Abstract
To study the effect of slow termination on the protein synthesizing machinery, we isolated suppressors to a temperature-sensitive release factor 1 (RF1). Of 26 independent clones, five complementation groups have been identified, two of which are presented here. The first mutation disrupts a base pair in the transcription terminator stem for the rplM-rpsI operon, which encodes ribosomal proteins L13 and S9. We have found that this leads to readthrough of the terminator and that lower levels of transcript (compared to the results seen with the wild type) are found in the cell. This probably leads to decreased expression of the two proteins. The second mutation is a small deletion of the yrdC open reading frame start site, and it is not likely that the protein is expressed. Both mutant strains show an increased accumulation of 17S rRNA (immature 16S rRNA). Maturation of 16S rRNA is dependent on proper assembly of the ribosomal proteins, a process that is disturbed when proteins are missing. The function of the YrdC protein is not known, but it is able to bind to double-stranded RNA; therefore, we suggest that it is an assembly factor important for 30S subunit biogenesis. On the basis of our findings, we propose that lesser amounts of S9 or a lack of YrdC causes the maturation defect. We have shown that as a consequence of the maturation defect, fewer 70S ribosomes and polysomes are formed. This and other results suggest that it is the lowered concentration of functional ribosomes that suppresses the temperature sensitivity caused by the mutant RF1.
Translation is one of the key processes in the cell, and its different steps are tightly coordinated. When, for instance, the cell is starved for an amino acid, elongation is stalled, the so-called stringent response is induced, and transcription of rRNA is stopped (8).
For protein synthesis, ribosomes are needed, the biogenesis of which is a complex process. In Escherichia coli the rRNA is transcribed as one transcript that is processed by RNaseIII to separate the different RNA species (18). The assembly of the subunit follows with ordered binding of the r-proteins (15, 24), maturation of rRNA by different endoribonucleases (18), a series of conformational changes, and addition of modifications to the rRNA (7). Final maturation of 23S rRNA requires protein synthesis and therefore occurs during the first rounds of translation (29). Protein synthesis may also be needed for final maturation of 16S rRNA (14).
The translation process as such is divided into four steps: (i) initiation, (ii) elongation, (iii) termination, and (iv) recycling. The mechanism for translation termination, the process studied here, has been investigated previously (12), but the impact of impaired termination on the total process has not been studied. In short, release factor 1 (RF1) is one of three release factors involved in translation termination. It recognizes and terminates at the stop codons UAG and UAA, while release factor 2 (RF2) terminates at UGA and UAA. The third release factor, RF3, releases RF1 and RF2 from the ribosome in a GTP-dependent manner (19). To address the issue of how impaired translation termination affects the total translation process, a temperature-sensitive RF1 (26) was used to isolate temperature-sensitive-positive (Ts+) suppressors. The result is a collection of mutations, half of which affect 16S rRNA maturation; two such mutants are described here. We also discuss possible mechanisms for the suppression.
MATERIALS AND METHODS
Media, strains, and plasmids.
Strains and plasmids used in this study are listed in Table 1 and Table 2, respectively. Media and genetic procedures were as described by Miller (22); preparation of phage λ was done according to the method of Chuang et al. (10). Where appropriate, the medium contained ampicillin (100 μg/ml), tetracycline (Tc) (25 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (Cm) (sublethal amount, 2 μg/ml; otherwise, 20 μg/ml).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype | Reference(s) or source |
|---|---|---|
| MG1655 | rph | 5, 16 |
| MK5 | ara Δ(gpt-lac)5 gyrA(nalA) aroE zhd-3169i::Tn10kan arg(UAG) rpoB(rif) thi aroE zhd-3169i::Tn10 | M.K. strain collection |
| MK33 | TA568 but prfA1 zcg -174::Tn10 | This work |
| MK35 | TA430 but prfA1 zcg -174::Tn10 | This work |
| MK37 | TA476 but prfA1 zcg -174::Tn10 | This work |
| MK39 | TA516 but prfA1 zcg -174::Tn10 | This work |
| MK47 | TA563 but prfA1 zcg -174::Tn10 | This work |
| MK49 | TA566 but prfA1 zcg -174::Tn10 | This work |
| MK51 | TA567 but prfA1 zcg -174::Tn10 | This work |
| MRA7 | MRA8 but zcg -174::Tn10 | M.R.A. strain collection |
| MRA8 | MG1655 but prfA1 | 33 |
| MRA30 | MG1655 but prfA1 recA | 33 |
| MRA76 | US477 but rpsIt2215 | This work |
| MRA100 | US477 but ΔyrdC | This work |
| TA563a | metB1 btuB3191::Tn10 | 3 |
| TA566a | metB1 btuB3191::Tn10 Δ(rrsA ileT alaT rrlA)I::cat+ = ΔAc | 3 |
| TA567a | metB1 btuB3191::Tn10 Δ(rrsA ileT alaT rrlA)1::cat Δ(purDH rrnE metA) = ΔEAc | 3 |
| TA568a | polA1 zih::Tn10 Δ(rrsA ileT alaT-rrlA)1::cat Δ(purDH rrnE metA) Δ(rrsB gltT rrlB)101 = ΔEBAc | 3 |
| TA430a | Δ(rrsA ileT alaT rrlA)1::cat Δ(purDH rrnE metA) Δ(rrsB gltT rrlB)101 Δ(rrsH ileV alaV rrlH)103 = ΔEBHAc | 3 |
| TA476a | Δ(rrsA ileT alaT rrlA)1::cat Δ(purDH rrnE metA) Δ(rrsB gltT rrlB)101 Δ(rrsH ileV alaV rrlH)103 Δ(rrsG gltW rrlG)30::lacZ+ = ΔEBHGzAc | 3 |
| TA516a | Δ(rrsA ileT alaT rrlA)34 Δ(rrsH ileV alaV rrlH)103 Δ(rrsB gltT rrlB)101 Δ(rrsG gltW rrlG)30::lacZ+ Δ(purDH rrnE metA) Δ(rrsD ileU alaU rrlD)25::cat+/pTRNA65 = ΔEBHGzADc/pTRNA65 | 3 |
| US475 | ara Δ(gpt-lac)5 zcg-174::Tn10 gyrA(nalA) arg(UAG) rpoB(rif) thi | MRA strain collection |
