Abstract
Most tumor-associated antigens (TAA) are self-molecules that are abnormally expressed in cancer cells and become targets of antitumor immune responses. Antibodies and T cells specific for some TAA have been found in healthy individuals and are associated with lowered lifetime risk for developing cancer. Lower risk for cancer has also been associated with a history of febrile viral diseases. We hypothesized that virus infections could lead to transient expression of abnormal forms of self-molecules, some of which are TAA; facilitated by the adjuvant effects of infection and inflammation, these molecules could elicit specific antibodies, T cells and lasting immune memory simultaneously with immunity against viral antigens. Such infection-induced immune memory for TAA would be expected to provide life-long immune surveillance of cancer. Using influenza virus infection in mice as a model system, we tested this hypothesis and demonstrated that influenza-experienced mice control 3LL mouse lung tumor challenge better than infection-naive control mice. Using 2D-Difference Gel Electrophoresis (2D-DIGE) and mass spectrometry, we identified numerous molecules, some of which are known TAA, on the 3LL tumor cells recognized by antibodies elicited by two successive influenza infections. We studied in detail immune responses against GAPDH, Histone H4, HSP90, Malate Dehydrogenase 2 and Annexin A2, all of which were overexpressed in influenza-infected lungs and in tumor cells. Lastly, we show that immune responses generated through vaccination against peptides derived from these antigens correlated with improved tumor control.
Keywords: tumor antigens, cancer vaccines, flu, DIGE
Introduction
Genetic mutations and epigenetic modifications can lead to cellular transformation and cancer (1). Tumor immunosurveillance is the mechanism by which the immune system recognizes and protects against abnormal cells (2). Successful tumor immunosurveillance leads to tumor elimination, which involves the recognition of tumor-specific or tumor-associated antigens (TAA) by antibodies and immune cells. T cells can recognize tumor antigens presented by the major histocompatibility complex (MHC) class I and II molecules and kill the tumor cells via lytic granule release (CD8+ T cells) or promote cellular and humoral responses through the production of cytokines (CD4+ T cells). TAA-specific antibodies can bind to and lyse tumor cells with the help of complement or facilitate the killing of tumor cells by T and natural killer (NK) cells through antibody-dependent cellular cytotoxicity (ADCC) (3, 4). Molecular and biochemical characterization of tumor antigens have yielded targets for immunosurveillance and for the development of immunotherapeutic strategies.
Cancer patients have circulating tumor-specific antibodies and T cells that have been used as reagents to characterize the individual's tumor antigens (5-8). Immune responses to several TAA have been correlated with favorable prognoses. Even when target antigens are not known, infiltration of tumors by activated T cells has been correlated with better prognosis and longer disease-free and overall survival (9). The promising new approaches in cancer treatment include immunotherapies that are directed towards regaining immune control by targeting both the cancer and the immune system (10).
DNA sequencing has shown that tumors have on average a dozen or more mutations that could generate new epitopes known as tumor-specific antigens (11). While tumors could express these epitopes as targets and adoptive transfer of T cells or antibodies could lead to their recognition and elimination, spontaneous immune responses to such epitopes (e.g. mutated Kras, EGFR or p53) have not been found in cancer patients as often as could be expected from the frequency of these mutations (12). Instead, the majority of the spontaneous antitumor immune responses are directed against the non-mutated self-antigens, which are expressed on tumor cells and are named TAA. They include molecules that are overexpressed on tumor cells [e.g. Her-2neu (13), MUC1 (14), CEA (15), Cyclin B1 (5)], molecules with dysregulated stage- or tissue-specific expression [e.g. oncofetal antigens α-fetoprotein (16), cancer-testis antigens NY-ESO-1 (17), Mage 1-7 (18)], or molecules with altered posttranslational modifications (glycosylation or phosphorylation)[e.g. hypoglycosylation of MUC1 (14) or aberrant phosphorylation of β-catenin (19)]. The aberrant expression of many of these antigens can be detected on premalignant precursors of various cancers early in tumor development (6, 20).
Results from several large epidemiological studies have indicated that individuals with a history of febrile childhood infections had a reduced life-time risk of various cancers (21-23). The mechanisms underlying this protective function are unknown. Healthy individuals with no previous history of cancer have been shown to have antibodies and/or T cells specific for several TAA (24, 25). For example, we found that the TAA MUC1 was expressed in the tumor form (overexpressed and hypoglycosylated) on salivary gland ducts during mumps parotitis infection (26), on breast ducts during lactation and in lactational mastitis (27), and in inflammatory bowel disease (20). Furthermore, we showed that the presence of anti-MUC1 IgG in women, who experienced early in life one or more of these events, correlated with a significantly lower risk for ovarian cancer (28).
In this study we present the first attempt to recapitulate these observations in an animal model. This experimental mouse model of influenza infection allows us to test the hypothesis that immunity and immune memory against abnormal self-antigens, known as TAA, is not elicited in response to their de novo expression on tumor cells or premalignant lesions, but rather it is elicited earlier in life in response to their expression during acute inflammations accompanying viral and other infections. When some of the same self-antigens are aberrantly expressed on premalignant lesions or tumor cells, they can be recognized by the infection-primed immune memory responses leading to tumor elimination or enhanced tumor control. We show that mice, which experienced two infections with two different influenza viruses, and which develop immunity to self-antigens abnormally expressed on infected lungs, have improved ability to control the growth of transplantable lung tumors expressing those same self-antigens. We analyzed in detail the infection-elicited immune responses to five such antigens: Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Histone H4, Malate Dehydrogenase 2 (MDH2), Annexin A2, and Heat Shock Protein 90 (HSP90). These antigens were all recognized in tumor cell lysates by post-infection sera. We show that they were overexpressed in tumor cells, as well as in influenza virus-infected lungs compared to healthy lungs, and that influenza virus infection induced antibody and CD8+T cells specific for these antigens. We demonstrate that immunization of mice with peptides derived from these antigens effectively protects them against tumor challenge.
Materials and Methods
Mice, tumor cell lines, and influenza virus
6-8 week old female C57BL/6 wildtype (WT) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and maintained in the University of Pittsburgh Animal Facility. All animal protocols were in accordance with IUCAC guidelines at the University of Pittsburgh. Lewis Lung Carcinoma cell line (3LL) derived from a murine lung epithelial tumor, was maintained in c-DMEM media containing 10% heat inactivated fetal calf serum (FCS), 1% Non-essential Amino Acid, 1% Penicillin/Streptomycin, 1% Sodium Pyruvate, 1% L-glutamine, 0.1% 2-mercaptoethanol. IG10, an epithelial tumor cell line derived from mouse ovarian epithelium, was cultured as described (29).
Influenza Virus Infection and Tumor Challenge
All mice were anesthetized with Ketamine (100mg/mL)/Xylazine (20mg/mL) solution. Mice were infected intranasally with 1.25x103 pfu of H1N1 Influenza A/Puerto Rico/8/34 (PR8) virus and re-infected 35 days later with 1.25x103 pfu of H3N2 Influenza A/Aichi/2/68 (Aichi) X-31 virus. Percent weight loss was used as a measure of successful infection, and mice were weighed at two-day intervals. On day 60 following the first infection, mice were injected subcutaneously in the right hind flank with 1x105 3LL tumor cells. Tumor length and width were measured every 2 days using calipers. Mice were sacrificed when the tumor diameter reached 20 mm, or the tumors became severely ulcerated, or otherwise advised by the University of Pittsburgh animal facility.
