Abstract
Aim:
Cytochrome P450 2C9 (CYP2C9) is a polymorphic enzyme that is responsible for the metabolism of approximately 15% of clinically important drugs. The aim of this study was to assess the catalytic characteristics of 37 CYP2C9 allelic isoforms found in Chinese Han population on the metabolism of tolbutamide in vitro.
Methods:
The wild-type and 36 CYP2C9 variants were expressed in sf21 insect cells using a baculovirus-mediated expression system. Then the insect microsomes were prepared for assessing the metabolic characteristics of each variant toward the CYP2C9-specific drug substrate tolbutamide.
Results:
Of 36 allelic variants tested, the intrinsic clearance values of 2 allelic isoforms (CYP2C9.36 and CYP2C9.51) were much higher than the wild-type CYP2C9.1 protein, 3 allelic isoforms (CYP2C9.11, CYP2C9.56 and N418T) exhibited similar intrinsic clearance values as the wild-type enzyme, whereas the other 31 variants showed significantly reduced intrinsic clearance values, ranging from 0.08% to 66.88%, for tolbutamide.
Conclusion:
Our study provides the most comprehensive data concerning the enzymatic activity of the CYP2C9 variants that are present in the Chinese Han population, and our data suggest that most of the carriers of these alleles might be paid more attention when using CYP2C9 mediated drugs clinically.
Keywords: CYP2C9, allelic variants, drug metabolism, tolbutamide, Chinese Han population
Introduction
As one of the major hepatic drug-metabolizing enzymes, cytochrome P450 (CYP) 2C9 is estimated to metabolize approximately 15% of the drugs that undergo phase I metabolism, including a number of drugs with narrow therapeutic indexes1,2. Similar to other superfamily members, the gene encoding the CYP2C9 enzyme is highly polymorphic, including functional variants of major pharmacogenetic importance, which are of major clinical importance. To date, 57 CYP2C9 alleles have been identified and named by the Human CYP Allele Nomenclature Committee (http://www.cypalleles.ki.se/cyp2c9.htm). Most of those alleles, especially for CYP2C9*2 (430C>T) and CYP2C9*3 (1075A>C), exhibited decreased enzymatic activity toward certain typical CYP2C9 substrates, such as diclofenac, tolbutamide, warfarin, phenytoin and losartan, in vitro and/or in vivo3,4.
Increasing evidences suggest that there are significant ethnic differences in the frequency of CYP2C9 variant occurrence. Two dominant variants, CYP2C9*2 and CYP2C9*3, are common in Caucasians at frequencies of approximately 8%–14% and 4%–16%, respectively, while significantly lower frequencies of these variants were found in African and Asian populations4,5. A large-scale genetic polymorphism study revealed that approximately 40% of Europeans carry these two mutated alleles6. In contrast, the *2 variant is rarely found, and the *3 variant is present in only at 1%–4% of East Asian populations7,8, and our previous study demonstrated that only 0.28% and 5.6% of Chinese people carry the CYP2C9*2 or CYP2C9*3 allele, respectively9.
Thus far, most of the CYP2C9 alleles are first identified in Asian populations, and the majority of these variants exhibit decreased enzymatic activity toward CYP2C9-specific substrates in vitro7,10,11,12,13,14. In a recent study, as many as 21 novel alleles (*36 to *56) were reported by our laboratory for the first time after screening 2127 healthy Chinese subjects, and 17 of the newly identified variants significantly influenced the protein's catalytic activity toward luciferin H and diclofenac in transfected COS-7 cells9. However, because of the low CYP2C9 expression levels, it is difficult to isolate enough CYP2C9 protein to determine the comprehensive catalytic characteristics of all the detected variants in a mammalian cell expression system. In this study, we highly expressed the CYP2C9 proteins in insect cells using baculovirus, and systematically analyzed the enzymatic activity of 37 CYP2C9 allelic isoforms that are present in the Chinese Han population toward the typical CYP2C9 phenotyping probe substrate tolbutamide in vitro. Our results demonstrated that 33 variants exhibited intrinsic clearance values toward tolbutamide that were significantly different than the wild-type protein. These data suggest that most of the carriers of these infrequently occurring alleles in China might be given different CYP2C9 substrate amounts orally in clinics than wild-type protein-expressing patients.
Materials and methods
Chemicals and material
Spodoptera frugiperda (Sf)21 insect cells, Sf-900™ III SFM insect culture medium, Antibiotic-Antimycotic, fetal bovine serum and the Bac-to-Bac Baculovirus Expression System were purchased from Invitrogen (Carlsbad, CA, USA). Restriction enzymes were purchased from New England BioLabs (Beverly, MA, USA). Baculosomes co-expressing human CYP2C9 and NADPH-cytochrome P450 oxidoreductase (OR) or cytochrome b5 and OR were purchased from BD Gentest (Woburn, MA, USA). Rabbit polyclonal anti-CYP2C9 antibody was from AbD Serotec (Kidlington, Oxford, UK). The mouse monoclonal anti-OR antibody was from Santa Cruz Biotechnology (Dallas, TX, USA). The Super Signal West Pico Trial kit was obtained from Thermo (Rockford, IL, USA). Tolbutamide was purchased from Sigma-Aldrich (St Louis, MO, USA). 4-Hydroxytolbutamide was purchased from Toronto Research Chemicals Inc (Toronto, Ontario, Canada). Chlorpropamide was from Tokyo Chemical Industry Co, Ltd (Tokyo, Japan). The NADPH-regenerating system was from Promega (Madison, WI, USA). High-pressure liquid chromatography-grade solvents were purchased from Fisher Scientific Co (Fair Lawn, NJ, USA). All of the other chemicals and solvents that were used were of the highest grade or analytical grade that was commercially available.