| US477 | US475 but prfA1 zcg-174::Tn10 | MRA strain collection |
The remaining genotype is F− ara Δlac thi.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Construction and/or reference and/or source |
|---|---|---|
| pmk19 | rpsI-sspA intergenic region with rpsIt2215; used for marker rescue experiments | PCR product amplified with oligonucleotides 5′ GGATCCCGTTTGTTGGCAGCGACAGCC 3′ and 5′ CTTAAGGCACGTCGTCGTCCGCAGTTCTCC 3′ from strain MRA76; cloned in pMOSBlue |
| pmk20 | rpsl-sspA wt intergenic region; used for mapping and marker rescue experiments | PCR product amplified with oligonucleotides 5′ GGATCCCGTTTGTTGGCAGCGACAGCC 3′ and 5′ CTTAAGGCACGTCGTCGTCCGCAGTTCTCC 3′ from strain MG1655; cloned in pMOSBlue |
| pmk40 | wt fragment spanning the region of ΔyrdC; used for marker rescue experiments | PCR product amplified with oligonucleotides 5′ GACGGGCCATCTGGTCCAGCGCCGC 3′ and 5′ CAGCCCCTTATCAACCGGACGCTG 3′ from strain US477; cloned in pMOSBlue |
| pmk41 | Fragment containing ΔyrdC; used for marker rescue experiments | PCR product amplified with oligonucleotides 5′ GACGGGCCATCTGGTCCAGCGCCGC 3′ and 5′ CAGCCCCTTATCAACCGGACGCTG 3′ from strain MRA100; cloned in pMOSBlue |
| pmk62 | wt 0.6-kb fragment containing the rplM gene; used for mapping experiments | Plasmid pmk71 was cleaved with restriction enzymes AccI and HindIII, and the 0.6-kb fragment was cloned in pSU19 |
| pmk71 | wt 5.0-kb fragment containing operons rplM-rpsI and sspAB and genes yhcL and yhcK; used for mapping experiments | λ Clone DD725 (10) was cleaved with HindIII, and the 5.0-kb fragment was cloned in pSU19 |
| pmk73 | wt yrdC gene; used for complementation experiments | PCR product amplified with oligonucleotides 5′ GCTCTAGAGTGAATAATAACCTGCAAAGAGACGCTATC 3′ and 5′ GGAATTCTTACCCCTGTCGAAACAGTTCACCCGTCAG 3′ from strain US477; cloned in pSU19 |
| pmk76 | wt 1.8-kb fragment containing the rplM-rpsI operon and part of the sspA gene; used for mapping experiments | Plasmid pmk71 was cleaved with restriction enzymes AvaI and HindIII, and the 1.8-kb fragment was cloned in pSU19 |
| pmk92 | wt 3.6-kb fragment with operons rplM-rpsI and sspAB and part of the yhcL gene; used for mapping experiments | Plasmid pmk71 was cleaved with restriction enzymes SphI and HindIII, and the 3.6-kb fragment was cloned in pSU18 |
| pmk94 | wt rpsI gene; used for complementation experiments | PCR product amplified with oligonucleotides 5′ CCCCTGTCGAAACAGTTCACCCGTC 3′ and 5′ GCTCTAGAATGGCTGAAAATCAATACTACGGCAC 3′ from strain US475; cloned in pSU19 |
| pMOSBlue | Amersham | |
| pSU18 | 6 | |
| pSU19 | 6 | |
| pUC18 | Pharmacia Biotech |
Construction of strains MK33, MK35, MK37, MK39, MK47, MK49, and MK51.
The transposon insertion btuB3191::Tn10 was removed from strains TA563, TA566, and TA567 by transduction with phage P1 grown on MG1655, selecting for growth without methionine and screening for Tcs. Strains MK33, MK35, MK37, MK39, MK47, MK49, and MK51 were constructed by transducing P1 grown on MRA7 (prfA1 zcg-174::Tn10) to TA568, TA430, TA476, TA516, TA563 (Met+ Tcs), TA566 (Met+ Tcs), and TA567 (Met+ Tcs), respectively, selecting for Tcr. The presence of prfA1 (coding for a mutated RF1) was verified by testing for temperature sensitivity at 42°C.
Construction of plasmids.
Procedures for plasmid constructions and transformation were as described by Sambrook et al. (27). For details, see Table 2.
PCR and sequencing.
PCR was done using either Ready-To-Go PCR Beads (Amersham Biosciences) or an Expand long-template PCR system (Roche), depending on the size of the fragment amplified. Sequencing was done using a BigDye terminator cycle sequencing ready reaction kit (Perkin Elmer Applied Biosystems) or a DYEnamic ET terminator cycle sequencing kit (Amersham Pharmacia Biotech).
Northern blot analysis of RNA.
Total RNA was prepared using TotallyRNA (Ambion Inc.). The RNA was separated on 1% agarose-6% formaldehyde (wt/vol) denaturing gels and transferred to a Hybond N membrane (Amersham) by capillary transfer. RNA was UV cross-linked to the membrane and hybridized (4); after the washing procedure, signals were detected and quantified with an IP reader/gauge (Fuji Photo Film).
When rRNA was analyzed, oligonucleotides labeled at the 5′ end with [γ-32P]ATP were used as probes. Unincorporated nucleotides were removed from the probe with Microspin G-25 columns (Amersham Biosciences). Data regarding the oligonucleotides used for probing rRNA are presented in Table 3.
TABLE 3.
Oligonucleotide probes used in Northern blot analysis
| Probe | Sequence 5′ to 3′ | Probe complementary to: |
|---|---|---|
| 683 | CGCATTTCACCGCTACA | Mature 16S rRNA |
| omk69 | CTCATTTTCATCAGACAATC | 3′ end of immature 16S rRNA, RNaseIII cleavage control |
| omk70 | GCACTGCAAAGTACGCTTC | 3′ end of immature 16S rRNA, RNaseX cleavage control |
| omk71 | CGTGTTCACTCTTGAGAC | 5′ end of immature 16S rRNA, RNaseG cleavage control |
| omk72 | GCGACGTTAAGAATCCG | 5′ end of immature 16S rRNA, RNaseE cleavage control |
| omk73 | GAGCAGTTGCGACGCGGC | 5′ end of immature 16S rRNA, RNaseIII cleavage control |
To obtain a stronger signal when probing mRNA, two measures were taken. First, the rRNA from total RNA samples was removed using MICROBExpress (Ambion Inc.). Second, long, hot RNA probes were transcribed using STRIP-EZ RNA T7 (Ambion Inc.) and [α-32P]dUTP (Amersham Biosciences). The primers used to make the DNA template are listed in Table 4. Unincorporated nucleotides were removed with Microspin G-50 columns (Amersham Biosciences). Hybridization was carried out with ULTRAhyb buffer (Ambion Inc.). After hybridization and detection, the membranes were stripped as described in the manual for STRIP-EZ RNA T7 (Ambion Inc.) and probed again against the ompA mRNA as an internal control.