Staining of tumor cells with pre- and post-infection sera
Four days prior to primary influenza infection, mice were bled to obtain their pre-infection sera antibody repertoire. Ten days following the second infection, mice were bled to obtain post-infection sera antibodies. Prior to staining, both sets of sera were diluted 1:62.5 in PBS. 2x105 3LL and IG10 tumor cells were plated in a 96-well plate and stained on ice for 1 hour with 100uL of the pre- or post-infection sera. Cells were then stained on ice for 30 minutes with FITC-conjugated Rat anti-mouse IgG2a (BD Bioscience) as the secondary antibody. Cells were fixed in 1.6% paraformaldehyde and samples were run on a LSRII flow cytometer.
Affinity purification of 3LL antigens
Total cell lysates were generated from 50x106 3LL cells in 300uL NP-40 lysis buffer (0.5% NP40, 0.5% Mega 9 (octylglucoside), 150 mM NaCl, 5 mM EDTA, 50 mM Tris pH 7.5, 2 mM PMSF, 5 mM iodoacetamide, and Protease Inhibitor (Roche)). Lysates were pre-cleared with the addition of Protein G Sepharose beads (Sigma-Aldrich, Inc, St. Louis, MO) and the mixture incubated for 1 hour at 4° C on an orbital shaker. Protein G beads were removed by centrifugation at 1200 rpm prior to affinity purification. Protein G HP Spin Trap Columns and Buffer Kits (GE Healthcare UK) were used following the manufacturer's protocol with the following modifications. Pre-infection and post-infection sera were pooled separately from mice (n=6) and each set of sera was poured over multiple protein G columns (100 µL per column). Columns were washed with the wash buffer and 50 mM dimethyl pimelimidate dihydrochloride (DMP) was added to covalently cross-link the bound antibodies from each set of sera to the protein G columns, as described in the manufacturer's protocol. This was done to ensure that only the bound protein fractions were eluted from the columns and not the antibodies. 400 µL of 3LL tumor lysate was then added to both the pre- and post-infection sera columns and incubated overnight at 4° C on an orbital shaker. The following day the columns were washed with TBS (50 mM Tris, 150 mM NaCl, pH 7.5) and the bound proteins were eluted off the columns with 0.1 M glycine provided from the kit with 2 M urea, pH 2.9. Pooled proteins from the pre-infection and post-infection antibody columns were concentrated and the elution buffers were changed to 2D-gel buffer (7 M urea, 2 M thiourea, 4% CHAPS, 10 mM DTT, 10m M HEPES, pH 8.0) using 5000 MWCO Vivaspin columns (Sartorious Stedim Biotech, Goettingen, Germany).
2D-DIGE and Liquid Chromatography/Mass Spectrometry (LC/MS) analysis
The immunoprecipitated proteins were subjected to Difference Gel Electrophoresis (DIGE) (30) to identify proteins largely or uniquely precipitated by post-infection sera. Protein labeling, Isoelectric Point Focusing (IEF), and 2nd dimension SDS-PAGE were conducted as described (31) with the following modifications. 2.5µg of pre-infection proteins and post-infection proteins were reduced in 10 mM Tris(2-carboxyethyl)phosphine (TCEP, Sigma) for 60 minutes in the dark at 37°C. 10 mM CyDye DIGE Fluor Cy3 or Cy5-maleimide saturation dyes (GE Healthcare, Uppsala, Sweden) diluted in Dimethylformamide (DMF) (Sigma), which label all available TCEP reduced cysteines on all proteins, were added to each sample for 30 minutes at 37° C. Labeling was quenched with 7M Dithiothreitol (DTT). Samples were then combined and immobilized pH gradient (IPG) buffer (GE Healthcare) was added at 1µL/40µL of sample. Labeling of the two samples was reversed (reciprocal labeling) and run concurrently on a 2nd gel to eliminate dye-dependent differences. Proteins were separated in the 1st dimension on 13cm pH3-10NL IPG strips on an IPGphor apparatus (GE Healthcare) for 35000 Volt-hours. The samples were then separated on the 2nd dimension SDS-PAGE in pre-cast 10-20% gradient polyacrylamide gels encased in low fluorescent glass (www.precastgels.com) in standard Tris-Glycine-SDS running buffer. Fluorescent images of reciprocal gels were taken as described (31). The Bioinformatics Analysis Core of the University of Pittsburgh Genomics and Proteomics Core Laboratories analyzed the resultant fluorescent images and selected spots that were then cut from the gels and identified via Nano LC-ESI-MS/MS, as described (32).
Western blot and densitometry analysis
Lung tissues were homogenized with a 2 mL dounce homogenizer and total lysates were obtained in NP-40 lysis buffer. The same procedure was applied to generate total cell lysates from 3LL and IG-10 tumor cells. Prior to Western blotting, protein concentrations were determined via Bradford assay. 50 µg of protein from various groups were separated on 10% TGX pre-cast gels (Bio-Rad) and immunoblotted onto PVDF membranes. The following antibodies were used to probe for their respective proteins on separate blots: anti-HSP 90α/β (1:100, Santa Cruz Biotechnology, Santa Cruz, CA), anti-Annexin II (1:100, Santa Cruz Biotechnology, Santa Cruz, CA), anti-Histone H4 (1:1000, Abcam, Cambridge, MA), anti-GAPDH (1:1000, Abcam, Cambridge, MA), anti-Malate Dehydrogenase 2 (1:100, Abcam, Cambridge, MA), anti-Actin (1:15000, Sigma-Aldrich, Inc, St. Louis, MO), goat anti-mouse HRP (1:5000, Santa Cruz Biotechnology, Santa Cruz, CA), goat anti-rabbit HRP (1:5000, Santa Cruz Biotechnology, Santa Cruz, CA). All Western blots were scanned on Kodak Image Station 4000MM and band densitometry analysis was performed on all blots using Image J (NIH, Bethesda, MD). All bands were normalized according to their actin control. Once normalized, all experimental bands and lanes were compared to a normal uninfected mouse lung.
ELISA
15 μg/mL of one of the following proteins were coated on Immulon 4HBX ELISA plates (Thermo scientific) in duplicate wells to examine the differences in antibody recognition between pre- and post-infection sera: MDH2 (Novus Biologicals, Littleton, CO), GAPDH (Abcam, Cambridge, MA), Histone H4 (New England Biolabs, Ipswich, MA), HSP90a (Abcam, Cambridge, MA), Annexin A2 (Novoprotein, Summit, NJ). Human proteins were used due to their high conservation between mouse and human. Duplicate wells that were not coated with antigen served as controls for non-specific binding. Plates were then placed on an orbital shaker overnight at 4° C. The next day pre- and post-infection sera were diluted (1:62.5) in PBS, added to ELISA plates, and placed on an orbital shaker for 2 hours at room temperature. Plates were washed and 0.3% Hydrogen Peroxide was added to the wells to block background peroxidase activity. Plates were washed and rat anti-mouse IgG-HRP (1:500) was added. Plates were again washed and TMB substrate was added for 15 minutes and 2N Sulfuric Acid was added to stop the developing signal. ELISA plates were then read at 450 nm on a Gen 5 plate reader. Data were represented using the average of duplicate antigen-coated wells after subtracting the value from the no antigen control wells.
