||
| Well differentiated | — | 110 (100·0) | 91 (44·0) | 178 (36·8) | — |
| Moderately differentiated | 569 (32·7) | — | 81 (39·1) | 171 (35·3) | — |
| Poorly differentiated | 1173 (67·3) | — | 35 (16·9) | 135 (27·9) | — |
| Macroscopic residual diseasec, N | 1151 | 64 | 148 | 370 | 290 |
| No | 403 (35·0) | 37 (57·8) | 109 (73·7) | 302 (81·6) | 239 (82·4) |
| Yes | 619 (53·8) | 21 (32·8) | 26 (17·6) | 29 (7·8) | 27 (9·3) |
| Unknown | 129 (11·2) | 6 (9·4) | 13 (8·8) | 39 (10·5) | 24 (8·3) |
| PR, N | 1661 | 101 | 195 | 460 | 363 |
| Negative | 1144 (68·9) | 43 (42·6) | 163 (83·6) | 150 (32·6) | 334 (92·0) |
| Weak | 393 (23·7) | 25 (24·8) | 15 (7·7) | 106 (23·0) | 18 (5·0) |
| Strong | 124 (7·5) | 33 (32·7) | 17 (8·7) | 204 (44·4) | 11 (3·0) |
| ER, N | 1691 | 104 | 197 | 475 | 381 |
| Negative | 326 (19·3) | 13 (12·5) | 156 (79·2) | 111 (23·4) | 307 (80·6) |
| Weak | 347 (20·5) | 17 (16·4) | 10 (5·1) | 78 (16·4) | 22 (5·8) |
| Strong | 1018 (60·2) | 74 (71·2) | 31 (15·7) | 286 (60·2) | 52 (13·7) |
| ER/PR, N | 1610 | 95 | 185 | 451 | 354 |
| Negative/Negative | 254 (15·8) | 9 (9·5) | 142 (76·8) | 83 (18·4) | 280 (79·1) |
| Negative/Positive | 52 (3·2) | 2 (2·1) | 5 (2·7) | 20 (4·4) | 6 (1·7) |
| Positive/Negative | 857 (53·2) | 32 (33·7) | 14 (7·6) | 64 (14·2) | 46 (13·0) |
| Positive/Positive | 447 (27·8) | 52 (54·7) | 24 (13·0) | 284 (63·0) | 22 (6·2) |
HGSC = high-grade serous carcinoma; LGSC = low-grade serous carcinoma; MC = mucinous carcinoma; EC = endometrioid carcinoma; CCC = clear cell carcinoma; IQR = interquartile range; FIGO = International Federation of Gynecology and Obstetrics.
Clinical follow-up data up to 10 years after diagnosis were included in the analyses.
Grade for CCC was not routinely collected at some sites because it was not used for clinical care.
Extent of residual disease following cytoreductive surgery was available for 7 sites (AOC, HAW, MAL, MAY+MAC, NOT, TOR, VAN).
The distribution of PR and ER expression differed markedly across ovarian cancer subtypes (Table 2). The proportion of tumors that stained positive (weak or strong) for PR was highest for EC (310/460=67·4%) and LGSC (58/101=57·4%), intermediate for HGSC (517/1661=31·1%), and lowest for MC (32/195=16·4%) and CCC (29/363=8·0%). A greater proportion of tumors stained positive for ER than PR for all subtypes: LGSC (91/104=87·5%), HGSC (1365/1691=80·7%), EC (364/475=76·6%), MC (41/197=20·8%), and CCC (74/381=19·4%). Among tumors that expressed ER, co-expression of PR was most likely for EC (284/348=81·6%), intermediate for MC (24/38=63·2%) and LGSC (52/84=61·9%) and least likely for HGSC (447/1304=34·3%) and CCC (22/68=32·4%). A substantial fraction of HGSC tumors expressed ER but not PR (857/1610=53·2%). The proportion of hormone receptor (PR and/or ER) positive tumors was much higher for LGSC (86/95=90·5%), HGSC (1356/1610=84·2%), and EC (368/451=81·6%) than for MC (43/185=23·2%), and CCC (74/354=20·9%).