Recombinant expression vector construction
A full-length human cDNA clone (Cat No SC100401) for the OR gene was purchased from Origene (Rockville, MD, USA). The ORF region of OR was then isolated by PCR amplification and inserted into the cloning sites downstream of the P10 promoter in the dual-expression baculovirus vector pFastBac Dual (Invitrogen, Carlsbad, CA, USA) to create the receptor vector pFastBac-OR.
ORF fragments from CYP2C9 *1, *2, *3, *13, *36–*56, and N418T variants were isolated from previously reported vectors using 2C9-EF (5′-GCCTGAATTCATGGATTCTCTTGTGGT-3′, introducing one EcoR I site) and 2C9-SR (5′-GAACGTCGACTCAGACAGGAATGAAGCA-3′, introducing one Sal I site) primer pairs. Other CYP2C9 variants were constructed by the overlap-extension PCR amplification method using the primers that are listed in Supplementary Table S1, as has been previously described9. Full-length PCR products for each of the variants were digested and ligated to the pFastBac-OR receptor vector to produce the ultimate dual-expression baculovirus vector pFastBac-OR-CYP2C9. To ensure that no errors were introduced during PCR amplification, all of the cDNA regions were confirmed by sequencing the plasmid constructs using the CEQ DTCS Quick Start Kit (Beckman Coulter, Inc, Brea, CA, USA) on the CEQ 8000 Genetic Analysis System.
Sf21 insect cells infection and microsomal fraction preparation
Recombinant baculoviruses were effectively generated in Sf21 insect cells according to the manufacturer's procedure. Sf21 cells were then infected with these viruses at a multiplicity-of-infection of 4 in Sf-900™ III SFM insect culture medium supplemented with 10% fetal bovine serum, 1×Antibiotic-Antimycotic and 4 μg/mL hemin. After 72 h of transfection, the cells were harvested and re-suspended in 0.1 mol/L phosphate buffer, pH 7.4 containing 0.25 mol/L sucrose, 1 mmol/L EDTA and 0.5 mmol/L PMSF. After centrifugation, the pellets were re-suspended, lysed by sonication for 50 s on ice and centrifuged at 14 000×g for 20 min at 4 °C to remove the precipitate. Then, the membrane fraction was prepared by ultra-centrifugation at 100 000×g for 1 h at 4 °C, and this pellet was re-suspended in stock solution (0.1 mol/L KPO4, pH 7.4, containing 20% glycerol) and stored at -80 °C before use.
Protein expression level determination
Western blotting was performed to determine CYP2C9 and OR protein expression levels as has been described previously9. Briefly, 2 μg microsomal protein was applied to 8% SDS-PAGE gels and electro-transferred onto PVDF membranes. Rabbit polyclonal anti-CYP2C9 and mouse monoclonal anti-OR antibodies (both at a 1:5000 dilution) were used as of CYP2C9 and OR primary antibodies for immunostaining, respectively. The signal was visualized with the Super Signal West Pico Trial kit, and the cytochrome P450 or OR content in the insect cell microsomes was quantified using a commercially available microsomal protein standard with ImageJ software (National Institutes of Health, Bethesda, MA, USA).
Conditions for analyzing enzymatic activity
Reaction mixtures contained 10 to 20 pmol CYP2C9 from insect microsomes, 20 to 40 pmol cytochrome b5 (CYP2C9/b5=1:2), and 10 to 1000 μmol/L tolbutamide in a final volume of 200 μL in 100 mmol/L Tris-HCl buffer (pH 7.5). The reactions were pre-incubated for 5 min and a NADPH-regeneration system was added to start the reaction at 37 °C and proceed for 60 min. The incubation condition has been optimized to ensure that the metabolic reaction is within the linear range of the given incubation time and protein concentration. The reactions were terminated by adding 40 μL 0.1 mol/L HCl and 50 μL 20 ng/μL chlorpropamide as an internal standard. The reaction samples were extracted with 800 μL ethyl acetate, and the organic layer was evaporated to dryness and reconstituted in 100 μL mobile phase.
Liquid chromatography-mass spectrometry analysis was performed using an LCMS/MS 1260-6410 (Agilent), and sample aliquots were applied onto a ZORBAX SB-C18 column (150 mm×4.6 mm, id 5 μm, Agilent Technologies, Palo Alto, CA, USA) that had been kept at 40 °C. The initial mobile phase was 60% H2O with 0.1% formic acid and 40% acetonitrile, and the proportion of acetonitrile linearly increased to 55% from 1.5 min to 1.6 min and kept for 2.4 min, and then the acetonitrile proportion was linearly decreased to 40% from 4 min to 4.1 min and kept unchanged till 6 min with a 1 mL/min flow rate. The quadrupole mass spectrometer was operated in the positive atmospheric pressure ionization-electrospray ionization mode using the following selected ion monitoring conditions: 350 °C gas temperature; 12 L/min gas flow rate. The MS/MS transitions and fragmentation conditions that were selected for the individual analytes are shown in Supplementary Table S2. Under these conditions, 4-Hydroxytolbutamide, tolbutamide, and chlorpropamide were eluted at 2.487, 4.487, and 4.138 min, respectively.
