Abstract
Aim:
To explore the mechanism of hepatocarcinogenesis associated with the hepatitis B virus X protein (HBx), we investigated the role of HBx in transformation using human liver L-O2 cells stably transfected with HBx as a model.
Methods:
Plasmids encoding HBx were stably transfected into immortalized human liver L-O2 cells and rodent fibroblast NIH/3T3 cells. The expression of alfa-fetoprotein (AFP), c-Myc, HBx, and survivin in the engineered cells was examined by Western blotting. The malignant phenotype of the cells was demonstrated by anchorage-independent colony formation and tumor formation in nude mice. RNA interference assays, Western blotting, luciferase reporter gene assays and flow cytometry analysis were performed. The number of centrosomes in the L-O2-X cells was determined by γ-tubulin immunostaining. The effect of HBx on the transcriptional activity of human telomerase reverse transcriptase (hTERT) and hTERT activity in L-O2-X cells and/or 3T3-X cells was detected by the luciferase reporter gene assay and telomerase repeat amplification protocol (TRAP).
Results:
Stable HBx transfection resulted in a malignant phenotype in the engineered cells in vivo and in vitro. Meanwhile, HBx was able to increase the transcription of the NF-κB, AP-1, and survivin genes and to upregulate the expression levels of c-Myc and survivin. Abnormal centrosome duplication and activated hTERT were responsible for the transformation.
Conclusion:
Stable HBx transfection leads to genomic instability of host cells, which is responsible for hepatocarcinogenesis; meanwhile, transactivation by the HBx protein contributes to the development of hepatocellular carcinoma (HCC). The L-O2-X cell line is an ideal model for investigating the mechanism of HBx-mediated transformation.
Keywords: HBx, transformation, hepatoma, genomic instability, stable transfection
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers in China, and approximately 90% of the HCC cases are associated with hepatitis B virus (HBV) infection. However, the complex mechanism by which HBV infection leads to the development of HCC remains unclear. The HBV genome is a partially double-stranded DNA molecule with four open reading frames (ORFs) termed S, C, P, and X1. Among them, the hepatitis B virus X protein (HBx), encoded by open-reading frame X, plays a central role in hepatocarcinogenesis. HBx is preferentially maintained in HCC and the surrounding liver parenchyma in the absence of other viral antigens2, 3. Previous experiments indicated that HBx regulates a wide variety of genes, suggesting that HBx not only upregulates HBV gene expression by transactivating the HBV enhancer but also modifies the environment by transactivating cellular genes in infected cells to facilitate viral replication. In addition, HBx affects a number of cellular processes, such as transcription, apoptosis, signaling, and cell growth1. However, HCC does not usually occur spontaneously upon HBV infection, indicating that onset of the disease requires other factors, such as oncogenes4. HBx itself is a poor transformer of human and rodent hepatic cells. Indeed, co-transfection with an oncogene, such as H-ras, c-fos, a p53 mutant or the SV40 T-antigen, is necessary for cellular transformation5, 6, 7, 8. These studies suggest that HBx may be unable to transform hepatocytes without the collaboration of other activated cellular factors. Interestingly, HBx is also able to induce abnormal mitotic spindle formation9, 10 and inhibit caspase 3 activity11, thus causing chromosome segregation defects, resulting in genetic instability12. Although extensive analyses on HBx functions in the stages of tumor progression have been performed13, 14, 15, 16, 17, the role of HBx in normal cells remains to be elucidated.
It has been suggested that HBx is a multifunctional oncoprotein that may be involved in malignant transformation of normal cells. A number of transgenic mouse lineages that express high levels of HBx have been shown to develop HCC18, 19. Previously, our data demonstrated that HBx upregulates both the expression and the activity of hTERT in hepatoma cells20. We also found that HBx is able to upregulate the expression of survivin, which suggests that survivin may be involved in the HBx-mediated carcinogenesis of HCC21. However, most of the previous studies were performed in mouse cell lines or human hepatoma cells. These models are not ideal for demonstrating the basic response of host cells to the HBx gene or protein.