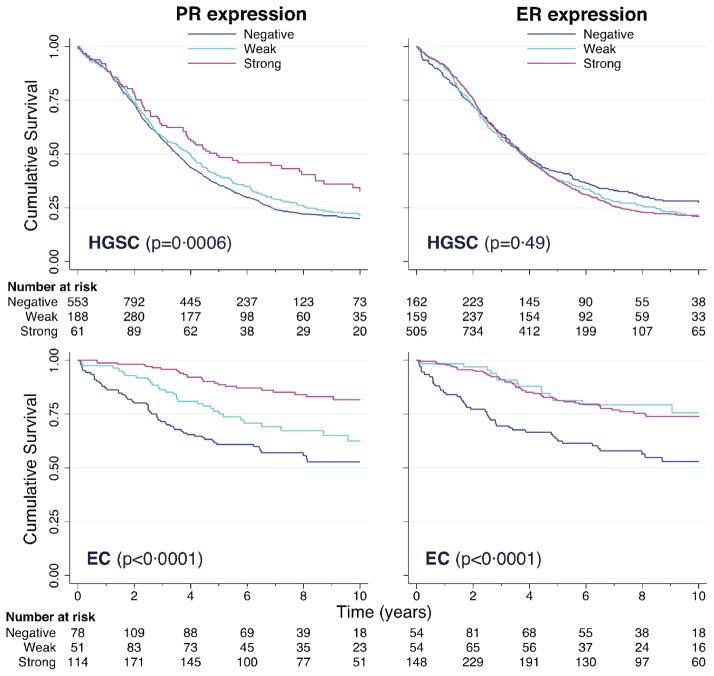
We observed distinct patterns of association between biomarker level and disease-specific survival across ovarian cancer subtypes (Figure 1, Supplementary Figure 2). PR (p=0·0006) but not ER (p=0·49) expression was strongly associated with better HGSC survival (Figure 1). In contrast, both PR (p <0·0001) and ER (p<0·0001) expression were associated with better EC survival. No significant survival differences by biomarker level were found for LGSC or MC (Supplementary Figure 2). ER expression appeared to be associated with worse CCC survival, but this association was of borderline significance after applying a Bonferroni correction for multiple comparisons, and did not persist after adjusting for other clinical covariates.
Figure 1.
Kaplan-Meier curves of disease-specific survival for high-grade serous (HGSC) and endometrioid (EC) carcinomas, according to hormone receptor status. The data were left truncated to avoid potential survival bias from the inclusion of patients (<10%) enrolled >1 year after diagnosis, and right censored at 10 years. P-values correspond to log-rank tests of the equality of survivor functions across hormone receptor expression levels, stratified by study site.
Strong PR expression was independently associated with significantly improved HGSC survival (HR, 0·71; 95% CI, 0·55–0·91; p=0·0080) compared with PR-negative tumors after accounting for site, age, stage, and grade, whereas no association with weak PR expression was found (Table 3). Similar associations were found after further adjustment for residual disease in a subset of 1151 HGSC patients (Supplementary Table 1). HGSC patients whose tumors stained strongly positive (124/1661=7·5%) versus negative or weakly positive for PR had a hazard ratio of 0·71 (95% CI, 0·55–0·91; p=0·0061) of dying from their disease (Supplementary Table 2). ER expression was not independently associated with HGSC survival (Table 3). No significant interaction between PR and ER effects was found (p=0·45). Strong PR expression was associated with better HGSC survival in both the presence (HR, 0·60; 95% CI, 0·36–1·02; p=0·059) and absence (HR, 0·74; 95% CI, 0·54–1·00; p=0·052) of strong ER expression, and no association was found for strong ER expression in the absence of strong PR expression (HR, 0·97; 95% CI, 0·85–1·11; p=0·64), compared with tumors lacking strong expression of either biomarker in analyses accounting for site, age, stage, and grade. LGSC results were similar to HGSC in that strong PR, but not ER, expression appeared to be associated with improved survival after accounting for site, age, stage, and grade (Table 3), but this association was not statistically significant after adjusting for residual disease in a subset of 64 LGSC patients (Supplementary Table 1).
Table 3.
Hormone receptor expression and disease-specific survival, by subtype.
| HGSC | LGSC | MC | EC | CCC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HRa (95% CI) | p | HRa (95% CI) | p | HRa (95% CI) | p | HRa (95% CI) | p | HRa (95% CI) | p | |
| PR expression | ||||||||||
| Negative | Referent | Referent | Referent | Referent | Referent | |||||
| Weak | 1·02 (0·89–1·18) | 0·74 | 0·53 (0·23–1·24) | 0·14 | 0·69 (0·20–2·34) | 0·55 | 0·61 (0·37–0·99) | 0·047 | 0·81 (0·36–1·83) | 0·60 |
| Strong | 0·71 (0·55–0·91) | 0·0080 | 0·39 (0·18–0·86) | 0·019 | 1·31 (0·50–3·45) | 0·58 | 0·38 (0·22–0·65) | 0·0005 | 1·13 (0·42–3·02) | 0·81 |
| ER expression | ||||||||||
| Negative | Referent | Referent | Referent | Referent | Referent | |||||
| Weak | 1·08 (0·89–1·31) | 0·44 | 1·18 (0·30–4·67) | 0·81 | 1·33 (0·34–5·17) | 0·68 | 0·40 (0·22–0·75) | 0·0041 | 1·94 (0·97–3·88) | 0·062 |
| Strong | 1·05 (0·89–1·24) | 0·56 | 0·84 (0·24–2·92) | 0·78 | 0·52 (0·23–1·21) | 0·13 | 0·41 (0·26–0·63) | 0·0001 | 1·11 (0·64–1·92) | 0·72 |
HGSC=high-grade serous carcinoma; LGSC=low-grade serous carcinoma; MC=mucinous carcinoma; EC=endometrioid carcinoma; CCC=clear cell carcinoma.
