Abstract
We used a DNA barcoding method to enable high-throughput sequencing of the cognate heavy- and light-chain pairs of expressed antibodies. We used this approach to elucidate the plasmablast antibody response to influenza vaccination. We show that >75% of the rationally selected plasmablast antibodies bind and neutralize influenza, and that antibodies from clonal families, defined by sharing both heavy chain VJ and light chain VJ sequence usage, do so most effectively. Vaccine-induced heavy chain VJ regions contained on average >20 nucleotide mutations as compared to their predicted germline gene sequences, and some vaccine-induced antibodies exhibited higher binding affinities for hemagglutinins derived from prior years’ seasonal influenza as compared to their affinities for the immunization strains. Our results show that influenza vaccination induces the recall of memory B cells that express antibodies that previously underwent affinity maturation against prior years’ seasonal influenza, suggesting that ‘original antigenic sin’ shapes the antibody response to influenza vaccination.
1. Introduction
Despite the availability of seasonable influenza vaccination, influenza continues be a major cause of morbidity and mortality in the US and worldwide [1, 2]. Even the healthy adult population is vulnerable, as highlighted by the significant morbidity and mortality during the 1918 and 2009 pandemics [3, 4]. Although influenza vaccination can reduce influenza-associated morbidity and mortality, a meta-analysis of available data indicate that the trivalent inactivated influenza vaccine (TIV) is only effective in approximately 60% of young adults [5], and the protection afforded is significantly lower in those over age 65 [6]. As a result, there is great need to better understand the mechanisms underlying effective vs. non-effective vaccination for influenza.
TIV induces neutralizing antibodies that mediate protection against infection [2, 7], and there are likely a variety of mechanisms underlying effective vs. non-effective vaccination. Because immune responses wane with advancing age, vaccination may not offer sufficient protection in the elderly, who account for >90% of influenza-related deaths [1]. The inadequacy of protection in the elderly is in part due to their mounting an inferior antibody response [8]. Compared to those in young adults, antibody repertoires in elderly individuals have a lower number of lineages and a higher pre-vaccination mutation load [9]. Deep sequencing of the immunoglobulin heavy chain RNA present in peripheral blood mononuclear cells following repeat yearly influenza vaccinations demonstrated direct genetic measurement of memory B-cell recall responses [10].
The immune history of individuals, specifically prior infections with related pandemic H1N1 influenza strains, can shape the specificity of the pandemic H1N1 influenza antibody responses to viral regions with shared homology [11]. Further, the antibody response to influenza vaccination can include antibodies that react to a greater extent with influenza strains present in prior years seasonal influenza to which an individual has been exposed (and not present in the current influenza strain), a phenomenon known as “original antigenic sin” (OAS) [12-15].
Secreted antibodies are produced by plasma cells and their B-cell precursors, termed plasmablasts [16-18]. After becoming activated and undergoing antigen-dependent affinity maturation, naïve and memory B cells differentiate into plasmablasts which proliferate, undergo affinity maturation, and then traffic via the blood to infected tissues and secondary lymphoid organs [19]. Although plasmablast frequencies in peripheral blood are low before and after an immune response (~0.1% of circulating B cells), they can account for 1 – 5% of peripheral blood B cells in response to influenza vaccination [17, 20] and over 30% of circulating B cells during an immune response to an infection [16, 21]. Further, it has been demonstrated that the majority of plasmablasts generated in response to influenza vaccination express influenza-reactive antibodies [14, 17, 20]. In this study we characterized the antibody repertoires of peripheral blood plasmablasts to provide a 'snapshot' of the ongoing antibody response.
To investigate antibody responses to vaccinations, microbial infections, in autoimmunity, and in other immune responses, we developed a novel DNA barcoding method that combines high-throughput sequencing and barcode-enabled pairing of cognate heavy- and light-chain antibody sequences expressed by individual B cells. This method enables the comprehensive analysis of antibody repertoires, followed by the direct and efficient production of endogenous monoclonal antibodies.
We describe use of this novel DNA barcoding method to characterize the plasmablast antibody response to influenza vaccination. We focused our analysis on the antibodies expressed by plasmablasts circulating in the bloodstream following influenza vaccination. We bioinformatically generated phylogenetic trees of the antibody response and rationally selected key antibodies for cloning, expression, and characterization of their binding and functional properties. Greater than 75% of the rationally selected plasmablast antibodies bound and neutralized influenza, and antibodies from clonal families (sharing heavy chain V(D)J and light chain VJ sequences) do so most effectively. Some vaccine-induced antibodies exhibited higher binding affinities for hemaglutinins derived from prior years’ seasonal influenza as compared to their affinities for the immunization strains.
2. Materials and Methods
2.1 Single-cell sorting of plasmablasts
We collected blood from three individuals (Suppl. Table 1) vaccinated 7 days earlier with the 2010/2011 seasonal trivalent influenza vaccine (Sanofi Pasteur), which consists of 3 strains of inactivated influenza: H1N1 A/California/7/2009, H3N2 A/Perth/16/2009, and B/Brisbane/60/2008. Samples were collected after obtaining informed patient consent and under human subject protocols approved by the Investigational Review Board (IRB) at Stanford University. PBMCs were stained with CD3-V450 (BD 560365), IgA-FITC (AbD Serotec STAR142F or Miltenyi #130-093-071), IgM-FITC (AbD Serotec STAR146F), IgM-APC (BD 551062) or IgM-PE (AbD Serotec STAR146PE), CD20-PerCP-Cy5.5 (BD 340955), CD38-PE-Cy7 (BD 335808), CD19-APC (BD 340437) or CD19-Brilliant Violet 421 (Biolegend 302233), and CD27-APC-H7 (BD 560222). We sorted cells with a BD FACSAria II or III, achieving purities of >80% from the first bulk sort. We gated on CD19+CD20−CD27+CD38++IgA−IgM− cells for the bulk plasmablast sort, and then single-cell sorted them into 96-well PCR plates containing a hypotonic buffer (10mM Tris-HCl pH 7.6) comprised of 2 mM dNTPs (NEB), 5 μM oligo(dT)20VN, and 1 unit/μL of Ribolock (Fermentas), an RNase inhibitor.