Statistical analysis
The kinetic parameters Km and Vmax were estimated using a software program designed for non-linear regression analysis using the hyperbolic Michaelis-Menten equation (Prism version 5, GraphPad Software, San Diego, CA, USA). The intrinsic clearance value of each variants is calculated as following formula: Clint=Vmax/Km. Kinetic data for each variant are presented as the mean±SD for three microsomal preparations that were derived from separate transfections and analyzed by one-way analysis of variance.
Results
Expression of wild-type CYP2C9 protein and 36 variants in insect cell microsomes
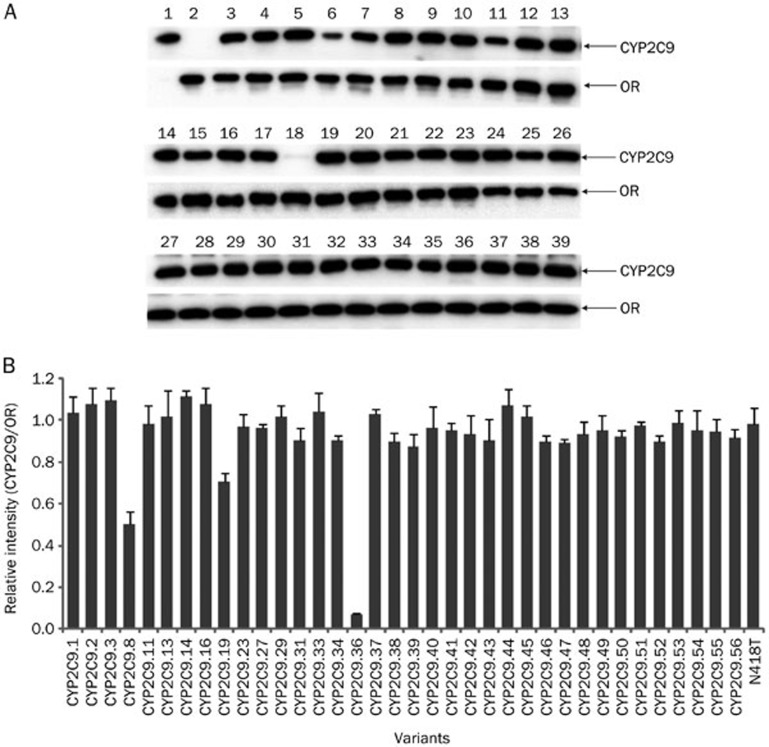
To obtain enough proteins for in vitro enzymatic activity analysis, full-length cDNAs encoding both cytochrome P450 reductase (OR) and wild-type or variant CYP2C9 protein were inserted into the dual-expression baculovirus transfer vector and used for simultaneous expression of the recombinant proteins in Sf21 insect cells. Then, the insect cell microsomes were extracted and immunoblotted to determine protein expression levels. As shown in Figure 1, OR protein expression levels were similar between the wild-type and the 36 variants that were assessed. However, protein expression levels of 3 allelic variants (CYP2C9.8, CYP2C9.19, and CYP2C9.36) were much lower than the wild-type enzyme CYP2C9.1 (P<0.05 for CYP2C9/OR by one-way ANOVA analysis), especially for the initiation codon mutation variant CYP2C9.36, and for another previously studied variant CYP2C9.8.
Figure 1.
Expression of wild-type CYP2C9 and 36 variants in insect cell microsomes. (A) Representative Western blottings for the CYP2C9 and OR proteins. Lane 1, commercially available Baculosomes co-expressing CYP2C9.1 (with high-expressied level of CYP2C9 and slightly expressed OR protein) and OR (BD Gentest); Lane 2, microsomes containing only OR; Lanes 3 to 39, microsomes co-expressing OR and CYP2C9.1, CYP2C9.2, CYP2C9.3, CYP2C9.8, CYP2C9.11, CYP2C9.13, CYP2C9.14, CYP2C9.16, CYP2C9.19, CYP2C9.23, CYP2C9.27, CYP2C9.29, CYP2C9.31, CYP2C9.33, CYP2C9.34, CYP2C9.36–CYP2C9.56, and N418T variants. (B) Relative CYP2C9/OR protein intensities. Each bar represents the mean±SD of three independent experiments.