Using a cDNA microarray assay, we found that only three genes are altered in both human hepatoma cells (H7402-X) and normal liver cells (L-O2-X cells) stably transfected with HBx22, 23. HBx may affect gene expression differently in hepatoma H7402 cells than in liver L-O2 cells because their genetic backgrounds are different. The gene expression profiles mediated by HBx in L-O2 cells may reflect alterations that occur in the early stages of carcinogenesis23.
Accordingly, in the present study, we demonstrated the role of stable HBx transfection in hepatocarcinogenesis using the human immortalized liver L-O2 cell line as a model. Our data show that HBx integration leads to malignant transformation, faster proliferation, overexpression of oncoproteins and genomic instability, which are required for hepatocarcinogenesis. Meanwhile, transactivation by the HBx protein contributes to the development of HCC. Thus, we demonstrate that the engineered L-O2 cell line is an ideal model for further exploration of the mechanisms involved in hepatocarcinogenesis mediated by HBx.
Materials and methods
Cell culture
The hepatocyte cell line L-O2 (Nanjing KeyGen Biotech Co Ltd, Nanjing, China), originating histologically from normal human liver tissue immortalized by stable transfection with the hTERT gene, has been used previously21, 24, 25, 26, 27. L-O2-P (a cell line derived from L-O2 cells stably transfected with an empty pcDNA3 plasmid) and L-O2-X (a cell line derived from L-O2 cells stably transfected with the HBx gene) were established as described previously28. L-O2, L-O2-P, and L-O2-X cells were cultured in RPMI Medium 1640 (GIBCO, NY, USA) containing 100 U/mL penicillin, 100 μg/mL streptomycin and 10% fetal calf serum (FCS, China). NIH/3T3 cells were cultured in Dulbecco's Modified Eagle's medium (DMEM, GIBCO-BRL, USA) containing 100 U/mL penicillin, 100 μg/mL streptomycin and 10% fetal calf serum (FCS, China). Cultures were incubated in a humidified atmosphere with 5% CO2 at 37 °C.
Establishment of the stably transfected cell lines
To replicate the role of HBx gene in another cell line, we attempted to establish a stably HBx-transfected NIH/3T3 cell line. NIH/3T3 cells were transfected with Lipofectamine (Invitrogen, USA) as described previously21. The plasmids pcDNA3 and pCMV-X were used in the transfection experiments. Briefly, 1 μg of each plasmid was mixed with 3 μL (6 μL in co-transfection) of Lipofectamine 2000 in serum-free media and incubated at 37 °C for 6 h. The media were then replaced with RPMI Medium 1640 or DMEM containing 10% FCS. After 48 h, the transfected cells were incubated in selection media containing 500 μg/mL G418 (Genview, Carlsbad, CA, USA). The selection was maintained for 3–4 weeks. Empty pcDNA3 vector plasmid was used as control. We termed the NIH/3T3 cells 3T3. The stable transfection of pCMV-X (termed 3T3-X) or the empty vector (termed 3T3-P) was confirmed by PCR using genomic DNA as the template and by western blotting.
Amplification of HBx gene
Genomic DNA was extracted from cultured cells using the QIAamp DNA mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Stable integration of the pCMV-X plasmid in the genome of NIH/3T3 cells was confirmed by PCR using genomic DNA from the HBx-transfected cells (termed 3T3-X). The following primers were used: the HBx gene (forward, 5′-GGCTCGAGATGGCTGCTAGGCTGTGC-3′ reverse, 5′-GGCGAATTCAGAAG TCGTCGTCGTCC-3′ 500 bp amplified fragment) and the human GAPDH gene (forward, 5′-GGTCATCCCTGAGCTGAACG-3′ reverse, 5′-TCCGTTGTCATACC AGGAAA T-3′ 298 bp amplified fragment). The cycling conditions for HBx amplification were 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 30 s, followed by an elongation cycle of 72 °C for 8 min. The resultant PCR products were analyzed on a 1% TBE agarose gel.
Soft agar assay
Anchorage-independent colony formation on these cell lines was determined as described previously29, with some modifications. Briefly, the cells were harvested and washed, and a total of 3×103 cells were resuspended in RPMI Medium 1640 containing 0.3% agarose. The suspensions were cultured in 24-well plates above a layer of solidified 0.5% agarose in RPMI Medium 1640 with 10% FCS. A soft agar colony formation assay was performed in triplicate for each cell line. After culturing for 3 weeks, each cell line was photographed. Plates were stained with 0.5 mL of 0.005% Crystal Violet for 1 h. The colonies were counted under a dissecting microscope.