2.2. Reverse transcription with well-ID-containing adaptor
We added 1 μM final concentration of the appropriate reverse transcription (RT) adapter molecule (each containing a unique well-ID barcode; listed in Suppl. Table 2) to each well, including the negative-control wells (containing no cells). We performed RT at 42°C for 120 minutes in buffer containing 6 mM MgCl2 with Ribolock and Superscript III (Life Technologies). Following RT, the cDNA products from all wells of each plate were pooled together, extracted with phenol-chloroform-isoamyl alcohol followed by chloroform, and concentrated and desalted them in 5-minute spins at 14 000g with Amicon Ultra-0.5 30 kDa (Millipore) units.
2.3. Polymerase Chain Reaction (PCR)
We used Phusion Hot Start II DNA polymerase (NEB/Fermentas) for both the first PCR (PCR1) and the nested PCR (PCR2). Primers and adapter molecules are listed in Suppl. Table 2. Reaction conditions were as follows: 200 μM of dNTPs, 0.2 μM of primers, 0.2 U of Phusion polymerase, 4% DMSO, and 2 μL of RT product. Forward (FW) primers used in PCR1 were the FW long primer1 and the FW short primer1. We added FW long primer1s containing different plate-IDs to different samples. We used gene-specific (GSP) reverse primers GSP1, lambda GSP1, and gamma GSP1, to amplify the kappa, lambda, and gamma chains, respectively. FW primer for PCR2 was FW primer2, and reverse primers were RV primer2 and GSP long primer2. We used kappa GSP long primer2, lambda GSP long primer2, and gamma long primer2 to amplify their respective amplicons. We added a different GSP long primer2s with plate-specific plate-IDs to each pooled samples from each plate.
2.4. 454 sequencing
For 454 sequencing, we pooled the amplified DNA, gel purified them, and then purified them with Ampure XP beads (Beckman Coulter). We determined DNA concentrations by using Picogreen DNA assay kits (Invitrogen) and sent the amplicons to Roche for 454 Titanium sequencing. Sequences are available at GenBank (accession numbers KF994033-KF994563).
2.5. Barcode assignment and assembly of sequences
Sequencing data were analyzed by 454 GS FLX data analysis software, and we received filter-passed, high-quality sequences. We made plate and well assignments for sequencing reads by comparing the sequence to the plate- and well-IDs. We used Newbler 2.6 to assemble all the sequence reads that were assigned the same well-ID and thereby obtain consensus sequences that correspond to the heavy- and light-chain mRNA sequences expressed by the sorted plasmablasts. Sff output files from 454 sequencing, containing sequences and quality scores for each nucleotide, were read into Python by using the Biopython package, and sequences were grouped and parsed into separate sff files on the basis of their compound ID (plate-ID + well-ID). Newbler assembled forward reads by using the "-cdna", "-ud" and "-urt" options, using a minimum threshold of 8 reads. Where multiple assemblies occurred per well, which is common with oversampling (>100 reads), an assembly was accepted if it contained >50% of all reads in a well, and was 3x more abundant than the next read. Otherwise, we assumed that the well contained more than one plasmablast, and we disregarded the sequence reads from that well.
2.6. VJ and clonal family assignment
We analyzed heavy- and light-chain sequences with HighV-QUEST[22], software that compares an antibody-chain sequence to a database of known alleles of germline sequences and predicts which germline alleles the antibody uses, how the germline sequences were recombined, and the number of mutations the antibody has relative to the germline sequence (including all mutations, silent mutations, and non-silent mutations). Clonal family antibodies were defined as antibodies with shared heavy chain VJ and light chain VJ usage and >20% shared mutations as compared to the corresponding germline sequence.
2.7. Clustering of sequences
Heavy- and light-chain sequences were binned according to their V-gene usage and then concatenated and aligned with Muscle[23]. They were clustered with PhyML[24] maximum-likelihood clustering, together with their germline V gene, and rooted by their germline V gene. Trees were drawn using ETE[25].
2.8. Cloning antibody genes
For selection of antibodies representative of clonal families, we bioinformatically aligned, compared and selected either the most frequent member and/or heavy and light chain pair most representative of the consensus sequence of the clonal family. Using PCR cloning or direct gene synthesis (Lake Pharma), we generated and inserted kappa and lambda light chains into vector pEE12.4 (Lonza), and the gamma heavy chain into vector pEE6.4 (Lonza). For PCR cloning, we used well-ID-specific forward primers and constant region-specific reverse primers to selectively amplify cDNA for insertion into the expression vectors.
2.9. Expression of monoclonal antibodies
We performed transient, dual transfections of paired light-chain-containing pEE12.4 and heavy-chain-containing pEE6.4 constructs in 293T cells by using Lipofectamine 2000 according to the manufacturer’s protocol. We purified the expressed antibodies from the culture supernatants by using Protein A Plus agarose beads (Pierce).
2.10. Influenza vaccine ELISA
We used ELISA to determine whether the monoclonal antibodies derived from vaccinated individuals bind to the influenza vaccine itself. We coated Nunc Maxisorp plates with the trivalent influenza vaccine diluted 100x in a pH 9.0 carbonate buffer, blocked the plates with PBS containing 1% BSA, added 100 ng/ml of expressed monoclonal antibodies, and detected antibody binding using an HRP-labeled detection antibody.