Functional characterization of wild-type and 36 allelic CYP2C9 variants activity toward tolbutamide
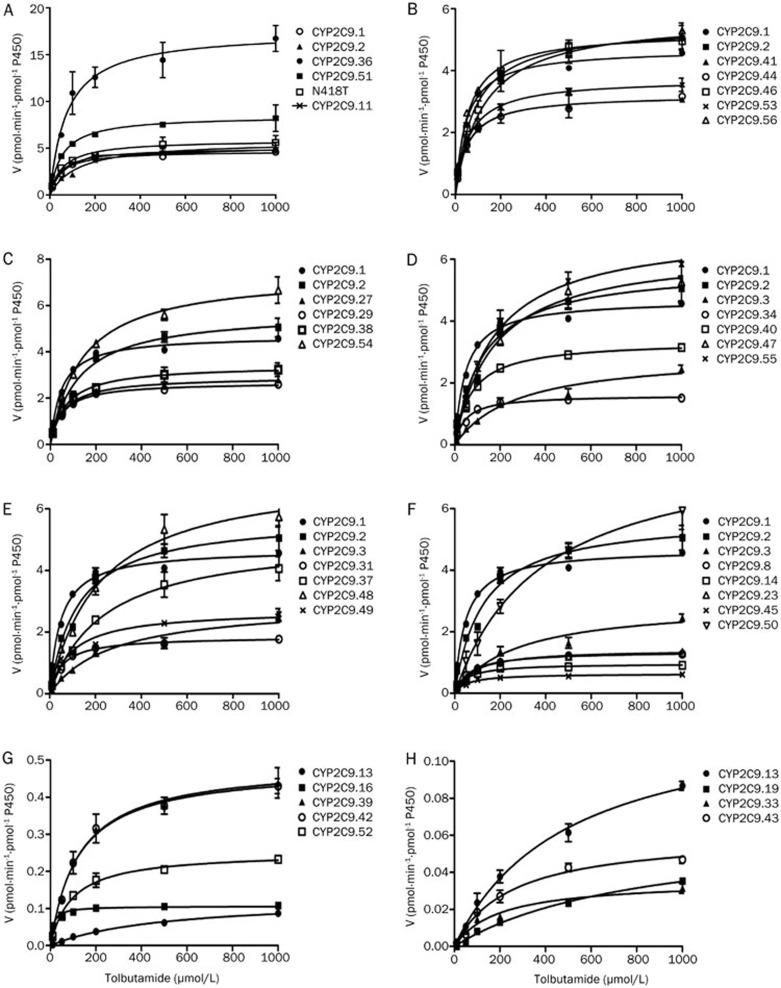
To determine the Michaelis-Menten kinetic parameters of the 37 reported CYP2C9 allelic isoforms in Chinese Han populations, one typical CYP2C9-specific substrate, tolbutamide, was chosen as the phenotypic analysis probe for in vitro protein enzymatic activity assays. As demonstrated in Figure 2 and Table 1, almost all of the variants exhibited changed Km or Vmax values compared with the values that were obtained for the wild-type protein, and only 3 of the variants (CYP2C9.11, CYP2C9.56, and N418T) exhibited similar intrinsic clearance (Clint) values as the wild-type enzyme. Specifically, the intrinsic clearance values of 2 allelic isoforms were much higher than the wild-type CYP2C9.1 protein; 8 variants had values that were lower than CYP2C9.1 but higher than the prototypical, moderately defective variant CYP2C9.2; 13 variants exhibited values that were lower than CYP2C9.2 but higher than another prototypical defective variant, CYP2C9.3; and 8 variants possessed the lowest intrinsic clearance values (Table 1).
Figure 2.
Michaelis-Menten curves for the enzymatic activity of recombinant wild-type CYP2C9 protein and 36 variants (each point represents the mean±SD of three or four separate experiments) towards tolbutamide. The variants were manually arranged into eight different groups with intrinsic clearance values: higher than wild-type (A), lower than wild-type, but higher than the moderately defective variant CYP2C9.2 (B, C), lower than the moderately defective variant CYP2C9.2, but higher than defective variant CYP2C9.3 (D, E, F), lower than the prototypical defective variant CYP2C9.3 (G, H).
Table 1. Kinetic values for tolbutamide activities of recombinant wild-type and mutant CYP2C9 proteins.
