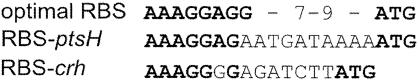Abstract
In Bacillus subtilis, carbon catabolite repression (CCR) of catabolic genes is mediated by ATP-dependent phosphorylation of HPr and Crh. Here we show that the different efficiencies with which these two proteins contribute to CCR may be due to the drastic differences in their synthesis rates under conditions that cause CCR.
In Bacillus subtilis, carbon catabolite repression (CCR) of many catabolic genes is mediated by ATP-dependent phosphorylation of Ser-46 of HPr and of its homologue Crh (4). Although these two proteins exhibit high sequence identity (45%) and are both efficiently phosphorylated by the HPr kinase/phosphorylase, their contributions to CCR differ (4). P-Ser-Crh can only partly substitute for P-Ser-HPr in CCR, whereas P-Ser-HPr can completely substitute for P-Ser-Crh in this signal transduction pathway. In order to understand the different behaviors of these two proteins, we compared the expression levels of the corresponding genes, crh and ptsH (encoding HPr), in the presence of different carbon sources by using transcriptional and translational lacZ reporter gene fusions.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
The plasmids and the bacterial strains used in this study are listed in Table 1. Escherichia coli DH5α was used as a general cloning host. Plasmid DNAs of the pMutin4 derivatives were prepared from E. coli BMH71-18 (recA+) prior to transformation into B. subtilis. E. coli, and B. subtilis strains were routinely grown in Luria-Bertani broth supplemented with the appropriate antibiotics when necessary (ampicillin at 100 μg/ml for E. coli and chloramphenicol at 5 μg/ml and erythromycin at 0.3 μg/ml for B. subtilis). Standard procedures were used to transform E. coli (15) and B. subtilis (7). Sequencing of PCR-derived DNA fragments in the final plasmid constructs was carried out by Genome Express (Meylan, France).
TABLE 1.
Plasmids and bacterial strains
| Plasmid or strain | Relevant structures or genotypea | Source or reference |
|---|---|---|
| Plasmids | ||
| pAC6 | Promoterless lacZ, cat encompassed by amyE-5′ and amyE-3′, bla | 17 |
| pMutin2 and -4 | MCS RBSspoVG-lacZ lacI bla ery rrnBt1t2-λt0 Pspac | 19 |
| pBGM6 | pMutin4 containing a 210-bp fragment of crh-5′ region (−120 to +90) in MCS | This work |
| pBGM8 | pMutin4 containing a Φ(crh-lacZ)(Hyb) fusion gene preceded by the RBS of crh | This work |
| pBGM31 | Bacillus deletion vector; cat followed by rrnBt1t2-λt0 inserted in EcoRI/ClaI of pBluescript SK− | This work |
| pBGM55 | Pspac in front of lacZ in pAC6 | This work |
| pBGM84 | glcT deletion vector; same as pBGM31 but cat rrnBt1t2-λt0 cassette encompassed by glcT 3′ region (+889 to +396) and glcT upstream region (−17 to −599) | This work |
| pLF1 | pMutin4 containing a Φ(ptsH-lacZ)(Hyb) fusion gene preceded by the RBS of ptsH | This work |
| pLF2 | pMutin4 containing a 365-bp fragment of ptsH-5′ region (−276 to +90) in MCS | This work |
| Strains | ||
| E. coli | ||
| BMH71-18 | Δ(lac-pro) thi supE F′[lacIqlacZΔM15 lacY+pro+] | 6 |
| DH5α | gyrA96 recA1 relA1 endA1 thi-1 hsdR17 glnV44 deoR Δ(lacZYA-argF)U169 [φ80dΔ(lacZ)M15] | 22 |
| B. subtilis | ||
| 168 | trpC2 | 1 |
| SG17 | trpC2 crh′::lacZ ery (::pMutin4) | pBGM6→168 |
| SG19 | trpC2 Φ(crh-lacZ)(Hyb) ery (::pMutin4) | pBGM8→168 |
| SG57 | trpC2 Φ(ptsH-lacZ)(Hyb) ery (::pMutin4) | pLF1→168 |
| SG58 | trpC2 ptsH′::lacZ ery (::pMutin4) | pLF2→168 |
| SG68 | trpC2 amyE::[Pspac-lacZ cat] | pBGM55/ScaI→168 |
| SG87 | trpC2 Φ(ptsH-lacZ)(Hyb) ery (::pMutin4) ΔglcT(−16 to +395)::cat | pBGM84/ScaI→SG57 |
| SG88 | trpC2 ptsH′::lacZ ery (::pMutin4) ΔglcT(−16 to +395)::cat | pBGM84/ScaI→SG58 |
MCS, multiple cloning site. Positions are relative to the initiation codon of the respective gene.