2.11. Surface plasmon resonance (SPR) determination of antibody affinities
We analyzed the binding of monoclonal antibodies to hemagglutinins at 25°C by using a ProteOn SPR biosensor (Bio-Rad Laboratories). We coupled the antibodies (25 nM in pH 4.5 acetate buffer) to a GLC sensor chip with amine coupling using EDAC-NHS chemistry, attaining a target density that yielded 500-800 resonance units (RU) in the test flow cells. We quenched unreacted active ester groups with ethanolamine. Recombinant hemagglutinins H3 (HA(ΔTM)(H3N2/Perth/16/2009), H3s from other strains used in experiments shown in Fig. 4, and H1 (HA(ΔTM)(A/California/07/2009)(H1N1)) from Immune Technology Corp. (New York, NY) were diluted to 100, 50, 25, 12.5, 6.25 nM and injected separately into the ProteOn System at a flow rate of 30 μL/min, with 120 seconds of contact time and 2000 seconds of dissociation time. As a control, we injected a buffer without H3 or H1. We analyzed binding kinetics with Bio-Rad ProteON Manager software and calculated affinities by using the bivalent analyte algorithm (because HA consists of several repeating units). We verified the accuracy of the fitted curves by ensuring that the χ2 values for each fit were <10% of the value of maximal binding (Rmax).
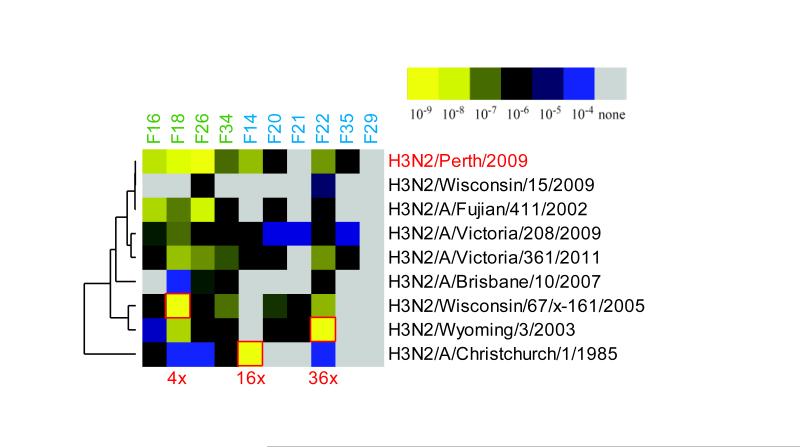
Figure 4.
Influenza vaccination induces plasmablasts expressing antibodies that bind to prior year’s hemagglutinins with high affinity. Plasmablasts responding to influenza vaccination develop from memory B cells and some show original antigenic sin. Heatmap of the affinity (KD value) of antibody binding to purified, recombinant H3 hemagglutinins from H3N2 influenza A strains prevalent in different influenza seasons, as measured by ProteOn SPR. Antibodies cloned from individual plasmablasts are listed along the top of the heatmap, and the different seasonal H3N2 influenza strains are listed to the right. Red boxes demarcate stronger antibody binding to H3 from that season’s influenza strain than to H3 from the influenza strain contained in the vaccine (H3N2/Perth/16/2009), with the numbers in red below the heatmap indicating the fold increase in binding.
2.12. Influenza microneutralization assay
Influenza microneutralization assays were performed by an external CRO, Virapur, LLC (San Diego). In brief, various amounts of monoclonal antibody were mixed with an equal volume of ~100 TCID50 infectious units of titered stock influenza H1N1 or H3N2 strains in quadruplicate in wells of a 96-well plate. Virus-antibody solutions were incubated for 2 hours and then the mixture was transferred to a 96-well plate containing 80% confluent MDCK cells. Cells, antibody, and virus were incubated for an additional 2 hours at 37°C, after which the virus was removed, the cells rinsed, and viral growth media added to each well. Influenza infection of the cells was assessed microscopically after 72 hours.
3. Results
3.1. Sequencing the paired heavy- and light-chain antibody sequences expressed by individual B cells derived from influenza-vaccinated individuals
Our approach consists of tagging all cDNA generated from an individual B cell with a unique DNA barcode before sequencing the cDNA; we then use these unique barcodes to match the heavy- and light-chain antibody sequences that derive from the same cell. In this way, we uncover the sequences of the specific heavy and light chains that make up each antibody, information that we use to generate phylogenetic trees of the antibody response and to clone and express the antibodies encoded by these sequences.
We collected blood from 3 individuals (Suppl. Table 1) 7 days after they received the trivalent influenza vaccine, a time at which the plasmablast response is at its peak [17], and single-cell sorted IgG+ plasmablasts into four to eight 96-well plates on the basis of cell-surface staining for CD19+CD20−CD27+CD38+IgA−IgM−. (Plasmablasts generated in response to influenza vaccination are predominantly IgG expressing [14, 17].) We performed reverse transcription with an MMLV H− reverse transcriptase that has 3'-tailing[26] and template-switching activity in order to synthesize cDNA and tag it with a ‘well-ID barcode’ sequence unique to each well (and hence unique to each cell). By adding a universal priming sequence 5' of the well-ID barcode, we obviate the need for degenerate V-gene primers and the attendant bias in amplification of V-region genes. We pooled the well-ID-tagged cDNAs from each plate and tag them with plate-ID barcodes using nested PCR and primers specific to the constant regions of the heavy and light chains; in this way we uniquely tagged all cDNA derived from the same plate and add titanium primers for 454 sequencing. Thus, each amplicon contains two barcodes, one identifying the plate from which the amplicon derives (the plate-ID) and another identifying the individual well within that plate (the well-ID). We pooled the double-barcoded amplicons from all plates and sequence them by 454 sequencing.