| Allelic protein | Vmax (pmol·min−1·pmol−1 P450) | Km (μmol/L) | Clearance (Vmax/Km) | Relative clearance*/.1 |
|---|---|---|---|---|
| CYP2C9.36 | 17.45±0.89b | 74.82±14.46b | (2.33±0.31)E-01b | 237.15%b |
| CYP2C9.51 | 8.44±0.30b | 54.17±8.13 | (1.56±0.33)E-01b | 158.50%b |
| N418T | 5.86±0.25b | 57.31±10.00 | (1.02±0.18)E-01 | 104.02% |
| CYP2C9.11 | 5.02±0.11 | 50.24±4.78 | (1.00±0.12)E-01 | 101.64% |
| CYP2C9.1 | 4.70±0.10 | 47.74±4.57 | (9.83±0.48)E-02 | 100.00% |
| CYP2C9.56 | 5.25±0.19 | 55.71±8.44 | (9.43±1.89)E-02 | 95.86% |
| CYP2C9.46 | 5.46±0.29 | 82.94±16.13b | (6.58±0.11)E-02b | 66.88%b |
| CYP2C9.53 | 3.73±0.08b | 59.98±5.55b | (6.21±0.03)E-02b | 63.17%b |
| CYP2C9.44 | 3.21±0.06b | 52.81±4.44 | (6.08±0.29)E-02b | 61.85%b |
| CYP2C9.41 | 3.22±0.14b | 54.08±10.25 | (5.95±0.72)E-02b | 60.51%b |
| CYP2C9.54 | 7.40±0.31b | 138.20±18.36b | (5.35±0.51)E-02b | 54.42%b |
| CYP2C9.29 | 2.69±0.08b | 52.00±6.32 | (5.17±0.20)E-02b | 52.60%b |
| CYP2C9.27 | 2.92±0.08b | 59.23±6.47b | (4.92±0.89)E-02b | 50.08%b |
| CYP2C9.38 | 3.42±0.15b | 69.77±11.85b | (4.89±0.12)E-02b | 49.77%b |
| CYP2C9.2 | 5.73±0.29 | 123.30±20.18b | (4.65±0.54)E-02b | 47.24%b |
| CYP2C9.40 | 3.36±0.11b | 74.07±9.27b | (4.53±0.09)E-02b | 46.08%b |
| CYP2C9.55 | 6.97±0.34b | 169.50±24.40b | (4.11±0.07)E-02b | 41.80%b |
| CYP2C9.47 | 6.32±0.48b | 168.30±37.76b | (3.75±0.63)E-02b | 38.16%b |
| CYP2C9.34 | 1.61±0.05b | 45.61±6.03 | (3.52±0.35)E-02b | 35.80%b |
| CYP2C9.31 | 1.85±0.05b | 54.71±6.15 | (3.38±0.41)E-02b | 34.40%b |
| CYP2C9.48 | 7.21±0.43b | 217.30±36.12b | (3.32±0.58)E-02b | 33.76%b |
| CYP2C9.49 | 2.71±0.15b | 95.14±18.46b | (2.85±0.09)E-02b | 28.96%b |
| CYP2C9.37 | 5.10±0.34 | 236.50±42.23b | (2.15±0.23)E-02b | 21.91%b |
| CYP2C9.50 | 8.19±0.73b | 383.00±79.38b | (2.14±0.47)E-02b | 21.75%b |
| CYP2C9.8 | 1.37±0.04b | 68.29±7.59b | (2.00±0.49)E-02b | 20.32%b |
| CYP2C9.14 | 0.97±0.03b | 51.68±6.91 | (1.87±0.26)E-02b | 19.03%b |
| CYP2C9.23 | 1.43±0.04b | 82.01±9.23b | (1.74±0.19)E-02b | 17.67%b |
| CYP2C9.45 | 0.64±0.03b | 54.66±11.38 | (1.17±0.050)E-02b | 11.90%b |
| CYP2C9.3 | 2.94±0.27b | 271.00±63.38b | (1.08±0.097)E-02b | 11.02%b |
| CYP2C9.16 | 0.11±0.0017b | 11.65±1.29b | (9.11±0.15)E-03b | 9.26%b |
| CYP2C9.42 | 0.48±0.02b | 122.20±20.44b | (3.94±0.92)E-03b | 4.01%b |
| CYP2C9.39 | 0.49±0.02b | 126.40±17.08b | (3.89±0.46)E-03b | 3.95%b |
| CYP2C9.52 | 0.25±001b | 94.72±12.41b | (2.66±0.51)E-03b | 2.70%b |
| CYP2C9.13 | 0.13±0.01b | 496.70±83.74b | (2.57±0.64)E-04b | 0.26%b |
| CYP2C9.43 | 0.06±0.0040b | 250.90±43.97b | (2.42±0.42)E-04b | 0.25%b |
| CYP2C9.33 | 0.04±0.0027b | 193.60±41.46b | (1.84±0.17)E-04b | 0.19%b |
| CYP2C9.19 | 0.06±0.01b | 827.00±151.70b | (7.74±0.89)E-05b | 0.08%b |
b Represent P<0.05 vs wild-type.
* Relative clearance value to wild-type.
Discussion
Thus far, several studies have been performed on CYP2C9 genetic polymorphisms in the Han populations of mainland China. However, only two groups focused on rare alleles in addition to two commonly investigated alleles, CYP2C9*2 and CYP2C9*3. One research group discovered a novel defective allele CYP2C9*1311, and the other group detected CYP2C9*8, CYP2C9*11, and CYP2C9*31, three rarely reported alleles15. Although the allelic frequencies of these rare alleles in the Chinese Han population are approximately 0.1%–1.8%, obtaining precise data regarding the other rare allele distribution in the Chinese population and exploiting the functional significance of these alleles is important and valuable in clinical practice because more than 1.3 billion people live in mainland China. In a recent study, we detected 14 previously reported alleles and 22 novel non-synonymous mutations after systematic genetic screening of the CYP2C9 gene in 2127 healthy Chinese subjects9. Combining all of the acquired data, 37 CYP2C9 allelic isoforms have been detected in the Chinese Han population, and functional characteristics of the majority of these variants have not yet been elucidated.