TABLE 4.
Oligonucleotides used for amplification of the template for transcription of RNA probes used in Northern blot analysis
| Oligo-nucleotide | Sequence (5′ to 3′) | RNA probe complemen- tary to: |
|---|---|---|
| omk110 | TTATACGACTCACTATAGGGAGGTT AACTCCGGCCCAGACGCATTTCAC | sspA |
| omk97 | CATCAGGTCCGCATTGTGCTGGCTG | sspA |
| omk111 | TTATACGACTCACTATAGGGAGGTT AACGTTTGGAGAACTGCGGACGAC | rpsI |
| omk49 | GCTGTAACCGGCAACAAGCGTACTG | rpsI |
| omk113 | TTATACGACTCACTATAGGGAGGTT AAGCCTGCGGCTGAGTTACAACG | ompA |
| omk114 | GCGTTTCTCCGGTCTTCGCTGGCGGT | ompA |
Ribosome preparation and sucrose gradients.
Culture (500 ml) grown in Luria broth (LB) to mid-log phase was chilled and harvested. The cells were washed and concentrated in cold R buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 6 mM β-mercaptoethanol, 10 mM MgCl2). Lysozyme (1 mg/ml) was added, and the cells were sonicated. The cell debris was pelleted, and the extract was loaded onto a 5 to 35% sucrose gradient in R buffer and centrifuged at 20,500 rpm for 15 h in an SW4ITI rotor (Beckman). Fractions (200 μl) were collected. rRNA from sucrose gradients was isolated by phenol and chloroform extractions.
RESULTS
Isolation and initial mapping of two suppressor mutations.
The prfA1 allele codes for an RF1 that makes the cell temperature sensitive at 42°C (26, 33). Another characteristic of the mutated factor is that the termination process is slowed down about 25% at 37°C (reference 25 and unpublished data). This mutant was used to select suppressors.
Independent cultures of strain US477 (prfA1) were started and grown overnight. Dilutions were spread on LB plates and incubated at 42°C. From the collection of isolated mutants, two independent clones were investigated further. The clones were called 2:3 (MRA76) and 5:4 (MRA100) and were shown by transduction not to be linked to prfA1. In separate experiments it was found that clone 2:3 was linked to aroE by about 5% on the side away from the ribosomal protein cluster (between 72.5 and 73 min on the E. coli map) and that clone 5:4 was linked to aroE by almost 100% (73.9 min on the map).
The strains MRA76 and MRA100 need at least 1 more day than the wild-type (wt) strain to give colonies with all tested temperatures (22, 30, 37, and 42°C) and tested media (LB and M9 minimal medium). Also, the colony size of the mutant strains never reaches that of the wt strain under any condition tested; still, the Ts+ phenotype at 42°C is clear. A strain carrying the prfA1 allele grows like the wt at 37°C and not at all at 42°C. The two suppressor phenotypes, slow growth and Ts+, have been used as markers in the genetic experiments.
We tried to separate both suppressors from the prfA1 mutation. Phage lysates were grown on strains MRA76 and MRA100 and transduced to strain MK5 (aroE zhd-3169::Tn10kan), selecting for Aro+. Transductants were purified and screened for slow growth. Besides transductants growing like the wt, we found transductants with which the restreaks were a heterogeneous mix of colony sizes. When small colonies were repurified from these streaks, heterogeneous growth was again observed. We tried this experiment several times with the same result. It seems that the isolated suppressor mutations are almost lethal in a wt background and that there is a strong selection pressure for reversion or suppression. This suggests some kind of mutual dependence between the prfA1 mutation and the suppressor mutations. In the following text, therefore, when we refer to strains with either mutation the prfA1 mutation is understood also to be present.
Map position of the suppressor mutation in strain MRA76 (clone 2:3).
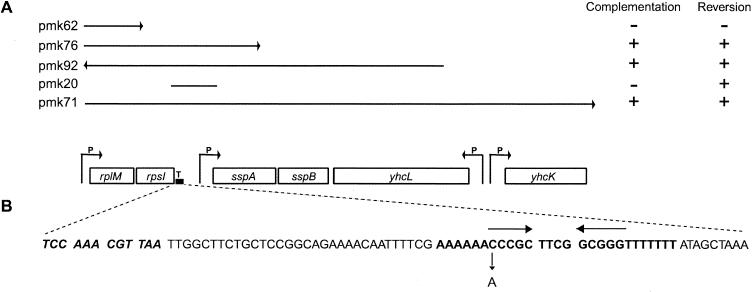
As mentioned above, the suppressor mutation in MRA76 was mapped by transduction between positions 72.5 and 73 min on the linkage map. To get the precise position of the mutation, DD725, a λ-clone that covers this region, was used (10). DD725 was digested with either EcoRI or HindIII, the fragments were cloned into pUC18, and a complementation test was done. It was found that a 5.0-kb HindIII fragment could complement the suppressor mutation, i.e., the cells behaved like those of strain US477 (prfA1), with normal growth at 37°C and temperature sensitivity at 42°C.
The 5.0-kb fragment was digested with different combinations of restriction enzymes into smaller fragments; together with the 5.0-kb fragment, these were cloned into pSU18 or pSU19. The two plasmids are derived from pACYC184, which has a lower copy number than pUC18 (6). In addition, a PCR amplified fragment was cloned into pMOSBlue. The clones were transformed into strain MRA76; the relevant ones are shown in Fig. 1A. When the transformation plates were inspected it was found that clones pmk71, pmk76, and pmk92 could complement the slow growth phenotype at 37°C. It should be pointed out that the slow growth was not fully complemented; it was not as fast as that seen with strain US477 (prfA1). A total of 12 colonies from each transformation were streaked and tested for growth at 42°C. Again, clones pmk71, pmk76, and pmk92 complemented the suppressor phenotype and the cells became temperature sensitive. As can be seen in Fig. 1A, these three plasmids carry fragments that cover the rplM-rpsI operon. It was also seen on the transformation plates that in the presence of clones pmk20, pmk71, pmk76, and pmk92, approximately 5% of the cells grew like strain US477 (prfA1) cells (Fig. 1A). A total of 12 such fast-growing colonies from each transformation were tested at 42°C, and they had all become temperature sensitive. Thus, these colonies behave as true revertants of the suppressor mutation and could have arisen through homologous recombination between the chromosome and the plasmid. The common sequence for the clones that can recombine is the intergenic region of approximately 400 bp between genes rpsI and sspA. The sequence starts with the last 29 nucleotides (nt) of the rpsI gene and extends 23 nt into the sspA gene. These results taken together suggest that the suppressor mutation in strain MRA76 affects the expression of the rplM-rpsI operon but that the mutation is outside the coding region.