Through in silico analysis of the sequences obtained, we identify cognate heavy- and light- chain pairs expressed by individual plasmablasts, generate phylogenetic trees representing the antibody response, and rationally select antibodies for functional characterization. This was achieved by first assigning the well-ID barcode and plate-ID barcode contained in each read, thereby assigning all reads to individual plasmablasts, and thereby identified the cognate heavy- and light-chain pairs expressed by individual plasmablasts. For all reads derived from a single plasmablast, we averaged the sequence for all heavy chain reads, and independently all light chain reads, to obtain consensus reads and thereby average-out errors arising from PCR or sequencing error. We determined their heavy and light chain VJ usage by analyzing them with IMGT HighV-QUEST [22]. Maximum likelihood clustering of heavy and light chains sequences grouped by heavy chain V-gene usage yielded phylogenetic trees that revealed clonal families of antibodies that use the same heavy-chain V(D)J and light-chain VJ sequences for each of the 3 influenza vaccine recipients (Figs. 1A-C).
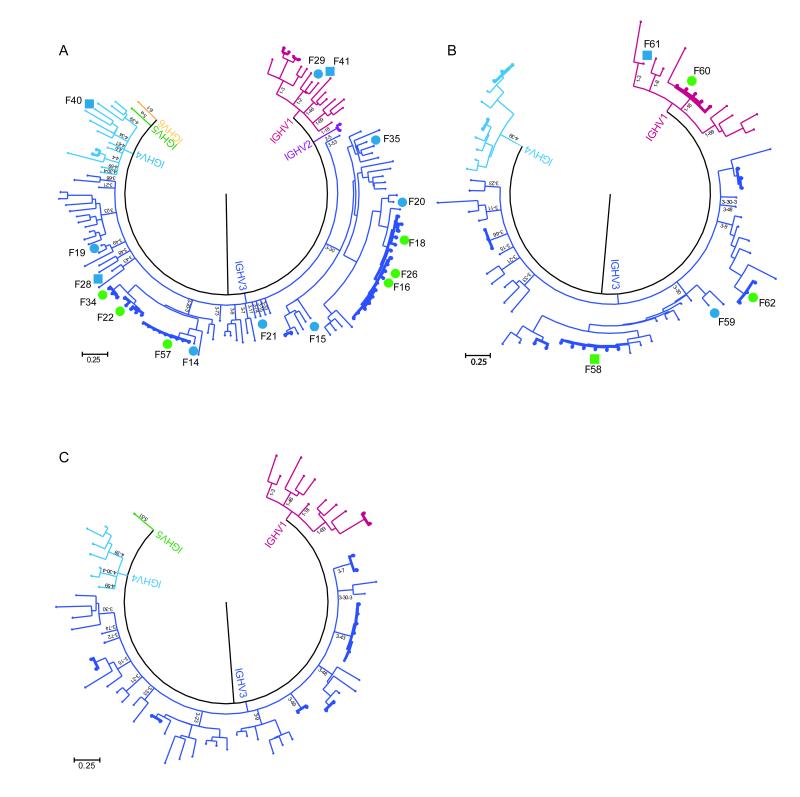
Figure 1.
Analysis of the plasmablast antibody repertoire of three individuals vaccinated against influenza. (A-C) The paired heavy-chain and light-chain antibody sequences expressed by individual plasmablasts were arranged by heavy chain V gene family usage and clustered to generate the displayed phylogenetic trees. Each phylogenetic tree represents an antibody repertoire from a different influenza vaccine recipient. Antibodies from clonal families are shown in bold. The letter F followed by a number denotes an antibody that was selected for cloning and expression; circles denote antibodies that bound components of the influenza vaccine, and squares denote antibodies that did not (see Fig. 3). The circles and squares are colored green if they denote antibodies from clonal families, and blue if they denote singleton antibodies.
The antibodies contained in each clonal family derive from distinct plasmablasts that are all the progeny of a single B cell; such antibodies therefore share mutations acquired through the multiple rounds of somatic hypermutation that occur in tandem with B-cell clonal expansion and lead to the generation of affinity-matured plasmablast. Indeed, sequence-alignment analyses confirmed that the amino acid and nucleotide sequences of antibodies from the same clonal family not only use the same V(D)J but also have a common set of mutations (data not shown). Missense mutations were concentrated in the complementarity determining regions, suggesting that the plasmablasts were functionally selected. The antibody response showed a high degree of clonality, which accounted for approximately one-third of the plasmablast antibodies sequenced in each of the individuals.
3.2. Singleton antibodies and antibodies from clonal families are both highly mutated
Relative to their predicted germline sequences, the plasmablast antibodies produced during the acute response to the influenza vaccine had means of >20 mutations in their heavy chains, >10 mutations in their light chains, and >30 mutations overall (Fig. 2). Mutations in the variable regions of antibody genes result from somatic hypermutation, a process that leads to affinity maturation of antigen-activated B cells in germinal centers[27]. Because the rate of somatic hypermutation in vivo is approximately 10−3 bp−1 division−1, with an assumed division time of 18 hours [28, 29], we estimate that, in the 7 days after vaccination, the heavy chains would undergo an average of 6 mutations and the light chains an average of 4 mutations. That only a small fraction of the antibodies had fewer than 20 mutations suggests that the influenza vaccination activated predominantly memory B cells, which are already affinity matured (and may undergo further affinity maturation in the process of forming plasmablasts). Memory B cells may dominate the response to vaccination because their activation threshold is lower than that of naïve B cells, and they also proliferate faster [30, 31].