Hazard ratio (HR) and 95% confidence interval (CI) estimated using Cox regression stratified by site, and adjusted for age, age-squared, and stage (localized, regional, advanced) at diagnosis. We also adjusted for grade in models for HGSC (moderately, poorly differentiated), MC and EC (well, moderately, poorly differentiated).
Positive (weak or strong) expression of PR and ER were associated with significantly improved EC survival compared with tumors that stained negative for each biomarker, respectively, independent of site, age, stage, and grade (Table 3). EC patients with tumors that stained positive for PR (310/460=67·4%) and ER (364/475=76·6%), respectively, had hazard ratios of 0·49 (95% CI, 0·32–0·75; p=0·0012) and 0·40 (95% CI, 0·27–0·61; p <0·0001) of dying from their disease compared with patients whose tumors stained negative for each biomarker. Similar associations were found before (data not shown) and after adjusting for extent of residual disease (Supplementary Table 1) in a subset of 370 EC patients, indicating that PR and ER effects on EC survival were not mediated or confounded by residual disease. Analyses of the joint effects of PR and ER on EC survival showed that the risk reductions associated with positive staining for one or both biomarkers versus neither were similar in magnitude (Table 4), and there was no significant interaction (p=0·19). The best prognostic model for EC, as determined by a comparison of model likelihoods, combined women with hormone receptor-positive tumors, defined by positive staining for PR and/or ER, into the low-risk group. EC patients with hormone receptor-positive versus negative tumors had a hazard ratio of 0·33 (95% CI, 0·21–0·51; p <0·0001) of dying from their disease, independent of other clinical prognostic factors (Table 4, Supplementary Table 3).
Table 4.
Joint effects of PR and ER expression on EC survival.
| Biomarker | N (events) | HRa (95% CI) | p | N (events) | HRb (95% CI) | p |
|---|---|---|---|---|---|---|
| ER/PR | ||||||
| Negative/Negative | 83 (37) | Referent | 62 (28) | Referent | ||
| Negative/Positive | 20 (5) | 0·40 (0·14–1·15) | 0·088 | 16 (4) | 0·32 (0·09–1·11) | 0·073 |
| Positive/Negative | 64 (19) | 0·35 (0·19–0·64) | 0·0007 | 46 (11) | 0·38 (0·19–0·79) | 0·0096 |
| Positive/Positive | 284 (47) | 0·31 (0·19–0·51) | <0·0001 | 216 (33) | 0·32 (0·18–0·58) | 0·0001 |
| Hormone receptor | ||||||
| Negative | 83 (37) | Referent | 62 (28) | Referent | ||
| Positive | 368 (71) | 0·33 (0·21–0·51) | <0·0001 | 278 (48) | 0·34 (0·20–0·57) | <0·0001 |
Hazard ratio (HR) and 95% confidence interval (CI) estimated using Cox regression stratified by site and adjusted for age, age-squared, grade (well, moderately, poorly differentiated), and stage (localized, regional, advanced).
Cox regression stratified by site and adjusted for age, age-squared, grade (well, moderately, poorly differentiated), stage (localized, regional, advanced), and macroscopic residual disease (no, yes, unknown) using data from 7 sites (AOC, HAW, MAL, MAY+MAC, NOT, TOR, VAN).
Sensitivity analyses to evaluate the influence of misclassification of the cause of death showed that treating deaths due to unknown causes as censored rather than disease-related yielded very similar disease-specific hazard ratios (Supplementary Table 4). Analyses of overall survival yielded consistent but slightly attenuated associations as expected because tumor hormone receptor status is unlikely to influence mortality from causes other than ovarian cancer (Supplementary Table 5). Examination of Schoenfeld residuals provided some evidence that the protective effect of PR expression on EC but not HGSC survival may wane over time. However, no significant (p >0·05) departures from the proportional hazards assumption were found in fully adjusted models of the effects of ER or hormone receptor positivity on EC survival, and therefore time-dependent covariates were not included in the final models. We found no evidence of study heterogeneity (p >0·10) of the estimated biomarker effects for HGSC or EC. To evaluate potential differential misclassification of EC tumors as HGSC, we examined WT1 expression, found in ~80% of HGSC and <5% of EC tumors3, in a subset of 966 HGSC tumors (data to be published separately). WT1 expression frequencies were similar (p=0.61) across HGSC tumors with strong (58/72=80.6%), weak (190/241=78.8%), and negative (499/653=76.4%) PR staining indicating that misclassification of EC as HGSC was not more likely among PR-positive tumors.