Peptide identification and MHC-I binding assays
Candidate peptide sequences were identified using the Immune Epitope Database (IEDB) MHC-I binding predictor program with a percentile rank of 5 or less (33). MDH251-260 (MAYAGARFVF), GAPDH300-310 (ALNDNFVKLIS), Annexin A2184-191 (SVIDYELI), H487-95 (VVYALKRQG), Histone H4 with an amino acid substitution (H4-sub VVYAFKRQG) peptides were synthesized by the Peptide Synthesis Core of the University of Pittsburgh Genomics and Proteomics Core Laboratories as described (34). MHC-I binding was verified performing RMA-S, a TAP-deficient cell line, MHC-Class I stabilization assays. In short, 2x105 RMA-S cells were plated in a 96-well plate. The cells were cultured overnight at 29° C. Unloaded RMA-S cells served as controls. Each peptide candidate was added in triplicates to the plate from 10-4 M to 10-7 M for 2 hours and 30 minutes in a 29° C incubator. Cells were placed in a 37° C incubator for 1 hour and 30 minutes. RMA-S cells were fixed in 1.6% Paraformaldehyde and then stained with anti-H2-Kb or anti-H2-Db (BD Bioscience). Samples were run on the LSR II Flow Cytometer (BD Bioscience) and analyzed using FACS Diva Software (Supplementary Figure S1).
Antigen-specific T cell detection
Animals were sacrificed six days following the second influenza infection. Lungs and spleens were harvested and cells isolated as described (35). 1x106 cells from each set of tissues were stained with anti-CD3, anti-CD4 and anti-CD8 antibodies (BD Bioscience). Dimer-X soluble dimeric mouse H-2Kb:Ig Fusion Protein and H-2Db:Ig Fusion Protein (BD Bioscience) were used according to the manufacturer's protocol to detect MDH251-260, GAPDH300-310, Annexin A2184-191 and H487-95 peptide-specific CD8+ T cells. In addition, the presence of flu-specific T cells was evaluated using peptides PA224-233 and NP147-155 purchased from GenScript USA, Piscataway, NJ. 100,000 events were collected and samples were run on the LSR II Flow Cytometer (BD Bioscience), gated (example in Supplementary Figure S2) and analyzed using FACS Diva Software.
Vaccination and Tumor Challenge
D1 dendritic cells (DC) are an established growth factor-dependent immature dendritic cell line. D1 DC were grown and maintained as described and used in all vaccinations (36). 1.25x106 D1 DCs/ mouse were cultured in 6-well plates, loaded separately with 100 μg of MDH251-260, GAPDH300-310, Annexin A2184-191, or H487-95, and matured with 12.5 μg/mL of Poly IC:LC adjuvant. 0.25x106 D1 DCs loaded with individual peptides were pooled together for a total number of 1x106D1 DCs. An additional 50 µg of each soluble peptide per mouse was added to the mixture and injected into mice in the right hind flank. Unloaded, Poly IC:LC matured D1 DCs were injected into control mice. Animals were vaccinated at weeks 0, 2, and 6, and challenged with 1x105 3LL cells in the right hind flank two days following the week 6 vaccination. Tumor length and width were measured at two-day intervals using calipers.
Data Analysis
Statistical analysis was performed using GraphPad Prism v6.0 software (GraphPad Inc. San Diego, CA). Results were represented as means ± standard error of the mean (SEM). Statistical means and significance were analyzed using unpaired two-tailed student's t test. Kaplan-Meier survival curves were analyzed with the log rank test. Significance for all experiments was defined as the following: * p<0.05, ** p<0.01, *** p<0.001.
Results
Influenza virus infection induces antibodies to multiple host cell antigens, some of which are known TAA
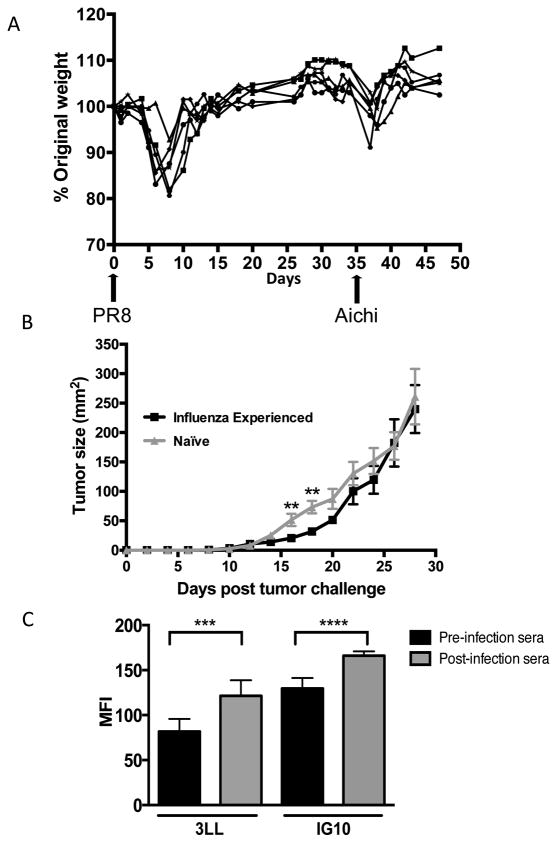
Mice were infected with influenza virus PR8 and then 35 days later with the second influenza strain Aichi, as described in Materials and Methods. Mice experienced the general signs of malaise and lost close to 20% of their starting body weight in the first week following each infection. By 7-8 days post infection, mice began to recover and by days 16-18 their weight returned to baseline (Figure 1A). Day 25 post second infection, animals were injected subcutaneously with 3LL tumor cells. Tumors became palpable eight days post injection (Figure 1B). At day 14, tumor growth kinetics between the influenza-experienced animals and the naïve mock-infected group began to diverge. Tumors in the influenza-experienced animals grew slower from days 14 to 22. On day 16 the average tumor size in the influenza-experienced mice was 20.67 mm2 versus 51.63 mm2 in the control group. The size difference was still significant at day 18 when the average tumor size in the influenza-experienced group was 31.78 mm2 compared to 73.25 mm2 in the control mice.
Figure 1. Influenza virus infection delays tumor growth at early time points.
(A) Animals were intranasally infected with PR8 and Aichi influenza viruses on day 0 and day 35, respectively. Mice were weighed every two days. (B) Influenza-experienced animals and naïve animals were challenged subcutaneously with 1x105 3LL tumor cells in the right hind flank. Tumor length and width were measured every two days using calipers. Data are representative of two experiments with at least 8 mice per group and are expressed as means ± SEM. (C) Sera from animals (n=7) four days prior to PR8 infection (pre-infection) and ten days after Aichi infection (post-infection) were diluted 1:62.5 and used to stain 3LL and IG10 tumor cells. Cells were subsequently stained with FITC-conjugated goat anti-mouse IgG2a antibody and analyzed on the LSRII flow cytometer. Results are shown as Mean Fluorescent Intensity (MFI). * p<0.05, ** p<0.01, *** p<0.001
We examined the post-infection sera for the ability to stain 3LL tumor cells, which would suggest that flu infections elicited antibodies against tumor cell surface proteins. In the experiment illustrated in Figure 1C, we obtained sera from mice (n=7) pre- and post-influenza infection. Tumor cell surface staining was performed with individual sera and the results were pooled into pre- and post-infection groups and represented as an average value. Average Mean Fluorescence Intensity (MFI) of staining of 3LL tumor cells with post-infection sera was significantly higher compared to that with pre-infection sera. The same result was obtained staining another mouse epithelial tumor cell line IG10 with the same sera.