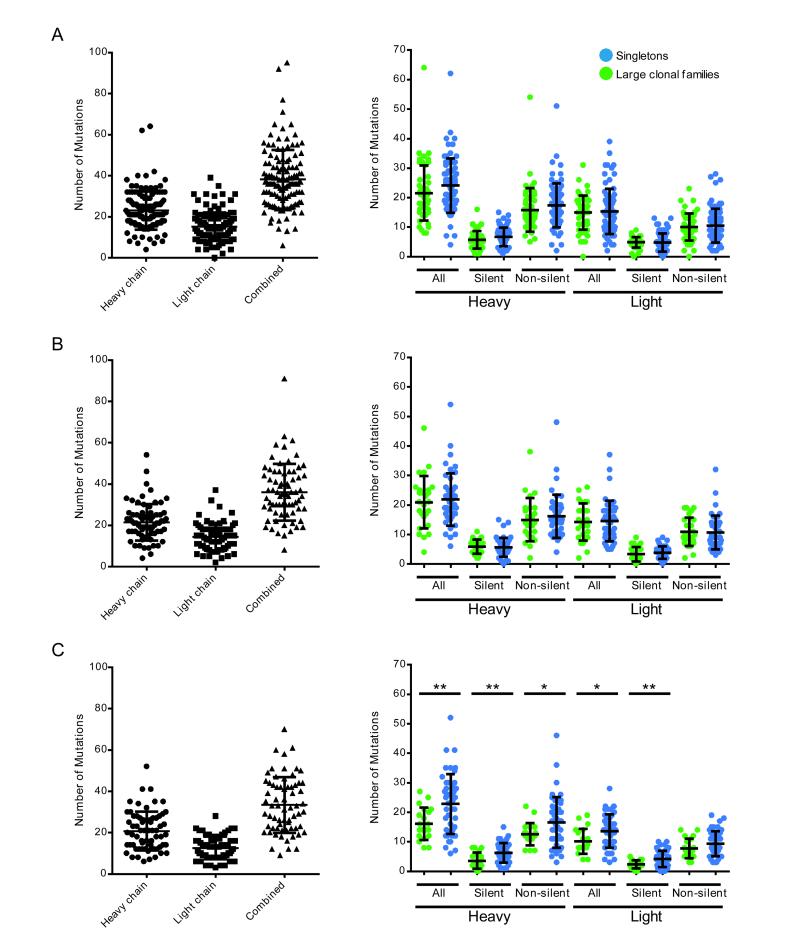
Figure 2.
Analysis of mutation rates in antibody repertoires of plasmablasts from influenza-vaccinated individuals. Number of mutations in the V and J genes relative to the predicted germline sequences in the heavy and light chains of the sequenced antibody repertoires presented (A) in Fig. 1A, (B) in Fig. 1B, and (C) in Fig. 1C. Left-hand panels show the total number of mutations in the V and J genes relative to the predicted germline sequences for the heavy chain, light chain, or both of the paired heavy and light chains (combined) of each sequenced plasmablast antibody. Right-hand panels show a breakdown of the number of mutations in the V and J genes into total (all), silent, and non-silent mutations, with green circles denoting antibodies from large clonal families and blue circles denoting singleton antibodies. *P < 0.05 by Student’s T-test.
Despite the fact that somatic hypermutation leads to affinity maturation, there were no significant differences in frequency of overall or of non-silent mutations between the antibodies from the identified clonal families, which had picomolar binding affinities for H3N2/Perth/16/2009 influenza, and the singleton antibodies, which had significantly lower affinities (Figs. 2, 3B). In fact, among antibodies from one of the vaccinated individuals, singleton antibodies had a higher frequency of overall mutations than antibodies from clonal families (Fig. 2C).
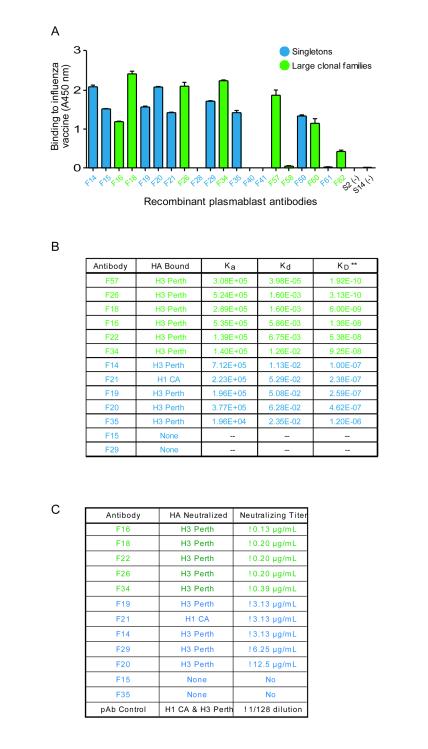
Figure 3.
Cloned plasmablast antibodies bind and neutralize influenza. (A) ELISA analysis of the binding of cloned plasmablast antibodies to a crude preparation of the components of the influenza vaccine. Data are shown as the mean with SEM and are representative of 3 independent experiments. (B) Affinities of antibody binding to purified, recombinant hemagglutinins H3 (from the H3N2/Perth/16/2009 influenza A strain) and H1 (from the H1N1/California/07/2009 influenza A strain), as assessed by surface plasmon resonance. Antibodies from clonal families (green lettering) had significantly higher affinity than did singleton antibodies (blue lettering); *P=0.016, Mann Whitney test. (C) Microneutralization assay analysis of the antibodies’ ability to neutralize influenza A strains contained in the vaccine. Antibodies from clonal families (green lettering) neutralize H3N2/Perth/16/2009 influenza at lower concentrations than do singleton antibodies (blue lettering).