In our previous study, 17 novel CYP2C9 variants exhibited altered catalytic activity compared with the wild-type protein when expressed in mammalian COS-7 cells9. Although mammalian cells have sufficient endogenous NADPH-CYP oxidoreductase and cytochrome b5 to support CYP activities and are much closer to the native state of the CYP protein, it is difficult to obtain enough protein to study CYP catalytic activity in vitro with these cells3. So far, besides mammalian cells, bacteria, yeast and baculovirus-mediated insect cells have been successfully used for the high expression of recombinant CYP2C9 proteins in vitro. Baculovirus-mediated expression of mammalian CYP was first carried out in 1989 and has been widely used for the in vitro assessment of allelic variants of various CYPs, because high levels of expression can be obtained with 300–1000 pmol/g total cell lysate3. In the present study, we highly expressed all 37 of the previously reported CYP2C9 allelic isoforms in insect cells and found that 3 allelic variants (CYP2C9.8, CYP2C9.19, and CYP2C9.36) have lower protein expression levels than the other variants (Figure 1). In contrast, as many as 8 of the allelic variants had lower protein expression levels in COS-7 cells compared with wild-type CYP2C9.1. As previously reported, CYP2C9*36 has one nucleotide mutation (1 A>G) in the starting codon and this mutation can influence the protein translation efficiency in COS-7 cells9. In insect microsomes, relatively lower expressed proteins could be detected for variant CYP2C9.36 when compared with which in COS-7 cells. Herman et al reported a similar phenomenon for the CYP2C9.24 variant16. As Maekawa et al proposed, this discrepancy might be because of the different degradation rates of improperly folded proteins in insect and mammalian cell expression systems8.
Using tolbutamide as the phenotyping probe, 23 of the 36 expressed allelic variants exhibited decreased Vmax values compared with the wild-type protein, and 24 allelic variants exhibited significantly increased Km values compared with CYP2C9.1. Thus, 31 CYP2C9 variants exhibited a significant reduction in the intrinsic clearance of tolbutamide in vitro (Table 1). The in vitro metabolism characteristics of CYP2C9.2 toward tolbutamide have been well studied previously by two research groups. However, they obtained conflicting results by expressing the proteins in HepG2 cells17 or yeast18. Using the insect cell expression system, we demonstrated that *2, a prototypical defective allele, exhibited significantly reduced intrinsic clearance value for tolbutamide (approximately 47.2% of wild-type levels). For *3, another commonly utilized defective allele, several studies have been conducted in yeast18,19,20 or COS-7 cells21, but none have been performed in the insect cell expression system. Here, we obtained similar results to those that have been previously described, and nearly 10-fold lower intrinsic clearance of tolbutamide was exhibited by CYP2C9.3 compared with CYP2C9.1.
The CYP2C9.13 variant was first identified in a Chinese subject who was unable to efficiently metabolize lornoxicam and tolbutamide11, and this variant has also been found in Korean22 and Japanese7 populations. A previous study demonstrated that the CYP2C9.13 variant had a higher Km, lower Vmax, and lower intrinsic clearance values for diclofenac, losartan and glimepiride in vitro. Our results were in good agreement with this previous work and demonstrated that this variant exhibited approximately 10-fold higher Km, 37-fold lower Vmax and 389-fold lower intrinsic clearance values for tolbutamide when compared with the wild-type CYP2C9.1 protein.
The CYP2C9.8 and CYP2C9.11 variants were both first identified and occur commonly in African Americans23,24. However, there are limited and confusing data regarding the functional significance of these alleles. For example, CYP2C9.8 reportedly increases the metabolism of tolbutamide in vitro23, but reduces the metabolism of phenytoin and warfarin in vivo and/or in vitro24,25. Our data revealed that the CYP2C9.8 variant significantly reduced the intrinsic clearance value, while the CYP2C9.11 variant has no effect on the enzymatic activity toward tolbutamide in vitro (Table 1). The CYP2C9.14, CYP2C9.16 and CYP2C9.19 variants were discovered in Southeast Asians12, and reportedly exhibit a moderately or markedly decreased intrinsic clearance of tolbutamide in vitro13. Our data demonstrated that these three alleles could significantly decrease metabolic activity of the protein toward tolbutamide, even for the variant CYP2C9.19, which was previously reported to be moderately defective13. We hypothesize that the difference in the protein's ability to fold properly in insect cells and in the bacterial expression system might explain the inconsistency of our study with previous reports. For other allelic variants including CYP2C9.23, CYP2C9.27, CYP2C9.29, CYP2C9.31, CYP2C9.33, CYP2C9.34, no functional studies have been reported using tolbutamide as a phenotypic assay probe7,14,26,27. Using the insect cell expression system, we found that the CYP2C9.27 and CYP2C9.29 variants exhibited a moderate (approximately 50%) decrease in catalytic activity toward tolbutamide, and CYP2C9.23, CYP2C9.31, CYP2C9.33, and CYP2C9.34 allelic variants caused a 64.2% to 99.8% decrease in the metabolic activity of the protein toward tolbutamide in vitro, which suggests that these alleles are severely defective.