Tumor formation in nude mice
The tumorigenicity of transfected cell lines was measured as follows. Cells were harvested by trypsinization, washed twice with sterile phosphate-buffered saline, and resuspended at 2×107 cells per mL. Aliquots (0.1 mL) were injected subcutaneously into 4- to 6-week-old nude mice (n=8 in each group). Mice were observed at periodic intervals. Photographs were taken at 3 weeks after injection.
Histologic analysis
Nude mice were killed by cervical dislocation. Half of the tumor tissues were snap frozen in liquid nitrogen for use in protein analyses. The remaining tumor tissues were used for histological analyses. For tumor cell examination, the tumor tissues from nude mice were fixed in 4% paraformaldehyde. Histological analysis was performed on 5-μm paraffin-embedded tissue sections that were stained with hematoxylin and eosin (H&E) (the slide from nude mice injected by L-O2-X cells). The sections were stained with H&E and were subjected to analysis under microscopy.
Flow cytometry analysis
The cells (1×106) were harvested by trypsinization and washed twice with PBS. Washed cells were resuspended in 0.6 mL PBS (pH 7.4), and fixed by the addition of 1.4 mL 100% ethanol at 4 °C overnight. The fixed cells were rinsed twice with PBS, and resuspended in propidium iodine (PI) solution, including 50 μg/mL PI and 50 μg/mL RNaseA (Sigma) in PBS without calcium and magnesium, and incubated at 37 °C for 30 min in the dark. Stained cells were passed through a nylon-mesh sieve to remove cell clumps and analyzed by a FACScan flow cytometer and Cell Quest analysis software (Becton Dickinson, San Jose, CA, USA). Flow cytometry analysis was repeated 3 times.
Luciferase reporter assay
L-O2 and NIH/3T3 cells were plated in 24-well plates (5×104 cells/well) and cultured in Dulbecco's modified essential medium (DMEM) containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA). The following luciferase reporters were used: pGL3-Survivin, pGL3-NF-κB, pGL3-hTERT, and pGL3-AP-1, pGL3-basic, pGL3-control and the Renilla Luciferase Reporter Vector pRL-TK vector. Cells were harvested after 48 h and lysed in 1×passive lysis buffer according to the manufacturer's protocol30. The luciferase activity was determined and normalized to a cotransfected control reporter by using the Dual-Luciferase Reporter® Assay System (Promega, USA) on a Luminometer (TD-20/20, Turner Desings) according to the manufacturer's instructions.
RNA interference (RNAi) targeting mRNA of HBx
The pSilencer3.0-X plasmid encoding double-chain HBx mRNA 21 was used to transfect 3T3-X cells as described above. Cultured 3T3-X cells were placed in 6-well dishes 24 h before transfection at a concentration that gave 80%−90% confluence. The pSilencer3.0-X and Lipofectamine (Life Technologies, Inc. USA) were added to 600 μL of Opti-MEM medium (GIBCO-BRL) and were then mixed and incubated for 20 min at room temperature. The cells were incubated with the pSilencer3.0-X/Lipofectamine/media mixture for 6 h at 37 °C. Transfected cells were lysed after 48 h. Western blot analysis was used to determine the expression levels of c-Myc, survivin and HBx.
Western blot analysis
L-O2 and NIH/3T3 cells were washed three times with ice-cold PBS and extracted directly in lysis buffer (62.5 mmol/L Tris-HCl, pH 6.8, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol). Equal amounts of protein (30 μg) were separated by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Following electro-transfer onto polyvinylidene difluoride membranes, proteins were hybridized with the following primary antibodies against AFP: (1:500 dilution, NeoMarkers, USA), c-Myc (1:500 dilution, Labvision, USA), survivin (1:1000 dilution, Labvision, USA), HBx (1:1000 dilution, Abcam, USA), Bcl-2 (1:1000 dilution, NeoMarkers, USA), PCNA (1:1000 dilution, NeoMarkers, USA) and β-actin (1:1000 dilution, NeoMarkers, USA) as described previously21, 31, and stained with an HRP-linked secondary antibody (GE Healthcare Bio-Science, USA). The protein bands were visualized by an enhanced chemiluminescence (ECL) kit according to the manufacturer's specifications (GE Healthcare Bio-Sciences).