FIG. 1.
The 5.0-kb fragment produced by cleavage of λ phage clone DD725 with HindIII. P, promoter sites; T, transcription terminator sequence. Arrows show the direction of gene expression. (A) Plasmids used for mapping and marker rescue experiments. Arrowheads show the direction of gene expression from the promoter of the vector. Whether the fragment can complement the temperature sensitivity phenotype and/or whether it can give Ts+ colonies through homologous recombination is indicated to the right. (B) Location of the rpsIt2215 mutation. Large arrows mark the transcription terminator stem; the small arrow shows the position of the mutation. Bold characters in sequences show the whole terminator sequence; bold and italic characters show the last four codons of the rpsI gene.
The 400-bp noncoding region between rpsI and sspA from strain MRA76 was sequenced, and a single-base alteration in the putative transcriptional terminator sequence of the rplM-rpsI operon was found (Fig. 1B). We have found that pmk76 (a plasmid covering the whole operon) could complement both suppressor phenotypes, while pmk62 (encoding L13 only) could not. To test whether expression of both L13 and S9 is needed for complementation or whether that of S9 alone is sufficient, we cloned the rpsI gene into pSU19 (pmk94) and transformed it into MRA76. When tested, complementation of both suppressor phenotypes by pmk94 was observed. These results suggest that decreased expression of ribosomal protein S9 is the cause of the suppression of the RF1 temperature-sensitive phenotype.
A marker rescue experiment was done to verify that the mutated base causes the suppressor phenotype. Two constructs containing the intergenic region with the suppressor mutation (pmk19) or the wt sequence (pmk20) were transformed into two strains: MRA8 (prfA1) and MRA30 (prfA1 recA). The transformants were plated and incubated at 37 and 42°C, respectively. At 42°C, neither of the strains should grow because of the prfA1 mutation and yet colonies do arise. In strain MRA8, Ts+ can occur by reversion of prfA1 or by recombination with the plasmid carrying the suppressor mutation; in strain MRA30, recombination is not possible because of the recA mutation. The result was clear; in both strains with the wt fragment and in strain MRA30 with the mutant fragment, the reversion frequency was about 10−7. In strain MRA8 with the mutant fragment, the reversion frequency was 2 × 10−5. This suggests that the base alteration in the terminator stem is the suppressor mutation. To confirm that recombination had taken place, the intergenic region on the chromosome in some MRA8 clones that had become Ts+ was sequenced. The suppressor mutation was present in all clones tested. This shows that a single-base alteration in the transcriptional terminator for the rplM-rpsI operon leads to suppression of the temperature sensitivity phenotype caused by the prfA1 mutation. The suppressor mutation was named rpsIt2215.
Map position of the suppressor mutation in strain MRA100 (clone 5:4).
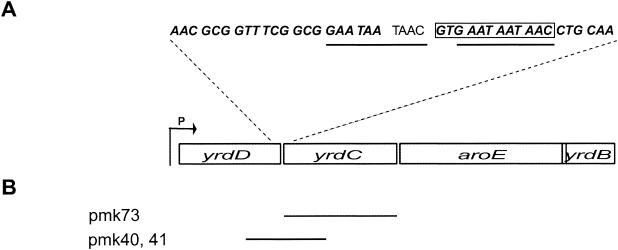
We had found that the mutation in strain MRA100 is closely (more than 99%) linked to aroE, but we could not determine on which side of the marker the mutation was located. Therefore, both the upstream region (approximately 2 kb) and the downstream region (approximately 1 kb) of the aroE gene were sequenced. A small deletion of 12 nt affecting the open reading frame (ORF) yrdC was found (Fig. 2A). This gene is located in an operon that is located between 73.8 and 73.9 min on the map and is suggested to consist of four genes: yrdD, yrdC, aroE, and yrdB (www.cifn.unam.mx/Computational_Genomics/regulondb). The YrdD protein has high-level homology to topoisomerase I, YrdC is homologous to the yeast Sua5 protein that has been suggested to have a role in translation (23), and yrdB is a short putative ORF. The aroE gene encodes dehydroshikimate reductase, which is involved in biosynthesis of aromatic amino acids.
FIG. 2.
The yrdD operon. P marks the promoter. The termination codon of the aroE gene overlaps the initiation codon of the yrdB gene. (A) Location of the ΔyrdC mutation. Bold and italic characters in sequences show the last seven codons of the yrdD gene and first eight codons of the yrdC gene, respectively. The yrdC gene initiates at GTG. The box marks the deletion. The perfect duplications are underlined. (B) Plasmids used for marker rescue and complementation experiments.
The deletion has occurred in a short perfect duplication of 10 nt, which can be extended to 13 when one mismatch is allowed. The initiation codon of the yrdC gene is deleted, and this may lead either to translation initiation at an AUG 43 codons further down in the gene or to no production of YrdC at all (Fig. 2A).
To verify that the deletion causes the suppression of prfA1, both a complementation test and a marker rescue experiment were performed. For the complementation test, the yrdC gene was cloned into the pSU19 vector (pmk73) and transformed into MRA100 (Fig. 2B). The phenotype of the double mutant changed into a phenotype like that of strain US477 (prfA1); i.e., temperature sensitivity and growth like the wt at 37°C.
For the marker rescue experiment, a small fragment covering the deletion and parts of the genes yrdD and yrdC from the mutant strain MRA100 and the (in this region) wt strain US477 was cloned into pMOSBlue and transformed into strain US477. US477 with the plasmid pmk40 (wt fragment) or pmk41 (mutant fragment) was grown and plated at 37 or 42°C, respectively. The reversion frequency in the presence of plasmid pmk40 (wt) was 1 × 10−7, while in the presence of plasmid pmk41 (mutant fragment) it was 4 × 10−2. This suggests that the small deletion is the suppressor mutation and that decreased amounts of functional YrdC protein suppress the prfA1 mutant phenotype. We named the suppressor mutation ΔyrdC.
The rpsIt2215 mutation affects the stability of the rplM-rpsI transcript.