Construction of transcriptional and translational fusions of crh and ptsH to lacZ.
To construct the fusion of lacZ to crh-5′, the 5′ region of crh (−120 to +90) was amplified by using the primers BG9 (crh [−120 to −103]) and BG10 (crh [+90 to +73]), digested at the HindIII and BamHI sites within the primers, and inserted between these sites in pMutin4, resulting in plasmid pBGM6. To construct the fusion of lacZ to ptsH-5′, the 5′ region of ptsH (−276 to +90) was amplified by using the primers LF1 (ptsH [−276 to −258]) and LF2 (ptsH [+90 to +73]), digested at the NotI and HindIII sites within the primers, and inserted between these sites in pMutin4, resulting in plasmid pLF2. Plasmid pBGM8 carrying a Φ(crh-lacZ)(Hyb) fusion gene was constructed by a three-fragment ligation. The entire crh gene with its ribosome binding site (RBS) was amplified by using the primers BG13 (crh [−20 to −3]) and BG14 (crh [+255 to +238]) and digested at the HindIII and XhoI sites within the primers. In parallel, the 5′ part of lacZ was amplified from pMutin4 as a template by using the primers BG15 (pMutin2 [382 to 399]) and BG16 (pMutin2 [698 to 681]) and digested with XhoI (located within primer BG15) and MstII (occurring naturally within lacZ). The two digested PCR fragments were ligated to the MstII-HindIII vector backbone of pMutin4 (lacking the RBSspoVG and the lacZ-5′ region). A CTC codon (Leu) was inserted together with the XhoI site between the crh and the lacZ parts of the fusion gene, and the Val initiation codon of lacZ had been removed. To obtain the isogenic construct pLF1, which carries a Φ (ptsH-lacZ)(Hyb) fusion gene instead of the Φ(crh-lacZ)(Hyb) fusion, the ptsH gene was amplified with primers LF3 (ptsH [−20 to −3]) and LF4 (ptsH [+264 to +247]), digested at the HindIII and XhoI sites within the primers, and used to replace the HindIII-XhoI fragment (encompassing crh) in pBGM8. The recombinant plasmids described above were used to transform B. subtilis 168 by a single crossover event by selection for erythromycin-resistant transformants. The resulting strains carried the fusions to lacZ described above in the respective chromosomal loci. For each strain, the correct structure of the plasmid insertion was verified by a set of seven different PCRs according to the strategy published by Pragai and Harwood (13).
Construction of the glcT Δ(−16 to +395) deletion mutants.
First, the generalized deletion vector pBGM31 was constructed by a three-fragment ligation. The cat gene from plasmid pAC5 (9) was isolated as an EcoRI-EheI fragment (1,096 bp). In parallel, a fragment (311 bp) encompassing the three transcriptional terminators rrnBt1t2-λt0 was isolated from pMutin2 by SmaI and PspI digestion. These fragments were ligated to the ClaI-EcoRI vector backbone of pBluescript SK−. The resulting plasmid, pBGM31, thus carries a cat-rrnBt1t2-λt0 cassette encompassed upstream by a SacI-SacII-NotI-SpeI-SmaI-BamHI multiple cloning site and downstream by a SalI-XhoI-ApaI-KpnI multiple cloning site. Thereafter, the glcT-3′ region (+396 to +889) was amplified with primers BG78 (glcT [+396 to +413]) and BG79 (glcT [+889 to +872]), digested at the BamHI and SacII sites within the primers, and inserted between these sites in plasmid pBGM31, resulting in the intermediate plasmid pBGM83. Next, the region upstream of glcT (−599 to −17) was amplified with the primers BG76 (glcT [−599 to −581]) and BG77 (glcT [−17 to −35]), digested at the ApaI and XhoI sites within the primers, and inserted between these sites in pBGM83. The resulting construct, pBGM84, was linearized by ScaI digestion and used to transform strains SG57 and SG58 by a double crossover with selection for chloramphenicol resistance. In the resulting strains, SG87 and SG88, the 5′ part of glcT (−16 to +395) was deleted and replaced by the cat-rrnBt1t2-λt0 cassette reading in the opposite direction of glcT transcription.