Exploratory analyses were performed to evaluate potential modification of biomarker effects by clinical prognostic factors. In EC tumors (Supplementary Table 6), hormone receptor positivity was associated with lower grade (punadjusted=0·0007) and absence of macroscopic residual disease (punadjusted=0·041), but not stage (punadjusted=0·57). Within each stratum of grade, and among patients with and without macroscopic residual disease, hormone receptor-positive tumors were associated with significantly (p <0·05) improved EC survival, after adjusting for other clinical prognostic factors; no effect modification by grade (p=0·82), residual disease (p=0·71), or stage (p=0·95) was found. In HGSC tumors (Supplementary Table 7), PR expression was associated with lower stage (punadjusted=0·0029), but not residual disease (p=0·11). Within each stratum of stage, strong PR expression was associated with better HGSC survival after accounting for site, age, and grade; no effect modification by stage was found (p=0·41). The protective association with strong PR expression appeared to be greater among HGSC patients without (HR, 0·23; 95% CI, 0·10–0·50; p=0·0002) versus with (HR, 0·91; 95% CI, 0·61–1·36; p=0·66) macroscopic residual disease following cytoreductive surgery (pinteraction=0·0012) in analyses accounting for site, age, stage, and grade. Exploratory analysis of a subset of 376 HGSC cases screened for deleterious germline BRCA1 or BRCA2 mutations did not reveal a significant association of carrier status and tumor PR expression (Supplementary Table 7). Strong PR expression was found in 4 (7·1%) of 56 mutation carriers and 21 (6·6%) of 320 non-carriers, respectively. Furthermore, inclusion of BRCA1/2 carrier status as a covariate in HGSC survival models did not appreciably attenuate the risk reduction associated with strong PR expression, or vice versa, suggesting that these two protective factors are likely to be independent (data not shown).
DISCUSSION
Ovarian cancer subtypes have distinct etiologies and variable clinical courses.20 We found that EC patients with hormone receptor-positive tumors have a hazard ratio of about 0.33 of dying from their disease compared to patients with hormone receptor-negative tumors. Furthermore, HGSC patients with tumors that stain strongly positive for PR have a hazard ratio of about 0.71 of dying from their disease compared to patients whose tumors stain negative or weakly positive for PR. The magnitude of these biomarker effects is comparable to the protective effect of germline BRCA mutations on ovarian cancer survival,16 and is stronger than the associations of grade and extent of residual disease with EC survival in these data. This study responds to the call for large-scale international collaboration2 and demonstrates the feasibility of this approach to robustly identify clinically important prognostic biomarkers for ovarian cancer.
This collaborative study had several important strengths. The large sample size enabled robust subtype-specific analyses. Centralized IHC analyses, biomarker scoring, data harmonization, and pooled analyses reduced technical sources of variation. Finally, representation of patients worldwide increased the generalizability of study findings. One limitation was potential misclassification of histological types, which generally would be expected to obscure subtype-specific associations. Histologic review by study pathologists with expertise in gynecologic cancers was conducted by most sites. Nonetheless, some HGSC tumors may have been misclassified as high-grade EC, although misclassification in the opposite direction is rare21, 22 and unlikely to have occurred differentially given the similar WT1 expression frequencies found across PR levels in HGSC tumors.3 High-grade EC tumors were less likely to be hormone receptor-positive than low-grade EC tumors which often harbor mutations in the β-catenin gene.23 However, hormone receptor positivity was associated with significantly improved EC survival within each stratum of grade, suggesting that histological misclassification could not explain the overall findings. A second limitation was variation in tissue handling in different clinical settings, which may cause false-negative biomarker tests and potential bias towards the null. However, the antigenicity of PR and ER has been shown to be relatively robust to variable time-to-fixation.24 We found little evidence of study heterogeneity, indicating that the observed associations of hormone receptor expression with improved ovarian cancer survival are likely to be robust and generalizable. Finally, quantitative assays and optimization of ER/PR scoring could yield stronger associations with ovarian cancer outcomes.
The finding of improved survival among EC and HGSC patients with tumors that express hormone receptors could be related to intrinsic biological characteristics or better response to treatment. ERα mediates the growth stimulatory effects of estrogen and is the principal biomarker for the response of breast cancers to tamoxifen treatment.25, 26 PR mediates the growth inhibitory effects of progesterone and has been associated with improved response to progestogen treatment for endometrial cancer,27 which shares genomic features with EC and HGSC.23, 28 A Cochrane review found no data from randomized or non-randomized studies on the effectiveness of tamoxifen for relapsed ovarian cancer, or whether hormone receptor status predicts response.29 However, a recent phase III trial of tamoxifen versus thalidomide in ovarian cancer patients with asymptomatic biochemical recurrence showed that tamoxifen was less toxic and associated with lower mortality than thalidomide.30 Limited data from clinical trials that have considered hormone receptor status suggest that ERα expression may be correlated with higher response to letrozole for recurrent ovarian cancer.4, 31 In principle, the improved survival in the subset of patients with hormone receptor-positive tumors could be partially explained by better response to endocrine therapy, although response rates for all ovarian cancers are generally modest.4, 29 In this cohort, the treatment data were limited, precluding an evaluation of hormone receptor status and response to treatment.