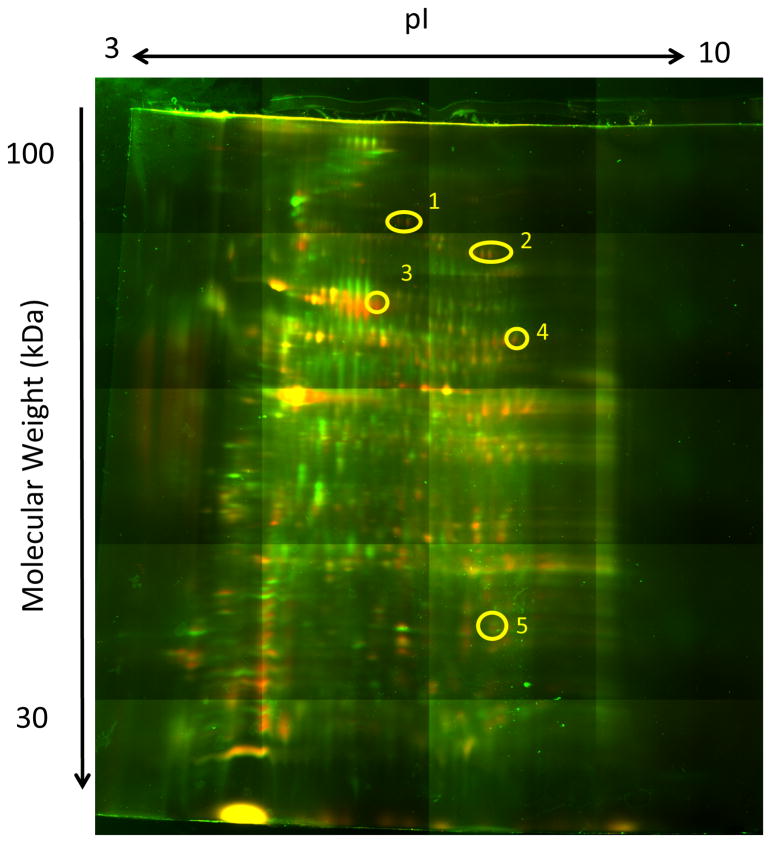
To identify molecules specifically recognized by post-infection sera, pre- and post-infection antibodies were bound to Protein G columns for affinity purification of proteins from 3LL tumor cell lysates. Tumor proteins bound to the antibody columns were eluted, labeled with two different Cyanine-based saturation dyes and resolved by 2D-DIGE as described in Materials and Methods. In a concurrently-run reciprocal gel, labeling was reversed such that the sample previously labeled with Cy3 was labeled with Cy5 and vice versa. Gel images were false-colored for analysis: green for Cy3, red for Cy5. Overlays were then created using Delta2D software, where proteins unique to one sample appeared either green or red, while proteins common to both samples appeared yellow (green and red combined). Figure 2 shows the gel used to identify antigens described below. Green dots mark proteins eluted from the pre-infection antibody columns while red dots mark proteins eluted from the post-infection antibody columns. Spots that were most remarkable (yellow circles) were cut out and subjected to mass spectrometry analysis and protein sequencing. Many proteins with a wide variety of functions were identified. They included voltage-dependent ion channels, proteasome subunits, mitochondrial and cytosolic enzymes, heat shock proteins and structural proteins (data not shown). We selected five identified proteins: Histone H4, Malate Dehydrogenase 2 (MDH2), Annexin A2, Glyceraldhyde-3-Phosphate Dehydrogenase (GAPDH) and Heat Shock Protein 90 (HSP90) for further study (Table 1). Several reports based on methods and approaches unrelated to ours had already identified these molecules as TAA in tumor-bearing mice or in cancer patients (37-41).
Figure 2. Influenza infection induces antibodies against specific proteins of tumor cells.
3LL tumor lysates were affinity purified on protein G columns to which post-infection and pre-infection antibodies were covalently bound and the eluted proteins compared by 2D-DIGE gels. Green spots mark proteins eluted from pre-infection antibody columns and red spots mark proteins from the post-infection antibody columns. Yellow color marks overlapping spots. Yellow circles indicate spots picked for sequencing. Spots were chosen according to image analysis provided by the University of Pittsburgh Bioinformatics Analysis Core (BAC) of the Genomics and Proteomics Core Laboratories.
Table 1. Biochemical characterization of selected tumor antigens detected by post infection antibodies.
| Spot Number | Accession Number | Protein Name | Peptide Sequence | X-Corr | Δ Score | Charge | M/Z[Da] | MH+[Da] | ΔM [ppm] | MW [kDa] | pI | % Sequence Coverage | Ions Matched |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | IP100885367.1 | Annexin A2 | TNQELQEINR QDIAFAYQR |
2.74 2.55 |
0.36 1.00 |
2 | 622.81384 556.28143 |
1244.62041 1111.55559 |
-2.08 2.22 |
19.6 | 5.96 | 10.80 | 14/18 14/16 |
| 3 | IPI00468203.3 | Annexin A2 | TPAQYDASELK AEDGSVIDYELIDQDAR SLYYYIQQDTK TNQELQEINR WISIMTER DIISDTSGDFR QDIAFAYQR |
3.52 3.46 3.30 3.08 2.67 2.53 2.41 |
0.65 0.68 0.47 0.47 1.00 1.00 0.60 |
2 2 2 2 2 2 2 |
611.80090 954.94586 711.35215 622.81384 518.26892 613.28820 556.28027 |
1222.59453 1908.88445 1421.69704 1244.62041 1035.53057 1225.56914 1111.55327 |
-0.43 1.16 1.62 -2.08 1.29 -0.36 0.13 |
38.7 | 7.69 | 22.71 | 15/20 17/32 16/20 15/18/ 13/14 16/20 15/16 |
| 4 | IPI00407339.7 | Histone H4 | ISGLIYEETR TVTAMDVVYALK TVTAmDVVYALK DAVTYTEHAK VFLENVIR |
3.39 3.22 2.88 2.61 2.56 |
0.68 0.74 0.42 0.52 0.36 |
2 2 2 2 2 |
590.81536 655.85559 663.85327 567.77410 495.29345 |
1180.62346 1310.70391 1326.69927 1134.54094 989.57964 |
2.19 1.06 1.39 -1.47 1.76 |
11.4 | 11.36 | 38.83 | 16/18 16/22 14/22 15/18 12/14 |
| 4 | IPI00622795.2 | GAPDH | LENPAKYDDIK | 2.36 | 8.03 | 2 | 653.33881 | 1305.67035 | 0.20 | 35.8 | 8.03 | 3.30 | 10/20 |