Construction of a control strain carrying a Pspac-lacZ cassette inserted in amyE.
A control strain (SG68) carrying a constitutively expressed Pspac-lacZ cassette on the chromosome was constructed. The Pspac promoter was amplified with primers BG61 (pMutin2 [143 to 161]) and BG23 (pMutin2 [428 to 406]) from pMutin2 (19) as a template, digested with MunI (site located within primer BG61) and BamHI (site located downstream of the Pspac promoter in pMutin2), and inserted between the EcoRI and BamHI sites located in front of lacZ in pAC6. The resulting plasmid, pBGM55, was linearized by ScaI digestion and used to transform B. subtilis 168 by a double crossover with the amyE gene with selection for chloramphenicol resistance.
β-Galactosidase assays.
Overnight cultures of the B. subtilis strains grown in C minimal medium (9) supplemented with l-tryptophan (100 μg/ml), potassium glutamate (8 g/liter), and one of the various carbon sources (0.5%, wt/vol) as indicated in Tables 2 and 3 were inoculated at an initial optical density at 600 nm (OD600) of 0.15 in the same medium. IPTG (isopropyl-β-d-thiogalactopyranoside) (1 mM) was added for induction of the Pspac promoter. The cultures were grown at 37°C with agitation (160 rpm), and cells were harvested when the cultures reached an OD600 of 0.6 to 0.8. β-Galactosidase activities were determined as described by Miller (11), using lysozyme and Triton X-100 treatment (12). The untransformed strain (B. subtilis 168) yielded values of less than 1 U. Pilot experiments were performed in which the β-galactosidase activities were determined at different time points during growth on succinate (lowest growth rate) and glucose (highest growth rate). These experiments revealed that a steady-state level of β-galactosidase activities was reached with all strains used when the cultures were in the OD600 range of 0.6 to 0.8. In order to test whether growth conditions (i.e., utilization of different carbon sources) may directly affect β-galactosidase activities, we used the control strain SG68. The β-galactosidase activities (in Miller units) obtained for SG68 were as follows: 18 for citrate, succinate, and gluconate; 23 for myo-inositol; 25 for fructose and glucose; 26 for malate; 27 for sucrose; 28 for mannitol; 29 for glucitol; and 30 for glycerol. Thus, the various growth conditions had only a marginal effect on expression or activity of β-galactosidase in our assays.
TABLE 2.
β-Galactosidase activities of strains carrying transcriptional fusions of crh or ptsH to lacZa
| Carbon source | β-Galactosidase activityb
|
β-Galactosidase activity ratio
|
|||
|---|---|---|---|---|---|
| crh′::lacZ (SG17) | ptsH′::lacZ (SG58) | ptsH′::lacZ ΔglcT (SG88) | ptsH′:: lacZ/crh′::lacZ | ptsH′::lacZ glcT+/ΔglcT | |
| Citrate | 44 | 42 | 40 | 1.0 | 1.1 |
| Succinate | 40 | 43 | 39 | 1.1 | 1.1 |
| myo-Inositol | 29 | 51 | 52 | 1.8 | 1.0 |
| Glycerol | 33 | 52 | 51 | 1.6 | 1.0 |
| Malate | 36 | 53 | 53 | 1.5 | 1.0 |
| Gluconate | 42 | 66 | 61 | 1.6 | 1.1 |
| Mannitol | 36 | 91 | 59 | 2.5 | 1.5 |
| Fructose | 33 | 91 | 57 | 2.8 | 1.6 |
| Sucrose | 33 | 254 | 66 | 7.7 | 3.9 |
| Glucose | 29 | 268 | 44 | 9.2 | 6.1 |
Bacteria were grown in minimal medium supplemented with the indicated carbon source (0.5%, wt/vol) to an OD600 of 0.6 to 0.8. For induction of expression of genes downstream of the plasmid insertion sites, 1 mM IPTG was added.