The finding that strong PR but not ER expression was associated with improved HGSC survival is consistent with previous studies combining all ovarian cancers, among which HGSC is the predominant subtype, that in aggregate support a favorable prognosis with elevated PR but not ER protein levels.14 PR activation induces apoptosis in ovarian cancer cells,4 which could contribute to the improved survival associated with PR-positive tumors. PR is transactivated by ERα,32 and PR expression may be a biomarker of improved prognosis because it indicates a functionally intact ER pathway and less aggressive tumor behavior.33 Loss of PR expression in ER-positive breast tumors is associated with poor prognosis and has been theorized to result from: nonfunctional ER pathway, low circulating levels of estrogen, loss or hypermethylation of the PR gene, and down-regulation of PR by crosstalk between ER and growth factor signaling pathways.33, 34 The Cancer Genome Atlas (TCGA) recently demonstrated that HGSC and triple-negative breast cancers, lacking expression of ER, PR, and HER2, share many molecular features, including frequent TP53 mutations, BRCA1 inactivation, and genomic instability.35 The finding that HGSC patients with tumors lacking strong PR expression had a worse prognosis, regardless of ER status, is reminiscent of the poor prognosis reported for breast cancer patients with triple-negative or ER-positive/PR-negative tumors.33, 34 Exploratory analyses suggested that strong PR expression was associated with improved survival primarily among HGSC patients without macroscopic residual disease. The median survival in HGSC patients with and without macroscopic residual disease was 2.6 and 6.9 years, respectively, and we speculate that the lethal effects of residual HGSC may dominate other protective factors. PR did not appear to be a marker of germline BRCA mutation carriage, which has been associated with improved survival in ovarian cancer patients.16 Thus, strong PR expression identifies a distinct group of HGSC patients with good prognosis who may benefit from endocrine therapy.
The findings of this study have potentially important implications for the clinical management of patients with ovarian cancer. IHC analysis of PR and ER expression in HGSC and EC tumors could help physicians counsel patients regarding their prognosis and distinguish patients who need aggressive chemotherapy from those who may benefit from less toxic endocrine therapy. Limited data from clinical trials to date show that a subset of ovarian cancer patients responds well to endocrine therapy.4 Additional clinical trials stratified by subtype and biomarker status are needed to determine whether PR and ER predict response to endocrine therapy and can guide personalized treatment for ovarian cancer. Future studies are also needed to improve our understanding of hormone signaling pathways and their different mechanisms of action across ovarian cancer subtypes in order to inform the development of new therapeutic interventions.
RESEARCH IN CONTEXT
Systematic review
To identify previous studies of tumor ER/PR protein expression and ovarian cancer survival, we searched the PubMed database for English language papers published during 1973–2012 using the terms: ovarian cancer, survival, prognosis, progesterone receptor, estrogen receptor, hormone receptor; and manually searched references in review articles. Previous studies have yielded inconsistent associations between ER/PR protein expression and ovarian cancer survival. However, most studies combined all disease subtypes, which can obscure subtype-specific associations, or had limited sample sizes.
Interpretation
This large international collaborative study provides the first robust subtype-specific analyses of hormone receptors as prognostic biomarkers for ovarian cancer. We found that hormone receptor expression was associated with significantly improved survival among women with invasive endometrioid or high-grade serous ovarian cancer, independent of other clinical prognostic factors. Evaluation of hormone receptor status in ovarian tumors could help clinicians counsel patients regarding their prognosis, and distinguish patients who need aggressive chemotherapy from those who may benefit from less toxic endocrine therapy. Clinical trials, stratified by subtype and biomarker status, are needed to determine whether hormone receptor status predicts response to endocrine therapy, and can guide personalized treatment for ovarian cancer.
Supplementary Material
Acknowledgments
This work was supported by the Carraressi Foundation through donations to the VGH & UBC Hospital Foundation (OTTA). It was also supported by the US Department of Defense DAMD17-01-1-0729, Cancer Council Victoria, Queensland Cancer Fund, Cancer Council New South Wales, Cancer Council South Australia, Cancer Foundation of Western Australia, Cancer Council Tasmania, National Health and Medical Research Council of Australia (Grants 400413 and 400281) (AOC); US National Institutes of Health (NIH) R01CA58598, N01CN55424, N01PC67001 (HAW); NIH R01CA61107, Danish Cancer Society research grant 94 222 52, Mermaid I project (MAL); NIH R01CA122443, P50CA136393, Mayo Foundation, Fred C. and Katherine B. Andersen Foundation (MAY, MAC); NIH Z01CP010126 (POL); Roswell Park Cancer Institute Alliance Foundation (RPX); Cancer Research UK C490/A10119 C490/A10124, National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre, Cambridge Experimental Cancer Medicine Centre (SEA); NIH U01CA71966, U01CA69417, R01CA16056, K07CA143047 (STA); Eve Appeal (Oak Foundation); NIHR University College London Hospitals Biomedical Research Centre (UKO); BC Cancer Foundation, VGH & UBC Hospital Foundation, Sanofi-Aventis Canada (VAN).