3.3. Recombinant plasmablast antibodies bind components of the influenza vaccine
We investigated whether the antibodies encoded by the sequences identified can bind to components of the trivalent influenza vaccine that elicited the antibody response. We used PCR or gene synthesis to clone, and transient transfection in 293T cells to express, the paired heavy and light chains of antibodies representative of clonal families, as well as ‘singleton’ antibodies, i.e., antibodies using heavy-chain V(D)J and/or light-chain VJ sequences that are not used by any other antibody in an individual’s plasmablast antibody repertoire (Fig. 1). Of the 8 antibodies from clonal families 7 bound to components of the influenza vaccine (87.5%), while only 8 of the 12 singleton antibodies bound (66%, Fig. 3A). That the majority of the antibodies exhibited reactivity to influenza is consistent with findings from previous studies examining the specificity of plasmablasts derived from influenza-vaccinated individuals [17].
3.4. Antibodies from clonal families bind hemagglutinin from the immunizing influenza strain with higher affinity than do singleton antibodies
All 6 analyzed clonal family antibodies, and 5 of the 8 singleton antibodies, bound hemagglutinin H3 (from the H3N2/Perth/16/2009 influenza A strain present in the trivalent vaccine) (Fig. 3B). None of the antibodies from clonal families, and only 1 of the singleton antibodies, bound hemagglutinin H1 (from the H1N1/California/07/2009 influenza A strain present in the trivalent vaccine) (Fig. 3B). Using surface plasmon resonance, we assessed the affinity with which the recombinant plasmablast antibodies bind to influenza hemagglutinin (a major target of antibodies that neutralize influenza A virus [32]), measuring the on/off rates and dissociation constants (Fig. 3B). Compared to singleton antibodies, antibodies from clonal families bound H3 with 10-1,000-fold higher affinities (P=0.0043, Mann Whitney test). This finding is consistent with the current paradigm that centrocytes (nondividing, activated B cells expressing membrane-bound antibodies) that have higher affinities for their antigen compete more successfully for positive selection in germinal centers and are therefore more likely to undergo antigen-driven clonal expansion[27].
3.5. Antibodies from clonal families neutralize influenza more effectively than do singleton antibodies
Testing the functionality of the antibody binding in a microneutralization assay, we found that 9 of the 12 antibodies tested could neutralize the H3N2/Perth/16/2009 influenza strain. In line with their differential affinities for influenza hemagglutinins, the antibodies from clonal families neutralized H3N2 at lower titers than did the singleton antibodies (Fig. 3C). Of the antibodies tested, only the singleton antibody that bound H1 hemagglutinin in the affinity-binding assay neutralized the H1N1/California/07/2009 influenza strain (Fig. 3B,C).
3.6. Influenza vaccination induces plasmablasts expressing antibodies that bind to prior year’s hemagglutinins with high affinity
To determine the binding affinity of the recall response to influenza vaccination, we evaluated the binding of the recombinant plasmablast antibodies to H3 hemagglutinins derived from different H3N2 influenza A strains. Of the antibodies from clonal families, some bound with greater affinity to H3 from the influenza strain present in the vaccine they were responding to (H3N2/Perth/16/2009), some bound with greater affinity to H3 from influenza strains prevalent in previous influenza seasons, and others bound to the H3N2/Perth/16/2009 H3 and to previous seasons’ H3 with similar affinity (Fig. 4). The singleton antibodies either bound with greater affinity to previous seasons’ H3 or did not bind to any of the H3s tested (perhaps because they are specific for an influenza antigen other than hemagglutinin H3, e.g., a different hemagglutinin subtype (as shown for antibody F21) or neuraminidase [32]) (Fig. 4).
Pre-vaccination serum from the subject whose plasmablast repertoire is shown in Fig. 1A was tested by ELISA against influenza hemagglutinin H3N2 A/Perth/16/2009 (vaccine strain) and H3N2/A/Christchurch/1/1985 (prior seasonal influenza strain). These strains are representative of cases where binding of a recombinant antibody to HA from a previous year (Christchurch/1985) had higher affinity than binding to the vaccine strain (Perth/2009) (Fig. 4), suggesting original antigenic sin shaped the response to influenza vaccination. ELISA analysis demonstrated that this individual did in fact have high pre-vaccination serum antibody reactivity to Christchurch/1985 (data not shown), which supports our hypothesis that vaccine-induced antibody responses are shaped by OAS.
4. Discussion
We describe use of a new method to sequence the paired heavy and light chains expressed by individual B cells, and its application to sequence the antibody repertoire of peripheral blood plasmablasts following influenza vaccination. Specifically, we combined cDNA barcoding with high-throughput sequencing to determine the sequences of the antibody heavy and light chains expressed by individual plasmablasts. The sequence datasets enable the identification of clonal families of antibodies that share HC V(D)J and LC VJ sequences. The identification of clonal families enabled rationale bioinformatic selection, and hence efficient production, of the antibodies that likely to be integral to the response.
We used this DNA barcoding technology to obtain a 'snapshot' of the functional antibody response to influenza vaccination. Focusing our analysis on the antibody repertoires of circulating plasmablasts, we generated phylogenetic trees of the santibody response, and bioinformatically selected and recombinantly produced antibodies that could bind to and neutralize influenza. We found that antibodies from clonal families bound influenza with greater affinity and neutralized it more effectively, consistent with the current understanding of affinity maturation, i.e., that B cells expressing antibodies with higher affinity for their antigen out-compete B cells expressing lower affinity antibodies in the germinal center, and are thus more likely to undergo antigen-driven clonal expansion[27]. This finding confirms the utility of the phylogenetic trees and the identification of clonal families in guiding selection of the critical antibodies for recombinant production. Moreover, the extent of clonality, the proportion of recombinant plasmablast antibodies that bound influenza, and the predominance of a recall response in our studies are all consistent with previous findings on the antibody response to influenza vaccination [17].