β-Galactosidase activities are expressed in Miller units, and each value is the average of three or four measurements of at least two independent cultures.
TABLE 3.
β-Galactosidase activities of strains carrying translational fusions of crh or ptsH to lacZa
| Carbon source | β-Galactosidase activity
|
β-Galactosidase activity ratio
|
|||
|---|---|---|---|---|---|
| Φ(crh-lacZ) (Hyb) (SG19) | Φ(ptsH-lacZ) (Hyb) (SG57) | Φ(ptsH-lacZ) (Hyb) ΔglcT (SG87) | Φ(ptsH-lacZ)/Φ(crh-lacZ) | Φ(ptsH-lacZ) glcT+/ΔglcT | |
| Citrate | 103 | 940 | 872 | 9 | 1.1 |
| Succinate | 94 | 988 | 867 | 11 | 1.1 |
| myo-Inositol | 49 | 988 | 1,225 | 20 | 0.8 |
| Glycerol | 38 | 1,130 | 1,126 | 30 | 1.0 |
| Malate | 38 | 1,215 | 1,163 | 32 | 1.0 |
| Gluconate | 59 | 1,465 | 1,312 | 25 | 1.1 |
| Mannitol | 40 | 1,884 | 1,097 | 47 | 1.7 |
| Fructose | 46 | 1,855 | 1,191 | 40 | 1.6 |
| Sucrose | 39 | 3,150 | 1,307 | 81 | 2.4 |
| Glucose | 34 | 3,534 | 1,022 | 104 | 3.5 |
See Table 2.
RESULTS AND DISCUSSION
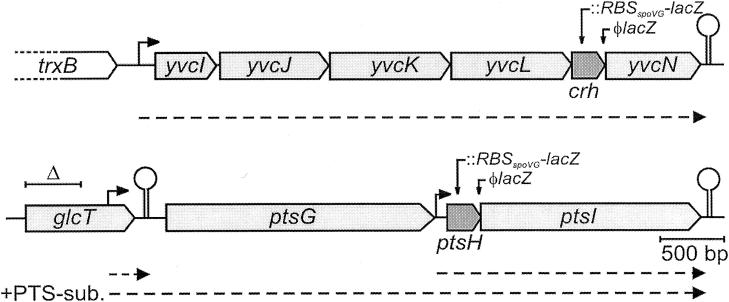
In order to compare the transcription rates, we constructed crh′::lacZ and ptsH′::lacZ transcriptional fusions. The resulting strains carry an RBSspoVG-lacZ reporter cassette starting 90 bp downstream of the first base of the respective open reading frame (ORF) at their natural chromosomal loci (Fig. 1). In these constructs, the complete ORFs of crh and ptsH are restored downstream of the lacZ cassette insertion sites. Their expression is directed by the IPTG-inducible Pspac promoter present in the inserted cassette in order to overcome polar effects caused by the insertions. The bacteria were grown in minimal medium supplemented with different carbohydrates, and β-galactosidase activities were determined (Table 2). The activities produced by the strain carrying the crh′::lacZ fusion varied only slightly with the carbon source (29 to 44 U) (Table 2). In contrast, the strain carrying the ptsH′::lacZ fusion showed remarkable differences in activities. High activities (91 to 268 U) were detectable when a substrate of the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) (mannitol, fructose, sucrose, or glucose) was the carbon source, whereas significantly lower activities (42 to 66 U) were obtained with the other carbohydrates, none of which is a substrate of the PTS (Table 2). In conclusion, transcription of ptsH relative to that of crh was found to be three- to ninefold stronger when a PTS substrate is utilized (Table 2). Control experiments (see Materials and Methods) excluded the possibility that the different growth rates may affect the β-galactosidase activities (21).
FIG. 1.
Organization of the crh- and ptsH-containing operons. The gene crh is part of an operon composed of six ORFs which are cotranscribed from a promoter located in front of the gene yvcI (reference 4 and our unpublished data). The gene ptsH can be transcribed from two promoters, one directly in front of ptsH and another in front of the preceding ptsG gene. The hairpin structures correspond to transcriptional terminators. The promoters are indicated by arrows and the corresponding RNAs are indicated by dotted lines, according to previous data (4, 5, 17) and this study. The positions of the transcriptional and translational fusions of lacZ to crh and ptsH, respectively, and of the glcT deletion (−16 to +395) constructed in this study are indicated.