We wish to thank all of the women, study centers and personnel who participated in this study. We gratefully acknowledge the contributions of the AOCS Group (http://www.aocstudy.org) (AOC); Karin Goodman, Ashley Pitzer, Mayo Clinic Medical Genome Facility (MAY, MAC); Kristy Angell, SEARCH team (SEA); Ian Jacobs, Andy Ryan, Jeremy Ford, Nayala Balogun (UKO); and the Cheryl Brown Ovarian Cancer Outcomes Unit (VAN). We assume full responsibility for the analyses and interpretation of these data.
Footnotes
CONTRIBUTORS
All authors contributed to the design and execution of this work and to the preparation of this report. Additionally, all had the opportunity to contribute to the interpretation of the results and to the redrafting of the report. Approval of the final report was obtained from all authors.
CONFLICTS OF INTEREST
We declare that we have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011 Mar-Apr;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Vaughan S, Coward JI, Bast RC, Jr, Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011 Oct;11(10):719–25. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köbel M, Kalloger SE, Boyd N, McKinney S, Mehl E, Palmer C, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS medicine. 2008 Dec 2;5(12):e232. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modugno F, Laskey R, Smith AL, Andersen CL, Haluska P, Oesterreich S. Hormone response in ovarian cancer: time to reconsider as a clinical target? Endocr Relat Cancer. 2012;19(6):R255–R79. doi: 10.1530/ERC-12-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gnant M, Harbeck N, Thomssen C. St Gallen 2011: Summary of the Consensus Discussion. Breast Care (Basel) 2011;6(2):136–41. doi: 10.1159/000328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogdall EV, Christensen L, Hogdall CK, Blaakaer J, Gayther S, Jacobs IJ, et al. Prognostic value of estrogen receptor and progesterone receptor tumor expression in Danish ovarian cancer patients: from the ‘MALOVA’ ovarian cancer study. Oncol Rep. 2007 Nov;18(5):1051–9. [PubMed] [Google Scholar]
- 7.Lee P, Rosen DG, Zhu C, Silva EG, Liu J. Expression of progesterone receptor is a favorable prognostic marker in ovarian cancer. Gynecol Oncol. 2005 Mar;96(3):671–7. doi: 10.1016/j.ygyno.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Munstedt K, Steen J, Knauf AG, Buch T, von Georgi R, Franke FE. Steroid hormone receptors and long term survival in invasive ovarian cancer. Cancer. 2000 Oct 15;89(8):1783–91. doi: 10.1002/1097-0142(20001015)89:8<1783::aid-cncr19>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 9.Burges A, Bruning A, Dannenmann C, Blankenstein T, Jeschke U, Shabani N, et al. Prognostic significance of estrogen receptor alpha and beta expression in human serous carcinomas of the ovary. Arch Gynecol Obstet. 2010 Mar;281(3):511–7. doi: 10.1007/s00404-009-1185-y. [DOI] [PubMed] [Google Scholar]
- 10.Sinn BV, Darb-Esfahani S, Wirtz RM, Budczies J, Sehouli J, Chekerov R, et al. Evaluation of a hormone receptor-positive ovarian carcinoma subtype with a favourable prognosis by determination of progesterone receptor and oestrogen receptor 1 mRNA expression in formalin-fixed paraffin-embedded tissue. Histopathology. 2011 Nov;59(5):918–27. doi: 10.1111/j.1365-2559.2011.04028.x. [DOI] [PubMed] [Google Scholar]
- 11.Darb-Esfahani S, Wirtz RM, Sinn BV, Budczies J, Noske A, Weichert W, et al. Estrogen receptor 1 mRNA is a prognostic factor in ovarian carcinoma: determination by kinetic PCR in formalin-fixed paraffin-embedded tissue. Endocr Relat Cancer. 2009 Dec;16(4):1229–39. doi: 10.1677/ERC-08-0338. [DOI] [PubMed] [Google Scholar]
- 12.Tangjitgamol S, Manusirivithaya S, Khunnarong J, Jesadapatarakul S, Tanwanich S. Expressions of estrogen and progesterone receptors in epithelial ovarian cancer: a clinicopathologic study. Int J Gynecol Cancer. 2009 May;19(4):620–7. doi: 10.1111/IGC.0b013e3181a44b62. [DOI] [PubMed] [Google Scholar]
- 13.Liu JF, Hirsch MS, Lee H, Matulonis UA. Prognosis and hormone receptor status in older and younger patients with advanced-stage papillary serous ovarian carcinoma. Gynecol Oncol. 2009 Dec;115(3):401–6. doi: 10.1016/j.ygyno.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 14.Zhao D, Zhang F, Zhang W, He J, Zhao Y, Sun J. Prognostic role of hormone receptors in ovarian cancer: a systematic review and meta-analysis. Int J Gynecol Cancer. 2013 Jan;23(1):25–33. doi: 10.1097/IGC.0b013e3182788466. [DOI] [PubMed] [Google Scholar]