Our previous study revealed that when expressed in COS-7 cells, CYP2C9.40 and CYP2C9.54 could increase the metabolism of diclofenac in vitro9. In contrast, two other variants (CYP2C9.36 and CYP2C9.51) exhibited higher metabolic activity, while CYP2C9.40 and CYP2C9.54 exhibited moderately decreased catalytic activity toward another drug substrate, tolbutamide, when expressed in insect cells. The *36 allele contains a mutation in the start codon, which could theoretically influence the translation initiation. In both COS-7 and insect cells, relatively lower CYP2C9.36 variant protein expression levels could be detected compared with wild-type CYP2C9.1, especially in the insect cell expression system. When expressed in COS7 cells, the CYP2C9.36 variant exhibited decreased catalytic abilities against the typical CYP2C9-sepecific probe diclofenac9. However, in the present study, the CYP2C9.36 variant demonstrated the highest intrinsic clearance value of tolbutamide in vitro because the highest Vmax values were detected for this variant toward tolbutamide, and less protein was required for the enzymatic reactions compared with the other allelic variants (Table 1). We believe that the intrinsic substrate-dependent character of the CYP proteins and the different protein-folding properties of the insect cell and mammalian cell expression systems might explain the inconsistent values of the 5 variants that were discussed above. When our data are compared with data collected from COS-7 cells9, more allelic variants exhibited lower metabolic activity in the insect cell microsomes than in intact mammalian cells (20 vs 17), but the activity pattern was similar for most of the allelic variants, especially for the variants that exhibited remarkably decreased intrinsic clearance values such as CYP2C9.39, CYP2C9.42, CYP2C9.43, CYP2C9.45, and CYP2C9.52.
In summary, this study functionally assessed the wild-type and 36 other CYP2C9 alleles, including 22 recently detected non-synonymous mutations that were discovered in Chinese Han populations in an insect cell expression system. Our data suggest that 33 allelic variants can change the intrinsic clearance value of tolbutamide in vitro, and 31 allelic isoforms can significantly decrease the metabolic activities of CYPs toward the sulfonylurea tolbutamide. However, the reported protein levels and catalytic activities of various CYP2C9 variants varied significantly in different in vitro cell systems studied, therefore, the reduction or increment of the intrinsic clearance of CYP2C9 substrate determined in the present study using insect cells may not directly reflect the same alterations in human. Whether individuals carrying these defective alleles should require lower oral doses for some sulfonylurea or other CYP2C9 substrate drugs remains to be confirmed by further in vivo studies.
Author contribution
Jian-ping CAI and Guo-xin HU conceived and designed the experiments; Da-peng DAI, Shuang-hu WANG, Yu-han WANG, Pei-wu GENG, and Li-ming HU performed the experiments; Da-peng DAI, Shuang-hu WANG, and Yu-han WANG analyzed data; Da-peng DAI and Jian-ping CAI wrote the paper.
Acknowledgments
This work was supported by the grant from the Ministry of Health of China (201302008).
Footnotes
Supplementary tables are available in the website of Acta Pharmacologica Sinica.
Supplementary Information
PCR primers used for the construction of the expression plasmids
References
- Van Booven D, Marsh S, McLeod H, Carrillo MW, Sangkuhl K, Klein TE, et al. Cytochrome P450 2C9-CYP2C9. Pharmacogenet Genomics. 2010;20:277–81. doi: 10.1097/FPC.0b013e3283349e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo SL, Zhou ZW, Yang LP, Wei MQ, Zhou SF. New insights into the structural features and functional relevance of human cytochrome P450 2C9. Part I. Curr Drug Metab. 2009;10:1075–126. doi: 10.2174/138920009790820129. [DOI] [PubMed] [Google Scholar]
- Hiratsuka M. In vitro assessment of the allelic variants of cytochrome P450. Drug Metab Pharmacokinet. 2012;27:68–84. doi: 10.2133/dmpk.dmpk-11-rv-090. [DOI] [PubMed] [Google Scholar]
- Wang B, Wang J, Huang SQ, Su HH, Zhou SF. Genetic polymorphism of the human cytochrome P450 2C9 gene and its clinical significance. Curr Drug Metab. 2009;10:781–834. doi: 10.2174/138920009789895480. [DOI] [PubMed] [Google Scholar]
- Kurose K, Sugiyama E, Saito Y. Population differences in major functional polymorphisms of pharmacokinetics/pharmacodynamics-related genes in Eastern Asians and Europeans: implications in the clinical trials for novel drug development. Drug Metab Pharmacokinet. 2012;27:9–54. doi: 10.2133/dmpk.dmpk-11-rv-111. [DOI] [PubMed] [Google Scholar]
- Sanchez-Diz P, Estany-Gestal A, Aguirre C, Blanco A, Carracedo A, Ibanez L, et al. Prevalence of CYP2C9 polymorphisms in the south of Europe. Pharmacogenomics J. 2009;9:306–10. doi: 10.1038/tpj.2009.16. [DOI] [PubMed] [Google Scholar]