Centrosome analysis
Because γ-tubulin was used as a marker for centrosomes10, we examined the expression of γ-tubulin in the engineered cell lines using immunofluorescence staining. After synchronization, engineered cells were fixed with 4% paraformaldehyde for 10 min and methanol for 20 min and then incubated in PBS for 5 min. Samples were blocked with 10% normal donkey serum at room temperature for 1 h, stained with mouse anti-γ-tubulin (1:100 dilution, Sigma, USA) at 37 °C for 1 h and stained with FITC-conjugated anti-mouse IgG at room temperature for 1 h. FITC staining was observed using a fluorescent microscope. L-O2 and L-O2-P cells were used as negative controls. We assessed the proportion of cells with either multipolar spindles or dipolar/monopolar spindles. A total of 60–80 cells were counted in 4 representative fields.
TRAP analysis
We assayed telomerase activity using the TRAP method with the TRAPeze telomerase detection kit (Intergen, Purchase, NY, USA). Cells stably expressing HBx and untransfected controls were pelleted, washed with ice-cold PBS, and then repelleted. Each cell pellet was then resuspended in the 3-cholamidopropyl dimethylammonio-1-propanesulfonate (CHAPS) lysis buffer supplied in the kit. After 30 minutes of incubation on ice, the lysate was centrifuged at 14 000 r/min for 30 minutes at 4 °C, and the supernatant was frozen and stored at -70 °C. We measured protein concentrations using the 2-Dquant kit (Amersham Biosciences Corp, USA); an aliquot of the extract containing 3 mg of protein was used for each TRAP assay in accordance with the manufacturer's suggested protocol. Products were separated on 12% SDS-PAGE and visualized after silver staining.
Statistical analysis
All values are presented as means±SEM. Each value is the mean of at least three separate experiments in each group. Data were analyzed to compare two groups (P<0.01 was considered significant) using Student's t test.
Results
Establishment of a stably HBx-transfected NIH/3T3 cell line
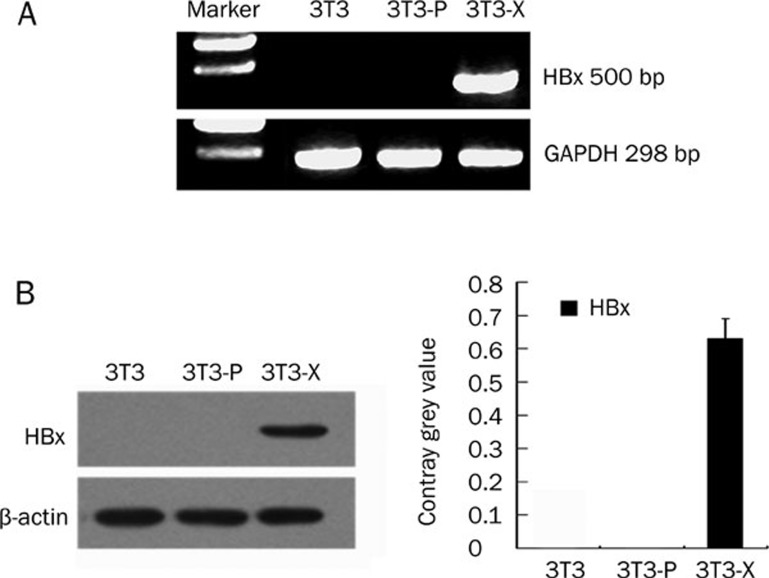
Previously, we successfully established a human liver L-O2 cell line conditionally expressing HBx23, 28. To investigate the different effects of HBx on L-O2 and NIH/3T3 cells, we established a stable HBx-expressing NIH/3T3 cell line (termed 3T3-X cells) via G418 screening. It has been reported that HBx alone is not able to induce tumor formation using rodent fibroblast NIH/3T3 cells as a model8, 32, 33, 34, 35. Here, we were interested in identifying the different roles of HBx in L-O2 cells and NIH/3T3 cells. We identified the integrated HBx gene in the genomic DNA of the engineered NIH/3T3 cells using genomic PCR. GAPDH was used as a loading control (Figure 1A). The data showed that the HBx gene had been successfully introduced into the host genome in 3T3-X cells. Western blotting showed that expression of the HBx protein was detectable in 3T3-X cells (Figure 1B). Our data suggest that a stably HBx-transfected NIH/3T3 cell line was successfully established.