The suppressor mutation rpsIt2215 is located at the bottom of a transcriptional terminator stem, and it is likely that the base alteration makes the stem less stable. If this is so, transcription readthrough should occur. To examine this, an RT-PCR over the terminator was done on mRNA from the double-mutant strain (MRA76) and the control strain (US475). A forward primer was placed at the end of the rpsI gene, and a reverse primer was placed just downstream of the terminator; only the MRA76 sample gave a 200-nt band, indicating transcription readthrough. As a control, the same forward primer was used and a reverse primer was placed between the rpsI gene stop codon and the terminator; as expected, each sample produced a fragment of about 120 nt (data not shown).
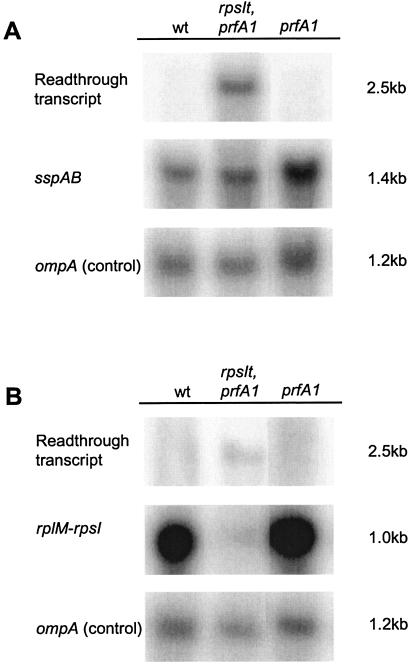
Readthrough of rpsIt may affect the abundance of the rplM-rpsI transcript and probably increases expression of the downstream sspAB operon. The sspA gene codes for a transcription factor (13, 32), and SspB is involved in degradation of tmRNA-tagged peptides (20). To examine the expression pattern for the two operons, a Northern blot analysis was done. RNA probes were transcribed using labeled dUTP to probe either the sspAB or the rplM-rpsI mRNA. Due to problems encountered when the rplM-rpsI transcript was probed, rRNA was removed from the samples of total RNA. The strains tested were the double-mutant strain MRA76, strain US477 (prfA1), and strain US475 (wt). As an internal control, ompA mRNA was probed. The results are shown in Fig. 3. First, it can be seen that expression of the sspAB transcript increased in the strains with the prfA1 mutation; when the sspAB/ompA ratio is set to 1 for the wt, the ratio is 1.5 for MRA76 and 1.7 for US477 (Fig. 3A). The reason for this remains unknown. When probing the sspAB transcript, the suppressor mutant showed (besides the expected wt mRNA) a longer product slightly smaller than 23S rRNA (2,900 nt). A transcript reaching from the rplM-rpsI promoter to the terminator of the sspAB operon would be about 2,500 to 2,600 nt in size. No such transcript was observed for the other two strains, indicating that the terminator is read through more readily in the suppressor strain MRA76 (Fig. 3A). To test whether overexpression of SspA and/or SspB suppresses the release factor temperature-sensitive phenotype, a plasmid with the sspAB operon under the arabinose promoter was transformed to strain US477. No suppression could be observed when the promoter was induced (data not shown).
FIG. 3.
Analysis of the expression of the rplM-rpsI and sspAB operons in strains US475 (wt), MRA76 (rpsIt2215 prfA1), and US477 (prfA1). (A) Northern blot analysis of total RNA samples, probing mRNA coding for sspA. The membrane was stripped after detection, and mRNA coding for ompA was probed as an internal control. (B) Northern blot analysis on mRNA samples, probing mRNA coding for rpsI. The membrane was stripped after detection, and mRNA coding for ompA was probed as an internal control.
When we probed the rplM-rpsI transcript it was found that strain MRA76 had significantly less transcript ending at the putative terminator than strains US475 and US477 and yet that the readthrough product was observable (Fig. 3B). The rpsI/ompA ratio values were essentially the same for strains US475 and US477, while strain MRA76 had a significantly smaller rpsI/ompA ratio value. This strongly suggests that expression of the ribosomal proteins L13 and S9 was affected. On the basis of this result and our finding that a cloned rpsI gene (but not a cloned rplM gene) can complement the suppressor phenotypes, we suggest that decreased levels of protein S9 lead to suppression of the temperature-sensitive RF1.
During experiments in which total RNA samples for Northern blot analysis were separated on gels and stained with ethidium bromide, an extra band just above 16S rRNA was observed in the sample from strain MRA76. This might be an immature 16S rRNA species (called 17S rRNA). We decided to investigate this further.
Does the extra band represent immature 16S rRNA?
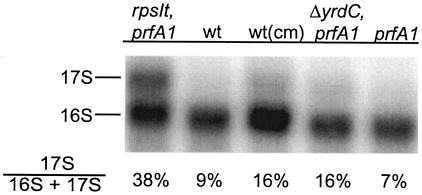
To test whether the larger band represents immature 16S rRNA, we probed total RNA with a probe complementary to the middle of 16S rRNA. As can be seen in Fig. 4, the probe does bind to the presumptive 17S band. The amount of 17S was quantified to 38% of total 16S rRNA in strain MRA76 and 9% in the wt US475 strain. The wt result is in good agreement with published results (18).
FIG. 4.
Northern blot analysis of 16S rRNA in strains MRA76 (rpsIt2215 prfA1), US475 (wt), US475 treated with Cm [wt(cm)], MRA100 (ΔyrdC prfA1), and US477 (prfA1). Total RNA samples were probed with oligonucleotide 683 against the middle part of 16S rRNA. The ratios of 17S/(16S + 17S) are shown under the figure.
The analysis was extended to strain MRA100 (ΔyrdC), since the YrdC homologue Sua5 from Saccharomyces cerevisiae has been implicated in protein synthesis (23). We also analyzed the wt strain treated with sublethal amounts of Cm. Cm is an antibiotic that binds to the 50S subunit and inhibits translation. It is known that addition of Cm leads to accumulation of precursor 16S rRNA (28). We have observed that sublethal amounts of Cm allow growth of minute colonies of strain US477 at 42°C (data not shown). Total RNA from strain MRA100, strain US475 (wt) treated with Cm, and the RF1 mutant strain US477 was prepared and analyzed by Northern blotting. The result can be seen in Fig. 4. Strains MRA100 and US475 treated with Cm both showed increased amounts of 17S (about 16% of the total 16S rRNA), while strain US477 behaved like the wt strain, with only 7% of 17S rRNA detected. Thus, the elevated amount of 17S is not caused by the prfA1 mutation.
Which step in 16S rRNA maturation is affected?