In contrast to previous findings [17], we found that a substantial proportion of the plasmablast antibodies bound with higher affinity to influenza strains prevalent during prior seasons than to the strain contained in the vaccine. It is possible that our technology enables identification of such antibodies by characterizing the antibody repertoire to a greater depth than previously attained. Nevertheless, we found that other antibodies, from clonal families specifically, exhibited high binding affinities for the immunizing hemagglutinin. Thus, vaccination against influenza activates a combination of B cells expressing high-affinity and low-affinity antibodies against the immunizing strain. This diversity of antibody specificity may partly explain why influenza vaccination induces antibodies that are broadly neutralizing [33-35].
According to the theory of original antigenic sin (OAS), the immune response to a newly encountered variant of a pathogen will be compromised if it is dominated by memory B cells whose antibodies have a higher affinity for a previously encountered variant [36]. OAS could be one potential explanation for the low affinity of certain highly mutated singleton antibodies for H3 from the H3N2/Perth/16/2009 immunizing strain. This is supported by our observation that singleton antibodies had a higher frequency of mutations than clonal family antibodies. Singletons may represent sequences that affinity matured in previous responses to influenza infection and/or vaccination, and that represent recall responses to the current year’s vaccine. Although previous findings suggest that OAS does not occur in response to influenza vaccination [17], it is possible that our method enables identification of OAS-constrained antibodies by characterizing the antibody repertoire to a greater depth than previously attained. Further in-depth studies are necessary for clarifying the contribution of OAS to the antibody response to influenza vaccination.
Several approaches to sequencing the immunoglobulin genes expressed by B cells have been described, but have significant limitations [37, 38]. Deep sequencing of immunoglobulin RNA expressed by bulk B cells does not allow for the accurate pairing of the heavy and light chains expressed by individual B cells [39, 40], while RT-PCR analysis of single B cells is low-throughput. Recently, several high-throughput methods were described for pairing of heavy- and light-chain immunoglobulin genes expressed by individual B cells, these approaches have shortcoming. One approach involves use of mass spectrometry to obtain the amino acid sequences of isolated antibodies, and is used in conjunction with combinatorial expression of heavy and light chain antibody genes and functional screening [40]. Another approach utilizes deep sequencing to analyze the paired CDR3s, but PCR cloning and Sanger sequencing with custom primers is then necessary to isolate the entire heavy and light chain V regions [41]. Further, these methods are unable to accurately assess the frequency of individual antibody clones due to PCR biases. These other methods also cannot readily distinguish between sequencing errors and closely related antibody sequences that differ due to somatic hypermutation that occurs during affinity maturation. Finally, these approaches utilize degenerate V-region PCR primers that fail to amplify all antibody genes and do not provide sequences encoding the entire V regions.
In contrast, the DNA barcoding approach used in this manuscript utilizes a universal 5’ adapter to provide full-length sequences of the V region and unbiased sequencing of antibody repertoires. In addition, the DNA barcodes enable repeat-sequencing of multiple independent barcoded amplicons derived from each individual plasmablast, thereby enabling the resulting sequences to be averaged to yield the true, error-free consensus sequence for the heavy and light chain antibody genes expressed by each plasmablast. Without the use of such barcodes, sequence differences arising from somatic hypermutation cannot be differentiated from artificial mutations arising from PCR or sequencing error, or re-sequencing of previously amplified antibody gene products due to PCR contamination. Finally, our barcoding approach enables accurate determination of clonal family size even in the presence of PCR bias that results in relative over- or under-amplification of the antibody cDNA in certain wells. Specifically, the unique barcodes attached to the heavy and light chain genes expressed by each individual B cell ensure that each antibody derived from an individual B cell is counted once and only once.
Limitations of the present study include the small number of individuals sequenced, the limited depth of sequencing performed, and the limited comparisons made between singleton antibodies and antibodies belonging to clonal families. Further studies will be necessary to fully assess the predictive utility of bioinformatic identification of clonal families in identifying functional antibodies that are key to the protective immune response against influenza.
In conclusion, we used a new DNA barcoding method to characterize the plasmablast antibody response generated in the immune response to influenza vaccination. We show that more than two-thirds of the rationally selected plasmablast antibodies we expressed bind and neutralize influenza, and that antibodies derived from clonal families of antibodies sharing heavy chain V(D)J and light chain VJ sequences do so most effectively. Some vaccine-induced antibodies exhibited higher binding affinities for hemaglutinins derived from prior years’ seasonal influenza as compared to their affinities for the immunization strains, suggesting that ‘original antigenic sin’ in part shapes the B cell and antibody response to influenza vaccination. At the sequence level, high-resolution analysis of the clonality and evolution of the antibody response could be useful for identifying, understanding, and monitoring protective immune responses induced by influenza vaccination and other microbial vaccinations.
Supplementary Material
Highlights.
Deep-sequencing of plasmablast antibody repertoires to influenza vaccination.
Single-cell barcoding enables recovery of natively paired heavy- and light-chains.
Identification of clonal families of antibodies with shared rearrangements.
Antibodies from clonal populations are enriched for binding and neutralization.
Some antibodies induced by influenza vaccination exhibit original antigenic sin.
Acknowledgments
Sequences are available at GenBank (accession numbers KF994033-KF994563).