- 15.Tavassoli FA, Devilee P. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Breast and Female Genital Organs. Lyon, France: IARC Press; 2003. [Google Scholar]
- 16.Bolton KL, Chenevix-Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012 Jan 25;307(4):382–90. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010 Jun 1;28(16):2784–95. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terry J, Torlakovic EE, Garratt J, Miller D, Kobel M, Cooper J, et al. Implementation of a Canadian external quality assurance program for breast cancer biomarkers: an initiative of Canadian Quality Control in immunohistochemistry (cIQc) and Canadian Association of Pathologists (CAP) National Standards Committee/Immunohistochemistry. Appl Immunohistochem Mol Morphol. 2009 Oct;17(5):375–82. doi: 10.1097/PAI.0b013e31819adacf. [DOI] [PubMed] [Google Scholar]
- 19.Malpica A, Deavers MT, Lu K, Bodurka DC, Atkinson EN, Gershenson DM, et al. Grading ovarian serous carcinoma using a two-tier system. Am J Surg Pathol. 2004 Apr;28(4):496–504. doi: 10.1097/00000478-200404000-00009. [DOI] [PubMed] [Google Scholar]
- 20.Berek JS, Hacker NF, editors. Gynecologic Oncology. 5. Philadelphia: Lippincott Williams & Wilkins; 2010. [Google Scholar]
- 21.Gilks CB, Ionescu DN, Kalloger SE, Kobel M, Irving J, Clarke B, et al. Tumor cell type can be reproducibly diagnosed and is of independent prognostic significance in patients with maximally debulked ovarian carcinoma. Hum Pathol. 2008 Aug;39(8):1239–51. doi: 10.1016/j.humpath.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Köbel M, Kalloger SE, Baker PM, Ewanowich CA, Arseneau J, Zherebitskiy V, et al. Diagnosis of ovarian carcinoma cell type is highly reproducible: a transcanadian study. Am J Surg Pathol. 2010 Jul;34(7):984–93. doi: 10.1097/PAS.0b013e3181e1a3bb. [DOI] [PubMed] [Google Scholar]
- 23.Cho KR, Shih Ie M. Ovarian cancer. Annu Rev Pathol. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumeister VM, Anagnostou V, Siddiqui S, England AM, Zarrella ER, Vassilakopoulou M, et al. Quantitative Assessment of Effect of Preanalytic Cold Ischemic Time on Protein Expression in Breast Cancer Tissues. J Natl Cancer Inst. 2012 Oct 22; doi: 10.1093/jnci/djs438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas C, Gustafsson JA. The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer. 2011 Aug;11(8):597–608. doi: 10.1038/nrc3093. [DOI] [PubMed] [Google Scholar]
- 26.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009 Sep;9(9):631–43. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 27.Sjoquist KM, Martyn J, Edmondson RJ, Friedlander ML. The role of hormonal therapy in gynecological cancers-current status and future directions. Int J Gynecol Cancer. 2011 Oct;21(7):1328–33. doi: 10.1097/IGC.0b013e31821d6021. [DOI] [PubMed] [Google Scholar]
- 28.Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013 May 2;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams C, Simera I, Bryant A. Tamoxifen for relapse of ovarian cancer. Cochrane Database Syst Rev. 2010;(3):CD001034. doi: 10.1002/14651858.CD001034.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurteau JA, Brady MF, Darcy KM, McGuire WP, Edmonds P, Pearl ML, et al. Randomized phase III trial of tamoxifen versus thalidomide in women with biochemical-recurrent-only epithelial ovarian, fallopian tube or primary peritoneal carcinoma after a complete response to first-line platinum/taxane chemotherapy with an evaluation of serum vascular endothelial growth factor (VEGF): A Gynecologic Oncology Group Study. Gynecol Oncol. 2010 Dec;119(3):444–50. doi: 10.1016/j.ygyno.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Smyth JF, Gourley C, Walker G, MacKean MJ, Stevenson A, Williams AR, et al. Antiestrogen therapy is active in selected ovarian cancer cases: the use of letrozole in estrogen receptor-positive patients. Clin Cancer Res. 2007 Jun 15;13(12):3617–22. doi: 10.1158/1078-0432.CCR-06-2878. [DOI] [PubMed] [Google Scholar]
- 32.Schultz JR, Petz LN, Nardulli AM. Estrogen receptor alpha and Sp1 regulate progesterone receptor gene expression. Mol Cell Endocrinol. 2003 Mar 28;201(1–2):165–75. doi: 10.1016/s0303-7207(02)00415-x. [DOI] [PubMed] [Google Scholar]