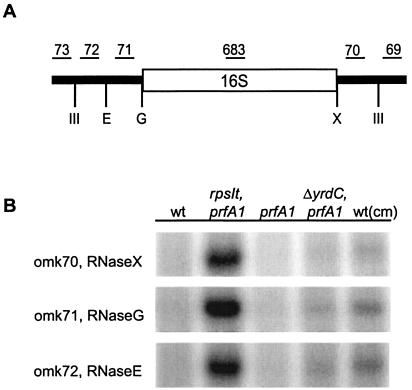
rRNA is transcribed as one transcript that is cut by RNaseIII to produce precursors for 16S, 23S, 5S, and a variety of tRNAs (18). 17S, the precursor for 16S rRNA, is trimmed at the 5′ end by RNaseE, leaving 66 nt. RNaseG then cleaves this transcript to the mature 5′ end. The extra 33 nucleotides at the 3′ end are processed by a still-unknown enzyme (21).
To determine which processing step is affected in our mutants and by the presence of Cm, a Northern blot analysis of total RNA was done. Oligonucleotides that probe the ends of the different intermediates leading to 16S rRNA were used (Fig. 5A and Table 3). When a particular RNase has cleaved the transcript, the probe will not hybridize to the RNA because its binding site on the transcript is lost. The experiment showed that only RNaseIII functions normally (probes omk69 and omk73 gave no band for any sample tested) and that the activities of all the other RNases are affected (Fig. 5B).
FIG. 5.
(A) A schematic drawing of maturation of 16S rRNA. The positions of oligonucleotides are shown above the line. The cleavage points for the different RNAses are shown below the line as follows: III, RNaseIII; E, RNaseE; G, RNaseG; X, unknown RNase. (B) Northern blot analysis of 16S rRNA maturation in strains US475 (wt), MRA76 (rpsIt2215 prfA1), US477 (prfA1), MRA100 (ΔyrdC prfA1), and US475 treated with Cm [wt(cm)]. Total RNA samples were probed with oligonucleotides omk69, omk70, omk71, omk72, and omk73. Oligonucleotide 683 was used as a positive control for 16S rRNA (data not shown). As omk69 and omk73 (RNaseIII) did not hybridize, those results are not shown.
30S subunit assembly is affected by the immature 17S in both suppressor mutants.
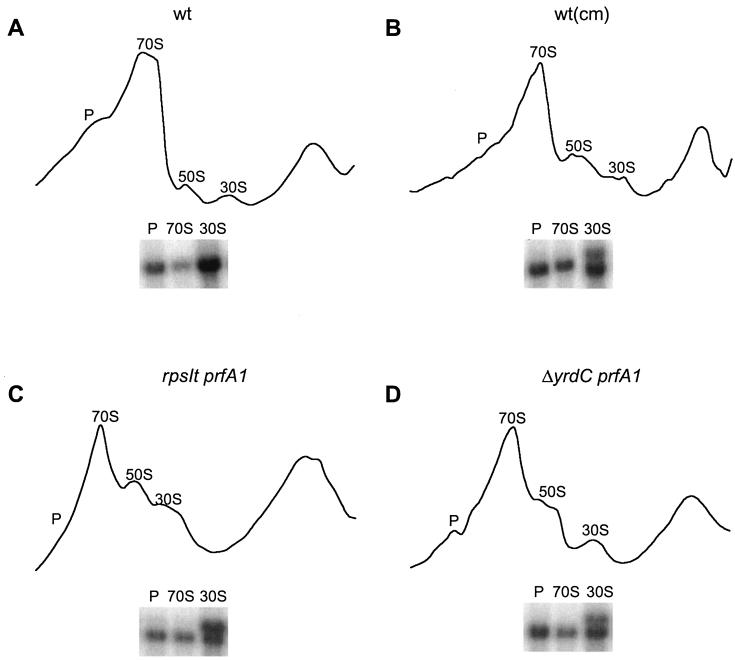
To investigate the effect of the maturation defect on ribosome assembly, ribosomes from strains MRA76, MRA100, US475, and US475 treated with Cm were prepared and separated on sucrose gradients. The preparation was done at 10 mM Mg2+, a concentration at which the 30S and 50S subunits are kept together in the 70S complex. In a typical gradient on wt ribosomes, almost all subunits were in 70S ribosomes and polysomes (Fig. 6A). A gradient on ribosomes from strain US477 (prfA1) looks like a wt profile (data not shown). In our experiments the polysomes can be seen as a shoulder on the 70S peak. If the 16S rRNA maturation deficiency were to affect 30S assembly or the formation of 70S particles, this would show as a relative increase of subunits compared to the results seen with 70S. The results can be seen in Fig. 6B to D. Both the suppressor mutants and the wt strain treated with Cm show an altered gradient pattern; the 30S and 50S peaks have increased relative to the 70S peak. This suggests that the ability of the 30S subunits to form 70S ribosomes is affected.
FIG. 6.
Sucrose gradients on ribosomes from strains US475 (wt) (A), US475 treated with Cm [wt(cm)] (B), MRA76 (rpsIt2215 prfA1) (C), and MRA100 (ΔyrdC prfA1) (D). P marks the polysome shoulder. The results of Northern analysis probing 16S rRNA from samples prepared from polysome, 70S, and 30S peaks are indicated beneath the gradient.
To investigate whether 17S rRNA can be incorporated into 70S ribosomes or polysomes, a Northern blot analysis was done on samples from the rRNA peaks that contain 16S rRNA: the polysomes, 70S, and 30S. 17S rRNA was observed in the 30S peak only with strain MRA76 (55% 17S), strain MRA100 (33% 17S), and the wt strain treated with Cm (31% 17S). The amount of 17S from the wt strain was below the detection limit. Immature 16S rRNA could not be detected in the polysomes or 70S ribosomes (Fig. 6).
A lowered amount of functional ribosomes suppresses the prfA1 mutation.
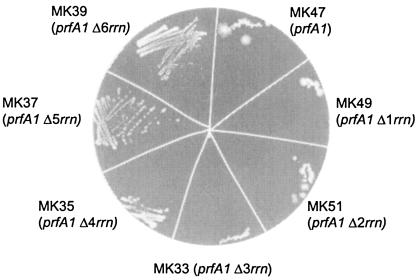
Our experiments so far suggest that slower maturation (and, thus, a decreased pool of functional ribosomes) is correlated to suppression of the temperature sensitivity phenotype caused by prfA1. To test whether this is a general phenomenon, we obtained a collection of strains with successive deletions of the rrn operons. The amount of rRNA in these strains decreases with fewer operons (3). The prfA1 mutation was transduced into the six strains, and the transductants were tested for the ability to grow at 42°C. A strain carrying the prfA1 mutation shows some growth in the beginning of the streak at 42°C; this also allows a few revertants to appear. We looked for continuous and homogenous growth and found that there is a gradient of increasing temperature resistance; the strains with up to three deletions are temperature sensitive and the strain with four deletions shows some resistance, whereas in the strains with five or six deleted rrn operons a Ts+ phenotype is seen (Fig. 7). These results strongly suggest that there is a link between a lowered concentration of active ribosomes and suppression of the temperature-sensitive RF1.