- Yin T, Maekawa K, Kamide K, Saito Y, Hanada H, Miyashita K, et al. Genetic variations of CYP2C9 in 724 Japanese individuals and their impact on the antihypertensive effects of losartan. Hypertens Res. 2008;31:1549–57. doi: 10.1291/hypres.31.1549. [DOI] [PubMed] [Google Scholar]
- Maekawa K, Harakawa N, Sugiyama E, Tohkin M, Kim SR, Kaniwa N, et al. Substrate-dependent functional alterations of seven CYP2C9 variants found in Japanese subjects. Drug Metab Dispos. 2009;37:1895–903. doi: 10.1124/dmd.109.027003. [DOI] [PubMed] [Google Scholar]
- Dai DP, Xu RA, Hu LM, Wang SH, Geng PW, Yang JF, et al. CYP2C9 polymorphism analysis in Han Chinese populations: building the largest allele frequency database Pharmacogenomics J 2013. doi: 10.1038/tpj.2013.2 [DOI] [PubMed]
- Imai J, Ieiri I, Mamiya K, Miyahara S, Furuumi H, Nanba E, et al. Polymorphism of the cytochrome P450 (CYP) 2C9 gene in Japanese epileptic patients: genetic analysis of the CYP2C9 locus. Pharmacogenetics. 2000;10:85–9. doi: 10.1097/00008571-200002000-00011. [DOI] [PubMed] [Google Scholar]
- Si D, Guo Y, Zhang Y, Yang L, Zhou H, Zhong D. Identification of a novel variant CYP2C9 allele in Chinese. Pharmacogenetics. 2004;14:465–9. doi: 10.1097/01.fpc.0000114749.08559.e4. [DOI] [PubMed] [Google Scholar]
- Zhao F, Loke C, Rankin SC, Guo JY, Lee HS, Wu TS, et al. Novel CYP2C9 genetic variants in Asian subjects and their influence on maintenance warfarin dose. Clin Pharmacol Ther. 2004;76:210–9. doi: 10.1016/j.clpt.2004.05.005. [DOI] [PubMed] [Google Scholar]
- DeLozier TC, Lee SC, Coulter SJ, Goh BC, Goldstein JA. Functional characterization of novel allelic variants of CYP2C9 recently discovered in southeast Asians. J Pharmacol Exp Ther. 2005;315:1085–90. doi: 10.1124/jpet.105.091181. [DOI] [PubMed] [Google Scholar]
- Maekawa K, Fukushima-Uesaka H, Tohkin M, Hasegawa R, Kajio H, Kuzuya N, et al. Four novel defective alleles and comprehensive haplotype analysis of CYP2C9 in Japanese. Pharmacogenet Genomics. 2006;16:497–514. doi: 10.1097/01.fpc.0000215069.14095.c6. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Wang M, Fang K, Xing Q, Feng G, Shen L, et al. A systematic genetic polymorphism analysis of the CYP2C9 gene in four different geographical Han populations in mainland China. Genomics. 2011;97:277–81. doi: 10.1016/j.ygeno.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Herman D, Dolzan V, Ingelman-Sundberg M. Characterization of the novel defective CYP2C9*24 allele. Drug Metab Dispos. 2007;35:831–4. doi: 10.1124/dmd.106.013722. [DOI] [PubMed] [Google Scholar]
- Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–9. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- Hanatani T, Fukuda T, Onishi S, Funae Y, Azuma J. No major difference in inhibitory susceptibility between CYP2C9.1 and CYP2C9.3. Eur J Clin Pharmacol. 2003;59:233–5. doi: 10.1007/s00228-003-0603-5. [DOI] [PubMed] [Google Scholar]
- Takanashi K, Tainaka H, Kobayashi K, Yasumori T, Hosakawa M, Chiba K. CYP2C9 Ile359 and Leu359 variants: enzyme kinetic study with seven substrates. Pharmacogenetics. 2000;10:95–104. doi: 10.1097/00008571-200003000-00001. [DOI] [PubMed] [Google Scholar]
- Guo Y, Wang Y, Si D, Fawcett PJ, Zhong D, Zhou H. Catalytic activities of human cytochrome P450 2C9*1, 2C9*3 and 2C9*13. Xenobiotica. 2005;35:853–61. doi: 10.1080/00498250500256367. [DOI] [PubMed] [Google Scholar]
- Bae JW, Kim HK, Kim JH, Yang SI, Kim MJ, Jang CG, et al. Allele and genotype frequencies of CYP2C9 in a Korean population. Br J Clin Pharmacol. 2005;60:418–22. doi: 10.1111/j.1365-2125.2005.02448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisdell J, Jorge-Nebert LF, Coulter S, Ferguson SS, Lee SJ, Chanas B, et al. Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics. 2004;14:527–37. doi: 10.1097/01.fpc.0000114759.08559.51. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jeong H, Takahashi H, Drozda K, Patel SR, Shapiro NL, et al. Decreased warfarin clearance associated with the CYP2C9 R150H (*8) polymorphism. Clin Pharmacol Ther. 2012;91:660–5. doi: 10.1038/clpt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allabi AC, Gala JL, Horsmans Y. CYP2C9, CYP2C19, ABCB1 (MDR1) genetic polymorphisms and phenytoin metabolism in a Black Beninese population. Pharmacogenet Genomics. 2005;15:779–86. doi: 10.1097/01.fpc.0000174787.92861.91. [DOI] [PubMed] [Google Scholar]
- Veenstra DL, Blough DK, Higashi MK, Farin FM, Srinouanprachan S, Rieder MJ, et al. CYP2C9 haplotype structure in European American warfarin patients and association with clinical outcomes. Clin Pharmacol Ther. 2005;77:353–64. doi: 10.1016/j.clpt.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Matimba A, Del-Favero J, Van Broeckhoven C, Masimirembwa C. Novel variants of major drug-metabolising enzyme genes in diverse African populations and their predicted functional effects. Hum Genomics. 2009;3:169–90. doi: 10.1186/1479-7364-3-2-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR primers used for the construction of the expression plasmids