| 4 | IPI00323592.2 | Malale Dehydrogenase 2 | IFGVTTLDIVR | 2.82 | 0.41 | 2 | 617.36487 | 1233.72246 | 0.41 | 35.6 | 8.68 | 3.25 | 14/20 |
| 5 | IPI00885367.1 | Annexin A2 | TNQELQEINR | 3.06 | 0.47 | 2 | 622.81384 | 1244.62041 | -2.08 | 19.6 | 5.96 | 5.68 | 14/18 |
Protein Accessions- Protein Accessions in IPI mouse database
Protein- Protein name
Peptide sequences- Primary amino acid sequence for identified peptides
Xcorr- XCorr is the cross-correlation of the experimental and theoretical spectra from SEQUEST search
Δ Score- A measure of the difference between the top two scores for the peptides identified by that spectrum
Charge- Charge stage of precursor ion
M/Z [Da]- Observed mass to charge ratio of precursor ion
MH+ [Da]- Monoisotopic mass for protonated molecular ion MH calculated based on observed m/z
ΔM [ppm]- Deviation from expected MH
MW-Molecular Weight
pi- Isoelectric Point
% Sequence Coverage- percentage of the database protein sequence covered by matching peptides
Ions Matched- The number of fragment ions matched out of the total number of fragment masses for the peptide
Overexpression of GAPDH, Histone H4, MDH2, Annexin A2, and HSP90 in tumor cells and influenza virus-infected lungs leads to specific immunity
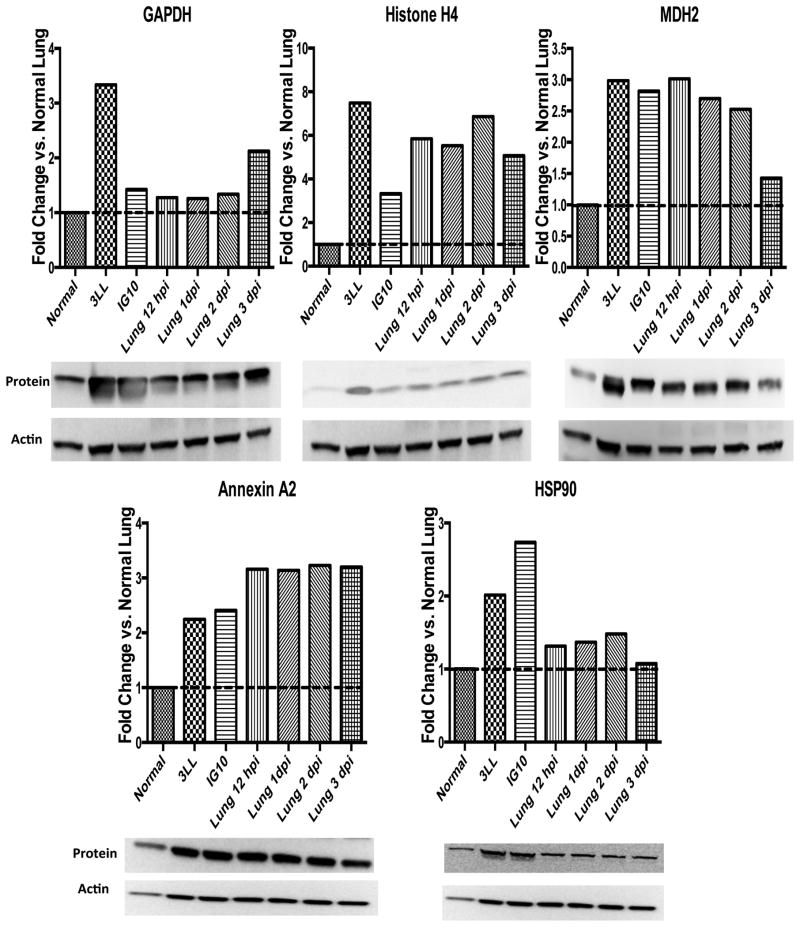
We hypothesized that post-infection antibodies against these proteins were elicited due to differences in their expression in infected versus normal lungs, analogous to their abnormal expression in tumors. Western blot analysis showed that these proteins were constitutively overexpressed in both epithelial tumor cell lines, 3LL and IG10, and also at various time points in the flu-infected lungs (2-7.5-fold higher compared to healthy lungs) (Figure 3). Expression of GAPDH in influenza-infected lungs appeared to be the highest at day 3 post-infection. Histone H4 protein levels were constitutively elevated in tumor cells and at all time points post flu-infection by 5 to 7-fold higher than in normal lung. MDH2 levels were elevated 12 hours post-infection and decreased to normal levels at day 3 post-infection. Annexin A2 remained 3-fold higher than in normal lung at all time points. HSP90 protein level was the highest at day 2 post-infection.
Figure 3. Histone H4, MDH2, GAPDH and HSP90 are elevated in epithelial tumor cells lines and influenza-infected lungs.
Whole cell lysates were generated from 3LL and IG10 cell lines and from normal and influenza-infected lungs. 50μg of protein was loaded on each gel, resolved by electrophoresis and transferred onto PVDF membranes. Membranes were blotted using antibodies against GAPDH, Histone H4, MDH2, Annexin A2, and HSP90. Actin loading controls were used to normalize each band. Densitometry analysis was performed using ImageJ. All lanes were compared to Normal Lung. Data are representative of two experiments.
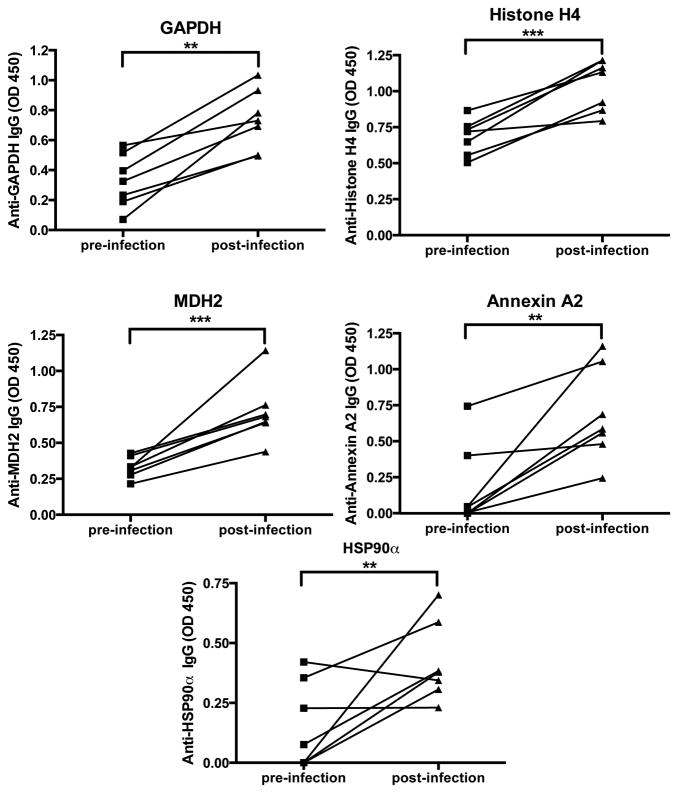
Even though these antigens were identified by affinity purification on post-infection sera bound to Protein G columns, we wanted to confirm in another assay the specificity of post-infection antibodies for each of these individual molecules. Figure 4 shows that in most mice there was an increase post-infection in IgG specific for GAPDH, Histone H4, MDH2, Annexin A2 and HSP90α as determined by ELISA.
Figure 4. Antibodies specific for GAPDH, Histone H4, MDH2, Annexin A2 and HSP90α increase following influenza infection.
Pre-infection sera (four days prior to the first infection) and post-infection sera (ten days post second infection) were assayed on ELISA plates in duplicate wells coated with individual proteins. Uncoated wells served as non-specific binding controls and their values were subtracted from the values in matching antigen-coated wells. Results are represented as mean optical density (OD) ± SEM of two experimental repeats with an n=7.