FIG. 7.
Temperature sensitivity test at 42°C of the prfA1 mutation combined with successive deletions of rrn operons.
More suppressor mutations.
A collection of 26 mutants, all of which suppress the temperature-sensitive RF1 (prfA1), have been selected in separate experiments. In this work we have presented data regarding two such suppressors, both of which show elevated amounts of 17S rRNA. To investigate how many of the other 24 suppressors are deficient in maturation of 16S rRNA, Northern blot analysis was used. Total RNA was prepared from all 24 mutants and probed as previously described. About half of the mutants show elevated amounts of 17S rRNA to various degrees. We are in the process of mapping these mutations and have so far found three additional complementation groups besides the described mutations. The high percentage of mutants showing elevated levels of 17S rRNA suggests that the suppressor mechanism described here is not an isolated event but is a common way to suppress the less-efficient RF1.
DISCUSSION
In this work we have presented data for two mutants, both of which suppress the temperature sensitivity caused by a mutated RF1. The suppressors were independently isolated and map at different locations on the linkage map, and yet both show increased amounts of immature 16S rRNA (17S).
The first mutation, rpsIt2215, is a point mutation in the stem of the transcription terminator for the rpsI-rplM operon which encodes the ribosomal proteins L13 and S9. The position of the mutation suggests that the stability of the stem might be decreased. We have shown that rpsIt2215 leads to both readthrough of the terminator and a decrease in the amount of the rplM-rpsI transcript. That a decrease in terminator stability can lead to decreased stability of the mRNA (and thus to a lower amount of protein product) has been shown previously (2). The fact that clones with either rplM-rpsI or rpsI only can complement the suppressor phenotypes indicates that (at least) protein S9 is expressed in amounts lower than those seen with the wt strain. Even though the rpsIt2215 mutation leads to a longer transcript (including the downstream sspAB operon), we have found no evidence that the SspA and/or SspB proteins are involved in the suppression of the RF1 mutation (data not shown).
Maturation of 16S rRNA and 23S rRNA is dependent on correct assembly of the subunits. Lesser amounts of S9 (and L13) may disturb this process and therefore slow down the maturation process. In our experiments we have only observed an accumulation of immature 16S rRNA; it is not possible to distinguish immature 23S rRNA from mature 23S rRNA on our gels, since immature 23S rRNA is only elongated with a few nucleotides at each end (30).
The second mutation, ΔyrdC, is a small deletion of the initiation codon for the yrdC gene. This probably means that the protein is not translated. The yrdC gene product is conserved, but its function is unknown. YrdC, YciO (another E. coli protein with unknown function), and a domain of the yeast protein Sua5 define a conserved motif: yrdC-yciO-SUA5. YrdC and YciO have been crystallized, their three-dimensional structures are similar (17), and both seem to have the capacity to bind nucleic acids (31). SUA5 was isolated as a suppressor of an aberrant AUG initiation codon upstream of the cyc1 gene, which encodes the iso-1-cytochrome c enzyme (23). The initiation codon was created by a point mutation at position −18, creating a short ORF with a stop codon prior to the normal start codon of the cyc1 gene. This cyc1-1019 mutant produces 2% of the wt enzyme level, a level that is increased to 60% by the mutated Sua5 protein. The suppression mechanism has been suggested to involve either alteration of the transcription rate of cyc1, mRNA stability, initiation codon recognition, or ribosome reinitiation. Judging on the basis of our results, we think that YrdC might be an assembly factor for the 30S subunit. It is possible that Sua5 has a similar function in yeast and that a deficient 40S subunit has changed properties with regard to initiation. This is something we are in the process of investigating.
Sublethal Cm concentrations cause increased amounts of precursor 16S (28) (Fig. 4) and suppress the temperature sensitivity phenotype caused by the prfA1 mutation (data not shown). Cm binds to the peptidyltransferase center of the 50S subunit. How this affects maturation of 16S rRNA is not understood. In a recent paper on the 50S subunit assembly factor SrmB, however, it was found that in a strain with the srmB gene deleted, 17S rRNA was accumulated (9), indicating a link between the two subunits with respect to maturation.
Processing of rRNA starts by RNaseIII cleavage, probably during transcription. We do not see any deficiency in this step. Thereafter, the proteins are loaded onto the transcript and final processing of the ends occurs late during assembly (18). The activities of RNaseE and RNaseG at the 5′ end of 16S rRNA and of an unknown RNase at the 3′ end are independent of each other, and we see that they are all affected. Therefore, we hypothesize that a step before the initiation of the activities of the RNaseE and RNaseG and the unknown RNase (possibly involving a structure needed for proper cleavage) is influenced in the suppressor mutants. We see only one band and no smear, indicating that maturation occur at both ends simultaneously. The fact that we have isolated at least five different mutations that give the same phenotype in subunit assembly indicates that it is a major rate-limiting step that is disturbed.
Our results with the sequential deletions of rrn operons indicate that a lowered concentration of functional ribosomes, and not the processing deficiency per se, suppresses the prfA1 mutation. It is known that the mutant release factor terminates more slowly at 37°C (reference 25 and unpublished data), and it is therefore likely that termination at 42°C becomes so slow that ribosomes get stuck on the mRNA. This may induce transcription termination, i.e., polarity, leading to less production of one or many proteins. Temperature sensitivity could be caused by starvation for ribosomes due to stacking on mRNA or by lowered expression of some proteins due to transcriptional polarity or both.
First, we looked at the possibility that a slow growth rate per se suppresses the temperature sensitivity phenotype. When the mutant strain US477 was grown on M9 minimal medium plates with glycerol as a carbon source, the strain was still temperature sensitive (data not shown), a result that indicates that slow growth is not the suppressor of the temperature sensitivity phenotype.
Fewer functional ribosomes will lead to less-frequent initiation. If the problem is ribosome queuing of (and, as a consequence, starvation for) ribosomes, it is possible that a slow initiation rate will balance the slow termination rate and hence lead to a Ts+ phenotype.