Thereafter, we aimed to compare HPr and Crh synthesis levels. We therefore constructed translational in-frame fusions of the entire crh and ptsH genes to lacZ and placed them at their natural chromosomal loci preceded by their natural RBSs. The strain which carries the chimeric Φ (crh-lacZ)(Hyb) fusion gene yielded β-galactosidase activities that varied only slightly with the carbon source, with the exceptions of citrate and succinate (Table 3). These substrates caused twofold-higher activities than the other tested substrates. The same effect, although less pronounced, was seen with the transcriptional crh′::lacZ fusion (Table 2). The strain which carries the Φ (ptsH-lacZ)(Hyb) fusion gene produced very high activities (940 to 3,534 U) (Table 3). This indicates that ptsH is approximately 10- to 20-fold more efficiently translated than crh, which is in agreement with previous observations suggesting that HPr is synthesized to very large cellular amounts (18). Indeed, the RBS of ptsH is very similar to the optimal B. subtilis RBS (14), whereas the RBS of crh matches the consensus less well (Fig. 2). The activities produced by the Φ (ptsH-lacZ)(Hyb) fusion were significantly higher when the bacteria were grown on a PTS substrate (1,855 to 3,534 U) than when they were grown on a non-PTS carbohydrate (940 to 1,465 U). This confirms our observation that utilization of a PTS substrate stimulates ptsH transcription (Table 2). The combined effects of stimulated transcription and highly efficient translation of ptsH amplify the differences in HPr and Crh synthesis levels up to 100-fold (Table 3).
FIG. 2.
Comparison of the RBSs of ptsH and crh. The optimal B. subtilis RBS is as described by Rocha et al. (14).
What may be the mechanism that accounts for the stimulatory effect of PTS substrates on ptsH transcription? The genes ptsH and ptsI are cotranscribed from a promoter located directly in front of ptsH (Fig. 1). This promoter is constitutively active and not subject to regulation by the carbon source (5). In the presence of glucose, ptsHI are, in addition, cotranscribed with the preceding gene ptsG (encoding the glucose-specific enzyme II [EIIGlc]) from a promoter located in front of ptsG (17). Induction of ptsG expression by glucose is mediated by antiterminator protein GlcT, which is in turn regulated by (de)phosphorylation via EIIGlc (2, 16). Upon transport of glucose, EIIGlc dephosphorylates and activates GlcT, which then prevents formation of a terminator present in the ptsG leader mRNA that otherwise blocks transcription elongation (8). This mechanism explains the high ptsH transcription rates that we observed in the presence of glucose (Table 2). Furthermore, it was shown that the expression of ptsG is also inducible by sucrose (17) and that this up-regulation is mediated by GlcT (8). To clarify whether cotranscription with ptsG also accounts for the higher ptsH transcription rates in the presence of various PTS substrates other than glucose, we deleted the left half of the glcT gene (−16 to +395) in the strains carrying the transcriptional and translational ptsH-lacZ fusions. This deletion abolishes GlcT antitermination activity but leaves the ptsG promoter-leader region unaffected (Fig. 1). As can be seen from the data (Tables 2 and 3), the stimulatory effect of the various PTS substrates was abolished in the ΔglcT strains. In contrast, the activities detected in the presence of the various non-PTS substrates were not affected by the ΔglcT deletion. These results establish (i) that in the absence of PTS substrates, ptsH is expressed exclusively from the promoter directly in front of it, (ii) that additional cotranscription with ptsG causes higher ptsH transcription rates in the presence of a PTS substrate in general, and (iii) that this cotranscription is mediated by antiterminator GlcT.
In conclusion, our data suggest that the highly efficient translation of ptsH and the increased transcription rate in the presence of PTS carbohydrates, i.e., under conditions of strong CCR, cause a higher cellular concentration of HPr than of Crh. These drastic differences in synthesis level, up to 100-fold, may explain why in previous studies (3, 4, 10) only a minor role of Crh in CCR was detected. Recently it was reported that citM, encoding a Mg2+-citrate transporter, is specifically repressed by Crh in a medium composed of glutamate, succinate, and citrate (20). Here we found that the differences in Crh and HPr synthesis levels are lowest (approximately 10-fold) when the bacteria utilize citrate or succinate (Table 3). This observation is in agreement with the proposal (20) for a specific role of Crh under these conditions.