- 33.Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005 Oct 20;23(30):7721–35. doi: 10.1200/JCO.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Thakkar JP, Mehta DG. A review of an unfavorable subset of breast cancer: estrogen receptor positive progesterone receptor negative. Oncologist. 2011;16(3):276–85. doi: 10.1634/theoncologist.2010-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012 Oct 4;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merritt MA, Green AC, Nagle CM, Webb PM. Talcum powder, chronic pelvic inflammation and NSAIDs in relation to risk of epithelial ovarian cancer. Int J Cancer. 2008 Jan 1;122(1):170–6. doi: 10.1002/ijc.23017. [DOI] [PubMed] [Google Scholar]
- 37.Goodman MT, Lurie G, Thompson PJ, McDuffie KE, Carney ME. Association of two common single-nucleotide polymorphisms in the CYP19A1 locus and ovarian cancer risk. Endocr Relat Cancer. 2008 Dec;15(4):1055–60. doi: 10.1677/ERC-08-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lurie G, Wilkens LR, Thompson PJ, McDuffie KE, Carney ME, Terada KY, et al. Genetic polymorphisms in the Paraoxonase 1 gene and risk of ovarian epithelial carcinoma. Cancer Epidemiol Biomarkers Prev. 2008 Aug;17(8):2070–7. doi: 10.1158/1055-9965.EPI-08-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glud E, Kjaer SK, Thomsen BL, Hogdall C, Christensen L, Hogdall E, et al. Hormone therapy and the impact of estrogen intake on the risk of ovarian cancer. Arch Intern Med. 2004 Nov 8;164(20):2253–9. doi: 10.1001/archinte.164.20.2253. [DOI] [PubMed] [Google Scholar]
- 40.Soegaard M, Jensen A, Hogdall E, Christensen L, Hogdall C, Blaakaer J, et al. Different risk factor profiles for mucinous and nonmucinous ovarian cancer: results from the Danish MALOVA study. Cancer Epidemiol Biomarkers Prev. 2007 Jun;16(6):1160–6. doi: 10.1158/1055-9965.EPI-07-0089. [DOI] [PubMed] [Google Scholar]
- 41.Goode EL, Chenevix-Trench G, Hartmann LC, Fridley BL, Kalli KR, Vierkant RA, et al. Assessment of hepatocyte growth factor in ovarian cancer mortality. Cancer Epidemiol Biomarkers Prev. 2011 Aug;20(8):1638–48. doi: 10.1158/1055-9965.EPI-11-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams E, Martin S, Moss R, Durrant L, Deen S. Co-expression of VEGF and CA9 in ovarian high-grade serous carcinoma and relationship to survival. Virchows Arch. 2012 Jul;461(1):33–9. doi: 10.1007/s00428-012-1252-9. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Closas M, Brinton LA, Lissowska J, Richesson D, Sherman ME, Szeszenia-Dabrowska N, et al. Ovarian cancer risk and common variation in the sex hormone-binding globulin gene: a population-based case-control study. BMC cancer. 2007;7:60. doi: 10.1186/1471-2407-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moysich KB, Baker JA, Menezes RJ, Jayaprakash V, Rodabaugh KJ, Odunsi K, et al. Usual adult body mass index is not predictive of ovarian cancer survival. Cancer Epidemiol Biomarkers Prev. 2007 Mar;16(3):626–8. doi: 10.1158/1055-9965.EPI-06-1052. [DOI] [PubMed] [Google Scholar]
- 45.Song H, Ramus SJ, Quaye L, DiCioccio RA, Tyrer J, Lomas E, et al. Common variants in mismatch repair genes and risk of invasive ovarian cancer. Carcinogenesis. 2006 Nov;27(11):2235–42. doi: 10.1093/carcin/bgl089. [DOI] [PubMed] [Google Scholar]
- 46.McGuire V, Felberg A, Mills M, Ostrow KL, DiCioccio R, John EM, et al. Relation of contraceptive and reproductive history to ovarian cancer risk in carriers and noncarriers of BRCA1 gene mutations. Am J Epidemiol. 2004 Oct 1;160(7):613–8. doi: 10.1093/aje/kwh284. [DOI] [PubMed] [Google Scholar]
- 47.Narod SA, Risch H, Moslehi R, Dorum A, Neuhausen S, Olsson H, et al. Oral contraceptives and the risk of hereditary ovarian cancer. Hereditary Ovarian Cancer Clinical Study Group. N Engl J Med. 1998 Aug 13;339(7):424–8. doi: 10.1056/NEJM199808133390702. [DOI] [PubMed] [Google Scholar]
- 48.Balogun N, Gentry-Maharaj A, Wozniak EL, Lim A, Ryan A, Ramus SJ, et al. Recruitment of newly diagnosed ovarian cancer patients proved challenging in a multicentre biobanking study. Journal of clinical epidemiology. 2011 May;64(5):525–30. doi: 10.1016/j.jclinepi.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 49.Prentice LM, Klausen C, Kalloger S, Kobel M, McKinney S, Santos JL, et al. Kisspeptin and GPR54 immunoreactivity in a cohort of 518 patients defines favourable prognosis and clear cell subtype in ovarian carcinoma. BMC Med. 2007;5:33. doi: 10.1186/1741-7015-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Köbel M, Reuss A, Bois A, Kommoss S, Kommoss F, Gao D, et al. The biological and clinical value of p53 expression in pelvic high-grade serous carcinomas. J Pathol. 2010 Oct;222(2):191–8. doi: 10.1002/path.2744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.