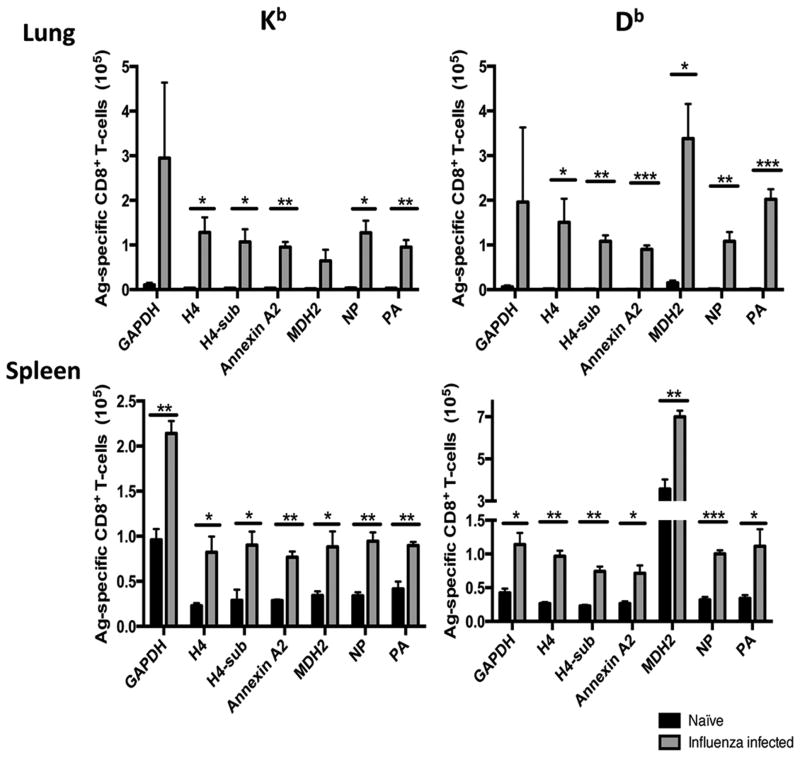
Influenza virus infection also induced an increase in CD8+ T cells specific for all of these antigens. Spleens and lungs were harvested from mice six days after the second influenza infection and from uninfected control mice. Peptides GAPDH300-310 (ALNDNFVKLIS), Annexin A2184-191 (SVIDYELI), MDH251-260 (MAYAGARFVF), Histone H487-95 (VVYALKRQG), Histone H4 with an amino acid substitution (H4-sub VVYAFKRQG) (38), were selected from the Immune Epitope Database (IEDB) and confirmed to bind to MHC-I in RMA-S stabilization assays (supplementary figure 1). Each peptide was loaded onto DimerX H-2Kb or H-2Db molecules and used to detect specific CD3+CD8+ T cells. There were no CD8 T cells specific for these peptides in the lungs and spleens of uninfected control mice but in flu-experienced animals they were present in similar numbers to the flu-specific T cells (Figure 5). The highest numbers both in the lungs and in the spleens were H2-Kb-restricted GAPDH-specific and H2-Db-restricted MDH2-specific T-cells.
Figure 5. Antigen-specific CD8+ T cells increase in the lungs and spleens following influenza infection.
Mice were infected with PR8 and Aichi Influenza viruses or mock PBS infected on day 0 and day 35, respectively. Six days after the second infection spleens and lungs were harvested and analyzed by Flow Cytometry for the presence of T cells staining with the Dimer-X reagent (as described in Methods and Materials). Data are representative of two experimental repeats of least n=3 animals per group and are expressed as means ± SEM.
Vaccination with DC loaded with the new TAA peptides delays tumor growth and promotes survival
We loaded the D1 dendritic cells with MDH2251-260, GAPDH300-310, Annexin A2184-191, and H487-95 and vaccinated mice as described in Materials and Methods. Control mice were vaccinated with unloaded D1 cells. Two days following the second boost, mice were challenged subcutaneously with 3LL tumor cells. In vaccinated animals tumor growth began to slow down significantly by day 12 (Figure 6A) resulting in all vaccinated animals still surviving at day 40, compared to only 2 animals surviving in the unloaded DC vaccinated controls (Figure 6B). On day 12, the average tumor size in peptide-loaded DC-vaccinated animals was 10.67 mm2 compared to 30.67 mm2 for the controls.
Figure 6. Vaccination with GAPDH, Histone H4, MDH2, and Annexin A2 peptides loaded on DC confers early protection and prolonged survival against 3LL tumor challenge.
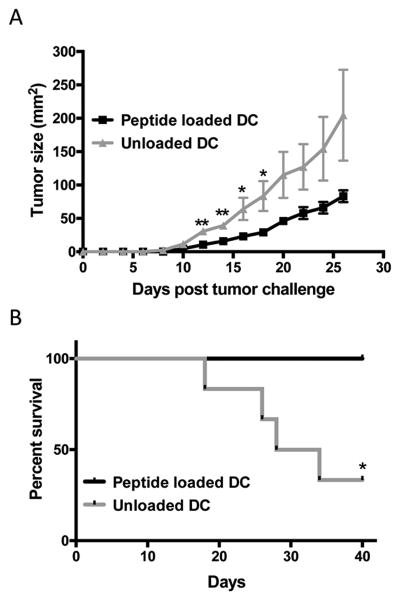
(A) Mice were vaccinated in the right hind flank with 1x106 D1 cells/mouse (a pool of 2.5x105 D1 dendritic cells/peptide) at week 0, week 2, and week 6. Control mice received the same number of unloaded D1 DC at the same time points. Two days after the week 6 boost, animals were challenged subcutaneously with 1x105 3LL tumor cells in the right hind flank. Tumor growth was measured with calipers at two-day intervals and expressed as length x width (mm2). (B) Survival post tumor challenge. Data are representative of two experiments with at least n=8 mice per group and are expressed as means ± SEM.
Discussion
The data we present here, add a new dimension to our understanding of the process of cancer immunosurveillance and its targets. We show that immune responses against abnormally expressed self-antigens, many of which have been characterized as TAAs, are generated during non-malignant infectious inflammatory events that occur much earlier in life than malignancies. We propose that immune memory for these antigens is later recruited for cancer immunosurveillance.
We used a mouse model to demonstrate that two bouts with influenza virus infection led to the ability of the host's immune system to slow down tumor growth. This effect was small and transient, which may be all that could be expected from a limited exposure of the mice to this one type of infection and to no other pathogens prior to influenza infection as the mice are kept in pathogen-free conditions. The significance of this small delay in tumor growth was confirmed by the induction of antibodies against multiple molecules in the tumor cell lysate. Focusing on five antigens identified by infection-elicited antibodies, GAPDH, Histone H4, MDH2, Annexin A2, and HSP90, we showed that these antigens were abnormally expressed (overexpressed) in flu-infected lungs and in mouse epithelial tumor cell lines, and that in addition to IgG the flu-infection induced antigen-specific CD8 T cells against these molecules. As predicted by our hypothesis, vaccination with peptides derived from GAPDH, H4, MDH2, and Annexin A2 led to a much more profound slowing down of tumor growth compared to that elicited by the virus infection, and to a prolonged survival of tumor-bearing animals.