Figure 1.
The identification of stable HBx transfection in NIH/3T3 cells. (A and B) Integration of the HBx gene in NIH/3T3. (A) Integration of the HBx gene in the engineered cells was identified by PCR using genomic DNA as a template. GAPDH was used as a loading control. (B) Western blotting showed the expression of HBx in 3T3-X. The histogram shows the results after analysis with the Glyco Band-Scan software. Results shown are representative of three independent experiments.
Engineered L-O2-X cells display a malignant phenotype
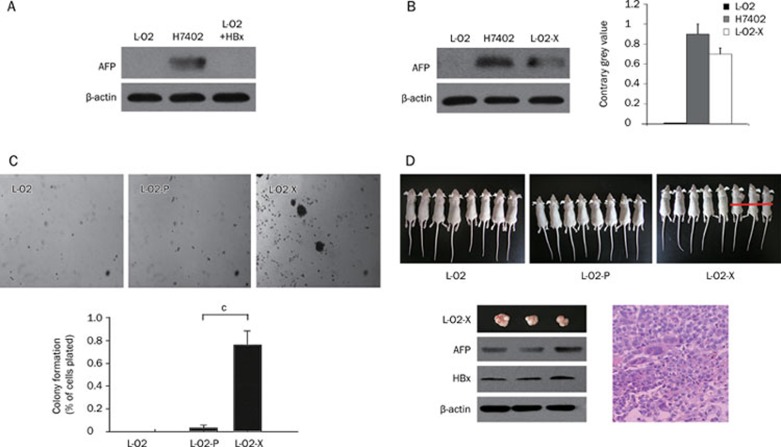
To investigate whether HBx could lead to transformation, we examined the expression of alfa-fetoprotein (AFP), a hepatoma maker, in the engineered L-O2-X cells. Our data show that AFP was detectable in human hepatoma H7402 cells (as a positive control), but not in L-O2 cells transiently transfected with HBx (L-O2+HBx) (Figure 2A). However, AFP could be detected in L-O2-X cells (Figure 2B). This suggests that, unlike transient HBx transfection, stable HBx transfection is able to transform L-O2 cells. We further confirmed the results using Glyco BandScan software (PROZYME, San Leandro, CA, USA). A soft agar assay showed that L-O2-X cells displayed a faster colony-forming rate than the controls (Figure 2C), suggesting that L-O2-X cells have characteristics of a malignant phenotype. Next, tumorigenicity assays showed that 3 mice (n=8) injected with L-O2-X cells developed tumors in 9 d, whereas L-O2 and L-O2-P cells did not cause any visible tumors (Figure 2D). The three primary tumors in the L-O2-X-injected mice reached volumes of 720, 805, and 645 mm3 and weights of 0.9, 1.0 and 0.8 g, respectively. In addition, we identified AFP and HBx in the tumor tissues by Western blot analysis (Figure 2D). Pathological observation of the primary tumors revealed a malignant phenotype by hematoxylin and eosin (H&E) staining (Figure 2D). Thus, our findings reveal that engineered L-O2-X cells exhibit a malignant phenotype.
Figure 2.