Funding
This research was supported by NIH R01 AR063676, a NIH NIAID U19 subcontract, and NIH NHLBI Proteomics Center N01-HV-00242 to W.H.R.; and A*STAR NSS (PhD) fellowship to Y-C.T. We thank Drs. Wayne Volkmuth, Tito Serafini, Daniel Emerling, Daniel Lu, Ruben Luo, and Harriet Robinson for useful discussions and input.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Financial Interests
Y-C.T. is an employee of and owns equity in Atreca, Inc.; J.S. and W.H.R. are consultants to and own equity in Atreca, Inc. All other authors have no disclosures.
References
- 1.Molinari NA, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007;25(27):5086–96. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 2.Yewdell JW, et al. Getting to the heart of influenza. Sci Transl Med. 2013;5(191):191ed8.1. doi: 10.1126/scitranslmed.3006735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry JM. Pandemics: avoiding the mistakes of 1918. Nature. 2009;459(7245):324–5. doi: 10.1038/459324a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed R, Oldstone MB, Palese, P. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat Immunol. 2007;8(11):1188–93. doi: 10.1038/ni1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osterholm MT, et al. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(1):36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 6.Goodwin K, Viboud C, Simonsen L. Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine. 2006;24(8):1159–69. doi: 10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- 7.Nabel GJ. Designing tomorrow's vaccines. N Engl J Med. 2013;368(6):551–60. doi: 10.1056/NEJMra1204186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki S, et al. Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies. J Clin Invest. 2011;121(8):3109–19. doi: 10.1172/JCI57834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang N, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med. 2013;5(171):171ra19. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vollmers C, et al. Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proc Natl Acad Sci U S A. 2013;110(33):13463–8. doi: 10.1073/pnas.1312146110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med. 2013;210(8):1493–500. doi: 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li GM, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A. 2012;109(23):9047–52. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu C, et al. Cross-reactive humoral responses to influenza and their implications for a universal vaccine. Ann N Y Acad Sci. 2013 doi: 10.1111/nyas.12012. [DOI] [PubMed] [Google Scholar]
- 14.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208(1):181–93. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ORIGINAL antigenic sin. N Engl J Med. 1958;258(20):1016–7. doi: 10.1056/NEJM195805152582014. [DOI] [PubMed] [Google Scholar]
- 16.Wrammert J, et al. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J Virol. 2012;86(6):2911–8. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fink K. Origin and Function of Circulating Plasmablasts during Acute Viral Infections. Front Immunol. 2012;3:78. doi: 10.3389/fimmu.2012.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radbruch A, et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6(10):741–50. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 20.He XS, et al. Plasmablast-derived polyclonal antibody response after influenza vaccination. J Immunol Methods. 2011;365(1-2):67–75. doi: 10.1016/j.jim.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moir S, Fauci AS. Insights into B cells and HIV-specific B-cell responses in HIV-infected individuals. Immunol Rev. 2013;254(1):207–24. doi: 10.1111/imr.12067. [DOI] [PubMed] [Google Scholar]
- 22.Alamyar E, et al. IMGT/HighV-QUEST: the IMGT(R) web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 2012;8(1):26. [Google Scholar]
- 23.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 25.Huerta-Cepas J, Dopazo J, Gabaldon T. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt WM, Mueller MW. CapSelect: a highly sensitive method for 5' CAP-dependent enrichment of full-length cDNA in PCR-mediated analysis of mRNAs. Nucleic Acids Res. 1999;27(21):e31. doi: 10.1093/nar/27.21.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–57. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 28.McKean D, et al. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 1984;81(10):3180–4. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sablitzky F, Wildner G, Rajewsky K. Somatic mutation and clonal expansion of B cells in an antigen-driven immune response. EMBO J. 1985;4(2):345–50. doi: 10.1002/j.1460-2075.1985.tb03635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Good KL, Tangye SG. Decreased expression of Kruppel-like factors in memory B cells induces the rapid response typical of secondary antibody responses. Proc Natl Acad Sci U S A. 2007;104(33):13420–5. doi: 10.1073/pnas.0703872104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tangye SG, et al. Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J Immunol. 2003;170(2):686–94. doi: 10.4049/jimmunol.170.2.686. [DOI] [PubMed] [Google Scholar]
- 32.Kaur K, Sullivan M, Wilson PC. Targeting B cell responses in universal influenza vaccine design. Trends Immunol. 2011;32(11):524–31. doi: 10.1016/j.it.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labrosse B, et al. Detection of extensive cross-neutralization between pandemic and seasonal A/H1N1 Influenza Viruses using a pseudotype neutralization assay. PLoS One. 2010;5(6):e11036. doi: 10.1371/journal.pone.0011036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei CJ, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329(5995):1060–4. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 35.He XS, et al. Heterovariant cross-reactive B-cell responses induced by the 2009 pandemic influenza virus A subtype H1N1 vaccine. J Infect Dis. 2013;207(2):288–96. doi: 10.1093/infdis/jis664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francis TJ. On the doctrine of original antigenic sin. Proceedings of the American Philosophical Society. 1960;104(6):572–578. [Google Scholar]
- 37.Fischer N. Sequencing antibody repertoires: the next generation. MAbs. 2011;3(1):17–20. doi: 10.4161/mabs.3.1.14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathonet P, Ullman CG. The Application of Next Generation Sequencing to the Understanding of Antibody Repertoires. Front Immunol. 2013;4:265. doi: 10.3389/fimmu.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddy ST, et al. Monoclonal antibodies isolated without screening by analyzing the variablegene repertoire of plasma cells. Nat Biotechnol. 2010;28(9):965–9. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- 40.Cheung WC, et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol. 2012;30(5):447–52. doi: 10.1038/nbt.2167. [DOI] [PubMed] [Google Scholar]
- 41.DeKosky BJ, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31(2):166–9. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.