Other viral infections may be capable of inducing TAA specific antibodies and T cells. Vaccinia Virus- (VV) and Lymphocytic Choriomeningitis Virus (LCMV)-infected mice were reported to have developed antibodies against many host cell antigens, some of which are orthologues of human TAAs (42). Human fibroblasts infected with varicella-zoster virus (VZV) or human cytomegalovirus (CMV) overexpress cyclin B1 in the cytoplasm in a similar fashion to tumor cells where cyclin B1 was identified as a TAA (43, 44). Cyclin B1 has been found in VZV virions (45). Many healthy individuals, presumably having experienced these infections, have cyclin B1-specific IgG and memory T cells (24). It has been reported that GAPDH and Annexin A2 are found in influenza virions produced by infected epithelial cell lines Vero and A549 (46). HSP90, Annexin A2, and GAPDH were also found within human CMV particles (47). A study examining the measles virus effect on presentation of self-peptides on MHC class I during infection showed that two abundant self-peptides on HLA-A*0201 measles-infected cells could induce auto-reactive CD8+ T cells. One of the peptides identified was HSP90β570-578 (ILDKKVEKV)(48). The HSP90β570-578 peptide has been found in melanoma cell lines (49). It is possible that these “auto-reactive” T cells contribute to increased tumor immune surveillance. None of these observations were followed by experiments to test the potential antitumor effects of either the viral infections or the immune responses against the identified molecules, with the exception of cyclin B1 that we showed was a target of antitumor immune responses (24).
The same protective effect of influenza virus-primed immunity specific for abnormally expressed self-antigens that we showed here, could be a collateral benefit of other viral, bacterial and parasitic infection or various acute inflammatory conditions of unknown etiologies. Therefore, we propose that the molecules abnormally expressed in these different disease states and also in cancer cells, that are currently referred to as TAAs, should be renamed disease-associated antigens (DAA) (50). Pre-existing immune responses to several known tumor antigens that are candidate DAAs have been reported to increase the odds of successfully eliminating spontaneously arising tumors (28). The arrival of memory DAA-specific T cells to the site of the tumor as a secondary immune response could promote priming of tumor-specific responses directed against individual mutations and epitope-spreading, adding to the efficacy of immunosurveillance. If DAA-specific immune memory is lacking or weak due to limited early exposures to infections, this may lead to establishment of chronic inflammation at the tumor site due to unopposed innate immune responses, which is likely to promote tumor development. Therefore, better understanding of the events early in life that prepare the immune system to protect an individual against known and unknown pathogens as well as future malignancies, will help direct vaccines and other immune manipulation towards strengthening rather than impairing the establishment of life-long immunosurveillance. In addition, these findings support the use of vaccines based on DAAs/TAAs for cancer prevention.
Supplementary Material
Acknowledgments
We would like to thank the University of Pittsburgh Genomics and Proteomics Core Laboratories for their assistance. This work used the Biomedical Mass Spectrometry Center and UPCI Cancer Biomarker Facility that are supported in part by award P30CA047904. This work was supported by the National Institutes of Health (CA056103 to OJF) and (GM083602 to TMR), the National Science Foundation (NSF-IDBR 1063236 to PTV and JSM) and the UNCF/Merck Graduate Student Dissertation Fellowship (UKI).
Bibliography
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 3.Weiner LM, Surana R, Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat Rev Immunol. 2010;10(5):317–27. doi: 10.1038/nri2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemenceau B, Vivien R, Berthome M, Robillard N, Garand R, Gallot G, et al. Effector memory alphabeta T lymphocytes can express FcgammaRIIIa and mediate antibody-dependent cellular cytotoxicity. J Immunol. 2008;180(8):5327–34. doi: 10.4049/jimmunol.180.8.5327. [DOI] [PubMed] [Google Scholar]
- 5.Kao H, Marto JA, Hoffmann TK, Shabanowitz J, Finkelstein SD, Whiteside TL, et al. Identification of cyclin B1 as a shared human epithelial tumor-associated antigen recognized by T cells. J Exp Med. 2001;194(9):1313–23. doi: 10.1084/jem.194.9.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki H, Graziano DF, McKolanis J, Finn OJ. T cell-dependent antibody responses against aberrantly expressed cyclin B1 protein in patients with cancer and premalignant disease. Clin Cancer Res. 2005;11(4):1521–6. doi: 10.1158/1078-0432.CCR-04-0538. [DOI] [PubMed] [Google Scholar]
- 7.Sahin U, Tureci O, Pfreundschuh M. Serological identification of human tumor antigens. Curr Opin Immunol. 1997;9(5):709–16. doi: 10.1016/s0952-7915(97)80053-2. [DOI] [PubMed] [Google Scholar]
- 8.Knuth A, Danowski B, Oettgen HF, Old LJ. T-cell-mediated cytotoxicity against autologous malignant melanoma: analysis with interleukin 2-dependent T-cell cultures. Proc Natl Acad Sci U S A. 1984;81(11):3511–5. doi: 10.1073/pnas.81.11.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pages F, Galon J, Dieu-Nosjean MC, Tartour E, Sautes-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29(8):1093–102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 10.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013 doi: 10.1038/nm.3161. Epub 2013/05/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 14.Vlad AM, Kettel JC, Alajez NM, Carlos CA, Finn OJ. MUC1 immunobiology: from discovery to clinical applications. Adv Immunol. 2004;82:249–93. doi: 10.1016/S0065-2776(04)82006-6. [DOI] [PubMed] [Google Scholar]
- 15.Nollau P, Scheller H, Kona-Horstmann M, Rohde S, Hagenmuller F, Wagener C, et al. Expression of CD66a (human C-CAM) and other members of the carcinoembryonic antigen gene family of adhesion molecules in human colorectal adenomas. Cancer Res. 1997;57(12):2354–7. [PubMed] [Google Scholar]
- 16.Evdokimova VN, Butterfield LH. Alpha-fetoprotein and other tumour-associated antigens for immunotherapy of hepatocellular cancer. Expert Opin Biol Ther. 2008;8(3):325–36. doi: 10.1517/14712598.8.3.325. [DOI] [PubMed] [Google Scholar]
- 17.Jager E, Nagata Y, Gnjatic S, Wada H, Stockert E, Karbach J, et al. Monitoring CD8 T cell responses to NY-ESO-1: correlation of humoral and cellular immune responses. Proc Natl Acad Sci U S A. 2000;97(9):4760–5. doi: 10.1073/pnas.97.9.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254(5038):1643–7. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 19.Depontieu FR, Qian J, Zarling AL, McMiller TL, Salay TM, Norris A, et al. Identification of tumor-associated, MHC class II-restricted phosphopeptides as targets for immunotherapy. Proc Natl Acad Sci U S A. 2009;106(29):12073–8. doi: 10.1073/pnas.0903852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beatty PL, Plevy SE, Sepulveda AR, Finn OJ. Cutting edge: transgenic expression of human MUC1 in IL-10-/- mice accelerates inflammatory bowel disease and progression to colon cancer. J Immunol. 2007;179(2):735–9. doi: 10.4049/jimmunol.179.2.735. [DOI] [PubMed] [Google Scholar]