Stable HBx transfection results in transformation of L-O2 cells. (A) Western blot analysis showed that AFP was detectable in human hepatoma H7402 cells (positive control), but not L-O2 cells transiently transfected with HBx (L-O2+HBx), suggesting that transient overexpression of HBx could not transform L-O2 cells. β-actin was used as loading control (lower panel). (B) Western blot analysis showed that AFP could be detected in both H7402 and L-O2-X cells, suggesting that stable HBx transfection promotes transformation of L-O2 cells. The histogram shows the results after analysis with the Glyco Band-Scan software. Results shown are representative of three independent experiments. (C) Representative photographs of soft agar colony formation were taken three weeks after culture of engineered cells. The diagram shows the mean colony counts from three petri dishes (error bars indicate SEM, n=3 experiments, cP<0.01, Student's t test). (D) Number of nude mice with tumor formation (n=8 per group) 3 weeks after injection with engineered cells. Only 3 of 8 mice injected L-O2-X cells generated tumors. Western blot analysis showed the expression levels of HBx and AFP in the tumor tissues. β-actin was used as loading control (lower panel). The histological analysis revealed the pathological features of the tumors (H&E. staining).
L-O2-X cells grow faster than controls
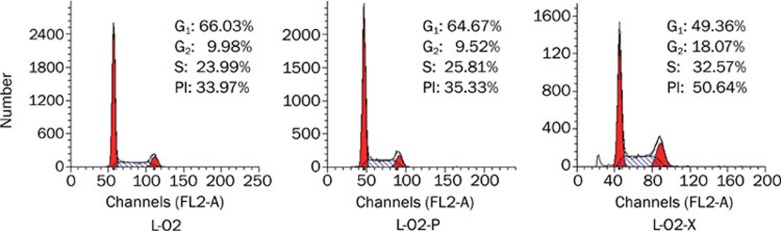
To investigate the effect of HBx on cell proliferation, we examined the cell proliferation of engineered cells by flow cytometry analysis. The data showed that L-O2-X cells grew faster than controls. The proliferation index (PI) is the sum of the S and G2/M phase activities of the cell cycle expressed as a fraction of the total cell population, that is PI=[(S+G2/M)/(G0/G1+S+G2/M)] ×100%30. A high PI value corresponds with fast cell proliferation. Cells were counted using a FACScan flow cytometer. The PIs of L-O2-X, L-O2-P, and L-O2 cells were 50.64%, 35.33%, and 33.97%, respectively (Figure 3). Thus, stable HBx transfection significantly accelerated proliferation of L-O2 cells.
Figure 3.
HBx is responsible for the proliferation of L-O2 cells. Flow cytometry analysis revealed that L-O2-X cells had higher proliferation rates than L-O2-P cells, as measured by the proliferation index (PI=S+G2M/G0/G1+S+G2M×100%) and the S phase fraction (SPF=S/G0/G1+S+G2M×100%) using flow cytometry analysis.
HBx promotes the transcriptional activities of NF-κB, AP-1 and survivin
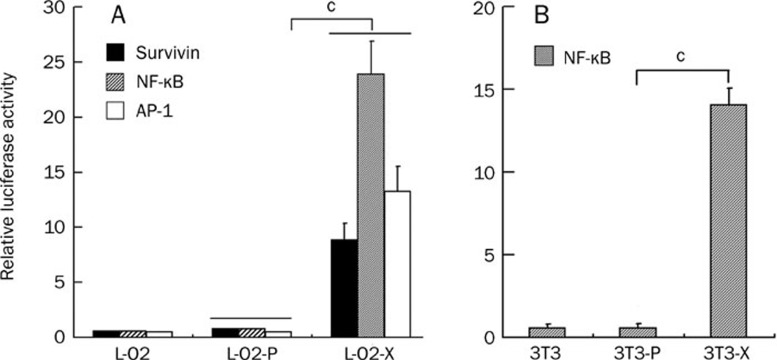
To further demonstrate the effect of HBx on cell proliferation, we examined whether the expression of HBx stimulated the transcriptional activities of NF-κB, AP-1, and survivin in the engineered cells. The data show that the transcriptional activities of NF-κB, AP-1, and survivin were remarkably increased in L-O2-X or 3T3-X cells relative to controls (P<0.01, Student's t test, Figure 4A and 4B), suggesting that HBx is a powerful transactivator.
Figure 4.
HBx promotes the transcriptional activities of NF-κB, AP-1, and survivin. A luciferase reporter gene assay revealed that the transcriptional activities of survivin, NF-κB and AP-1 were increased in L-O2-X/3T3-X cells relative to controls (cP<0.01 vs L-O2-P or 3T3-P, Student's t test).