Acknowledgments
This research was supported by the CNRS, the Université d'Aix-Marseille II, and the Ministère de la Recherche “ACI-jeunes-chercheurs.”
We are grateful to F. Denizot for critical reading of the manuscript and to J. Stülke for gifts of plasmids and for helpful discussion.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachem, S., and J. Stülke. 1998. Regulation of the Bacillus subtilis GlcT antiterminator protein by components of the phosphotransferase system. J. Bacteriol. 180:5319-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galinier, A., J. Deutscher, and I. Martin-Verstraete. 1999. Phosphorylation of either Crh or HPr mediates binding of CcpA to the Bacillus subtilis xyn cre and catabolite repression of the xyn operon. J. Mol. Biol. 286:307-314. [DOI] [PubMed] [Google Scholar]
- 4.Galinier, A., J. Haiech, M. C. Kilhoffer, M. Jaquinod, J. Stülke, J. Deutscher, and I. Martin-Verstraete. 1997. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl. Acad. Sci. USA 94:8439-8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzy-Tréboul, G., M. Zagorec, M. C. Rain-Guion, and M. Steinmetz. 1989. Phosphoenolpyruvate:sugar phosphotransferase system of Bacillus subtilis: nucleotide sequence of ptsX, ptsH and the 5′-end of ptsI and evidence for a ptsHI operon. Mol. Microbiol. 3:103-112. [DOI] [PubMed] [Google Scholar]
- 6.Kramer, W., V. Drutsa, H. W. Jansen, B. Kramer, M. Pflugfelder, and H. J. Fritz. 1984. The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res. 12:9441-9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langbein, I., S. Bachem, and J. Stülke. 1999. Specific interaction of the RNA-binding domain of the Bacillus subtilis transcriptional antiterminator GlcT with its RNA target, RAT. J. Mol. Biol. 293:795-805. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Verstraete, I., M. Debarbouille, A. Klier, and G. Rapoport. 1990. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J. Mol. Biol. 214:657-671. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Verstraete, I., J. Deutscher, and A. Galinier. 1999. Phosphorylation of HPr and Crh by HprK, early steps in the catabolite repression signalling pathway for the Bacillus subtilis levanase operon. J. Bacteriol. 181:2966-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Nicholson, W. L., and P. Setlow. 1990. Genetic analysis, p. 443. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley, Chichester, United Kingdom.
- 13.Pragai, Z., and C. R. Harwood. 2000. YsxC, a putative GTP-binding protein essential for growth of Bacillus subtilis 168. J. Bacteriol. 182:6819-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocha, E. P., A. Danchin, and A. Viari. 1999. Translation in Bacillus subtilis: roles and trends of initiation and termination, insights from a genome analysis. Nucleic Acids Res. 27:3567-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Schmalisch, M. H., S. Bachem, and J. Stülke. 2003. Control of the Bacillus subtilis antiterminator protein GlcT by phosphorylation: elucidation of the phosphorylation chain leading to inactivation of GlcT. J. Biol. Chem. 278:51108-51115. [DOI] [PubMed] [Google Scholar]
- 17.Stülke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65-78. [DOI] [PubMed] [Google Scholar]
- 18.Vadeboncoeur, C., M. Frenette, and L. A. Lortie. 2000. Regulation of the pts operon in low G+C Gram-positive bacteria. J. Mol. Microbiol. Biotechnol. 2:483-490. [PubMed] [Google Scholar]
- 19.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 20.Warner, J. B., and J. S. Lolkema. 2003. A Crh-specific function in carbon catabolite repression in Bacillus subtilis. FEMS Microbiol. Lett. 220:277-280. [DOI] [PubMed] [Google Scholar]
- 21.Warner, J. B., and J. S. Lolkema. 2002. LacZ-promoter fusions: the effect of growth. Microbiology 148:1241-1243. [DOI] [PubMed] [Google Scholar]
- 22.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]