- 21.Hoption Cann SA, van Netten JP, van Netten C. Acute infections as a means of cancer prevention: opposing effects to chronic infections? Cancer Detect Prev. 2006;30(1):83–93. doi: 10.1016/j.cdp.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Kolmel KF, Pfahlberg A, Mastrangelo G, Niin M, Botev IN, Seebacher C, et al. Infections and melanoma risk: results of a multicentre EORTC case-control study. European Organization for Research and Treatment of Cancer. Melanoma Res. 1999;9(5):511–9. [PubMed] [Google Scholar]
- 23.Urayama KY, Ma X, Selvin S, Metayer C, Chokkalingam AP, Wiemels JL, et al. Early life exposure to infections and risk of childhood acute lymphoblastic leukemia. Int J Cancer. 2011;128(7):1632–43. doi: 10.1002/ijc.25752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vella LA, Yu M, Fuhrmann SR, El-Amine M, Epperson DE, Finn OJ. Healthy individuals have T-cell and antibody responses to the tumor antigen cyclin B1 that when elicited in mice protect from cancer. Proc Natl Acad Sci U S A. 2009;106(33):14010–5. doi: 10.1073/pnas.0903225106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erfurt C, Sun Z, Haendle I, Schuler-Thurner B, Heirman C, Thielemans K, et al. Tumor-reactive CD4+ T cell responses to the melanoma-associated chondroitin sulphate proteoglycan in melanoma patients and healthy individuals in the absence of autoimmunity. J Immunol. 2007;178(12):7703–9. doi: 10.4049/jimmunol.178.12.7703. Epub 2007/06/06. [DOI] [PubMed] [Google Scholar]
- 26.Cramer DW, Vitonis AF, Pinheiro SP, McKolanis JR, Fichorova RN, Brown KE, et al. Mumps and ovarian cancer: modern interpretation of an historic association. Cancer Causes Control. 2010;21(8):1193–201. doi: 10.1007/s10552-010-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jerome KR, Kirk AD, Pecher G, Ferguson WW, Finn OJ. A survivor of breast cancer with immunity to MUC-1 mucin, and lactational mastitis. Cancer Immunol Immunother. 1997;43(6):355–60. doi: 10.1007/s002620050344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cramer DW, Finn OJ. Epidemiologic perspective on immune-surveillance in cancer. Curr Opin Immunol. 2011;23(2):265–71. doi: 10.1016/j.coi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Budiu RA, Elishaev E, Brozick J, Lee M, Edwards RP, Kalinski P, et al. Immunobiology of human mucin 1 in a preclinical ovarian tumor model. Oncogene. 2012 doi: 10.1038/onc.2012.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18(11):2071–7. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 31.Minden JS. Two-dimensional difference gel electrophoresis. Methods Mol Biol. 2012;869:287–304. doi: 10.1007/978-1-61779-821-4_24. [DOI] [PubMed] [Google Scholar]
- 32.Balasubramani M, Nakao C, Uechi GT, Cardamone J, Kamath K, Leslie KL, et al. Characterization and detection of cellular and proteomic alterations in stable stathmin-overexpressing, taxol-resistant BT549 breast cancer cells using offgel IEF/PAGE difference gel electrophoresis. Mutat Res. 2011;722(2):154–64. doi: 10.1016/j.mrgentox.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, et al. The immune epitope database 2.0. Nucleic Acids Res. 2010;38(Database issue):D854–62. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner MS, Cohen PA, Finn OJ. Lack of effective MUC1 tumor antigen-specific immunity in MUC1-transgenic mice results from a Th/T regulatory cell imbalance that can be corrected by adoptive transfer of wild-type Th cells. J Immunol. 2007;178(5):2787–93. doi: 10.4049/jimmunol.178.5.2787. [DOI] [PubMed] [Google Scholar]
- 35.Schneider-Ohrum K, Giles BM, Weirback HK, Williams BL, DeAlmeida DR, Ross TM. Adjuvants that stimulate TLR3 or NLPR3 pathways enhance the efficiency of influenza virus-like particle vaccines in aged mice. Vaccine. 2011;29(48):9081–92. doi: 10.1016/j.vaccine.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winzler C, Rovere P, Rescigno M, Granucci F, Penna G, Adorini L, et al. Maturation Stages of Mouse Dendritic Cells in Growth Factor–dependent Long-Term Cultures. The Journal of Experimental Medicine. 1997;185(2):317–28. doi: 10.1084/jem.185.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schek N, Hall BL, Finn OJ. Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in human pancreatic adenocarcinoma. Cancer Res. 1988;48(22):6354–9. [PubMed] [Google Scholar]
- 38.Savage PA, Vosseller K, Kang C, Larimore K, Riedel E, Wojnoonski K, et al. Recognition of a ubiquitous self antigen by prostate cancer-infiltrating CD8+ T lymphocytes. Science. 2008;319(5860):215–20. doi: 10.1126/science.1148886. [DOI] [PubMed] [Google Scholar]
- 39.Lokman NA, Ween MP, Oehler MK, Ricciardelli C. The role of annexin A2 in tumorigenesis and cancer progression. Cancer Microenviron. 2011;4(2):199–208. doi: 10.1007/s12307-011-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pick E, Kluger Y, Giltnane JM, Moeder C, Camp RL, Rimm DL, et al. High HSP90 expression is associated with decreased survival in breast cancer. Cancer Res. 2007;67(7):2932–7. doi: 10.1158/0008-5472.CAN-06-4511. [DOI] [PubMed] [Google Scholar]
- 41.Liu Q, Harvey CT, Geng H, Xue C, Chen V, Beer TM, et al. Malate dehydrogenase 2 confers docetaxel resistance via regulations of JNK signaling and oxidative metabolism. Prostate. 2013 doi: 10.1002/pros.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludewig B, Krebs P, Metters H, Tatzel J, Tureci O, Sahin U. Molecular characterization of virus-induced autoantibody responses. J Exp Med. 2004;200(5):637–46. doi: 10.1084/jem.20040358. jem.20040358 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez V, McElroy AK, Spector DH. Mechanisms governing maintenance of Cdk1/cyclin B1 kinase activity in cells infected with human cytomegalovirus. J Virol. 2003;77(24):13214–24. doi: 10.1128/JVI.77.24.13214-13224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leisenfelder SA, Moffat JF. Varicella-zoster virus infection of human foreskin fibroblast cells results in atypical cyclin expression and cyclin-dependent kinase activity. J Virol. 2006;80(11):5577–87. doi: 10.1128/JVI.00163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leisenfelder SA, Kinchington PR, Moffat JF. Cyclin-dependent kinase 1/cyclin B1 phosphorylates varicella-zoster virus IE62 and is incorporated into virions. J Virol. 2008;82(24):12116–25. doi: 10.1128/JVI.00153-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008;4(6):e1000085. doi: 10.1371/journal.ppat.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol. 2004;78(20):10960–6. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herberts C, van Gaans-van den Brink J, van der Heeft E, van Wijk M, Hoekman J, Jaye A, et al. Autoreactivity against induced or upregulated abundant self-peptides in HLA-A*0201 following measles virus infection. Human immunology. 2003;64(1):44–55. doi: 10.1016/s0198-8859(02)00707-3. [DOI] [PubMed] [Google Scholar]
- 49.Jarmalavicius S, Welte Y, Walden P. High immunogenicity of the human leukocyte antigen peptidomes of melanoma tumor cells. The Journal of biological chemistry. 2012;287(40):33401–11. doi: 10.1074/jbc.M112.358903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finn OJ. Immunological weapons acquired early in life win battles with cancer late in life. J Immunol. 2008;181(3):1589–92. doi: 10.4049/jimmunol.181.3.1589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.