If the problem is premature transcription termination due to stalling, this can be overcome by faster translation termination. One way to do this is to increase the concentration of RF1. It is known that overexpression of mutated RF1 will give a Ts+ phenotype (26). Thus, if the suppressor mutations were to lead to an increased expression of the prfA1 allele, the cells should become Ts+. It has been shown that rrn operon transcription is induced when the amount of actively translating ribosomes decreases. This phenomenon has been called feedback regulation (11). In accordance with this, expression of the rrn operons should be induced in the two suppressor strains (since they have fewer functional ribosomes). The P1 promoter for the hemA-prfA operon shares several characteristics with the P1 promoter for rrn operons. Hence, it is possible that not only rRNA transcription but also expression of the hemA-prfA operon is induced in the suppressor mutants. This mechanism may also explain why the suppressor mutations seem to be dependent on the prfA1 mutation and cannot be isolated in a wt background; either of the two mutations would lead to overexpression of RF1—which is lethal to the cell (1). We have just initiated experiments to investigate these different possibilities.
Acknowledgments
We thank Mitch Dushay for comments on the manuscript.
This work was supported by the M. Bergvall Society.
REFERENCES
- 1.Adamski, F. M., K. K. McCaughan, F. Jorgensen, C. G. Kurland, and W. P. Tate. 1994. The concentration of polypeptide chain release factors 1 and 2 at different growth rates of Escherichia coli. J. Mol. Biol. 238:302-308. [DOI] [PubMed] [Google Scholar]
- 2.Aiba, H., A. Hanamura, and H. Yamano. 1991. Transcriptional terminator is a positive regulatory element in the expression of the Escherichia coli crp gene. J. Biol. Chem. 266:1721-1727. [PubMed] [Google Scholar]
- 3.Asai, T., C. Condon, J. Voulgaris, D. Zaporojets, B. Shen, M. Al-Omar, C. Squires, and C. L. Squires. 1999. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 181:3803-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M. (ed.). 2003. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 6.Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 7.Björk, G. R. 1996. Stable RNA modification, p. 861-886. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 8.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 9.Charollais, J., D. Pflieger, J. Vinh, M. Dreyfus, and I. Iost. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 48:1253-1265. [DOI] [PubMed] [Google Scholar]
- 10.Chuang, S.-E., D. L. Daniels, and F. R. Blattner. 1993. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175:2026-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, J. R., C. L. Olsson, J. W. B. Hershey, M. Grunberg-Manago, and M. Nomura. 1987. Feedback regulation of rRNA synthesis in Escherichia coli: requirement for initiation factor IF2. J. Mol. Biol. 198:383-392. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenberg, M., and T. Tenson. 2002. A new beginning of the end of translation. Nat. Struct. Biol. 9:85-87. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, A.-M., H. Lehnherr, X. Wang, V. Mobley, and D. J. Jin. 2003. Escherichia coli SspA is a transcription activator for bacteriophage P1 late genes. Mol. Microbiol. 48:1621-1631. [DOI] [PubMed] [Google Scholar]
- 14.Hayes, F., and M. Vasseur. 1976. Processing of the 17-S Escherichia coli precursor RNA in the 27-S pre-ribosomal particle. Eur. J. Biochem. 61:433-442. [DOI] [PubMed] [Google Scholar]
- 15.Herold, M., and K. H. Nierhaus. 1987. Incorporation of six additional proteins to complete the assembly map of the 50S subunit from Escherichia coli ribosomes. J. Biol. Chem. 262:8826-8833. [PubMed] [Google Scholar]
- 16.Jensen, K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia, J., V. V. Lunin, V. Sauvé, L.-W. Huang, A. Matte, and M. Cygler. 2002. Crystal structure of the YciO protein from Escherichia coli. Proteins Struct. Funct. Genet. 49:139-141. [DOI] [PubMed] [Google Scholar]
- 18.King, T. C., R. Sirdeshmukh, and D. Schlessinger. 1986. Nucleolytic processing of ribonucleic acid transcripts in prokaryotes. Microbiol. Rev. 50:428-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kisselev, L. L., and R. H. Buckingham. 2000. Translational termination comes of age. Trends Biochem. Sci. 25:561-566. [DOI] [PubMed] [Google Scholar]
- 20.Levchenko, I., M. Seidel, R. T. Sauer, and T. A. Baker. 2000. A specificity-enhancing factor for the ClpXP degradation machine. Science 289:2354-2356. [DOI] [PubMed] [Google Scholar]
- 21.Li, Z., S. Pandit, and M. P. Deutscher. 1999. RNaseG (CafA protein) and RNaseE are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18:2878-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor, N.Y.
- 23.Na, J. G., I. Pinto, and M. Hampsey. 1992. Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics 131:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nomura, M. 1973. Assembly of bacterial ribosomes. Science 179:864-873. [DOI] [PubMed] [Google Scholar]
- 25.Roesser, J. R., and C. Yanofsky. 1988. Ribosome release modulates basal level expression of the trp operon of Escherichia coli. J. Biol. Chem. 263:14251-14255. [PubMed] [Google Scholar]
- 26.Rydén, M., J. Murphy, R. Martin, L. Isaksson, and J. Gallant. 1986. Mapping and complementation studies of the gene for release factor 1. J. Bacteriol. 168:1066-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Shen, V., and H. Bremer. 1977. Chloramphenicol-induced changes in the synthesis of ribosomal, transfer, and messenger ribonucleic acids in Escherichia coli. J. Bacteriol. 130:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava, A. K., and D. Schlessinger. 1988. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl. Acad. Sci. USA 85:7144-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srivastava, A. K., and D. Schlessinger. 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 44:105-129. [DOI] [PubMed] [Google Scholar]
- 31.Teplova, M., V. Tereshko, R. Sanishvili, A. Joachimiak, T. Bushueva, W. F. Anderson, and M. Egli. 2000. The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding. Protein Sci. 9:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams, M. D., T. X. Ouyang, and M. C. Flickinger. 1994. Glutathione s-transferase-sspA fusion binds to E. coli RNA polymerase and complements deleted sspA mutation allowing phage P1 replication. Biochem. Biophys. Res. Commun. 210:123-127. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, S., M. Rydén-Aulin, L. A. Kirsebom, and L. A. Isaksson. 1994. Genetic implication for an interaction between release factor one and ribosomal protein L7/L12 in vivo. J. Mol. Biol. 242:614-618. [DOI] [PubMed] [Google Scholar]