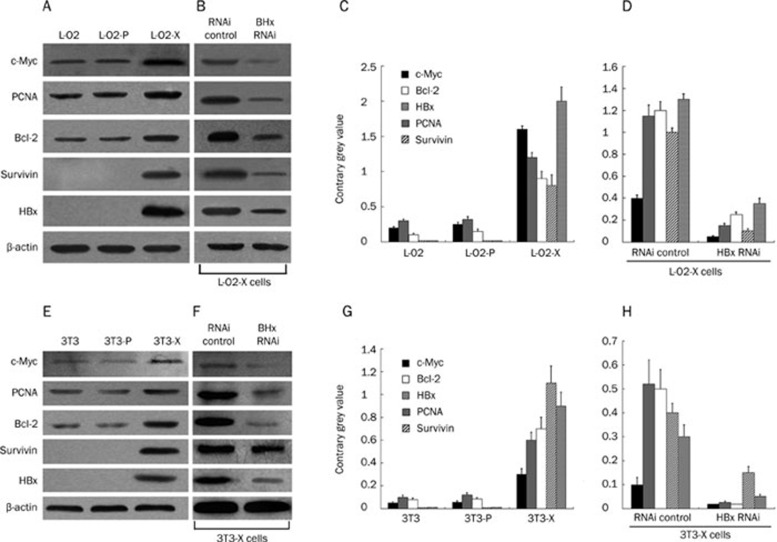
HBx upregulates expression of c-Myc and survivin
In order to further clarify the molecular mechanism involved in the transformation mediated by HBx, we investigated several proteins associated with carcinogenesis, including c-Myc, proliferating cell nuclear antigen (PCNA), Bcl-2 and survivin. Western blot analysis revealed that expression of each of these proteins was remarkably upregulated in L-O2-X cells relative to the controls (Figure 5A and 5C). RNA interference (RNAi) targeting HBx mRNA could abolish the upregulation 48 h after transfection (Figure 5B and 5D), suggesting that HBx was responsible for the upregulation of these proteins involved in carcinogenesis. Similar results were observed in the engineered NIH/3T3 cells (Figure 5E–5H).
Figure 5.
HBx upregulates expression of c-Myc, PCNA, Bcl-2, and survivin in the engineered cells. (A) Western blot analysis showed that expression levels of c-Myc, PCNA, Bcl-2, and survivin were upregulated in L-O2-X cells. (B) RNAi targeting HBx mRNA abolished this upregulation. (C, D) The histogram shows the results after analysis using the Glyco Band-Scan software. Results shown are representative of three independent experiments. (E–H) Similar results were found in 3T3-X cells. The findings suggest that HBx transforms the cells by upregulating c-Myc, PCNA, Bcl-2, and survivin.
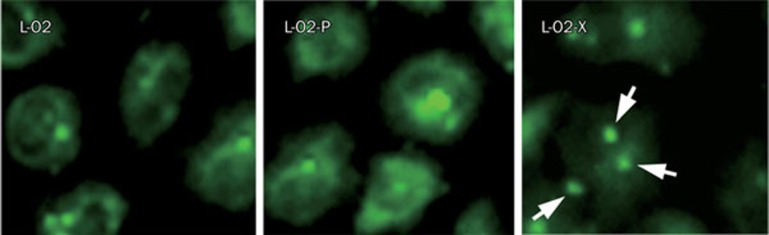
HBx leads to genetic instability by centrosome hyperamplification
Excessive centrosome production and multipolar mitotic spindles cause chromosome segregation defects, which result in genetic instability. Chromosomal instability is a characteristic feature of HCC, and it plays an important role in liver carcinogenesis. Here, we examined centrosome production in the engineered L-O2 cells by immunofluorescence analysis. Abnormal centrosome duplication was observed in L-O2-X cells, where three or more centrosomes were found, but few cases of centrosome duplication were found in other control cells (Figure 6). The data show that the proportions of L-O2, L-O2-P, and L-O2-X cells with either multipolar spindles or dipolar/monopolar spindles were about 3.0%, 4.0%, and 93.0%, respectively, suggesting that stable HBx transfection leads to genetic instability of L-O2 cells.
Figure 6.
HBx stimulates centrosome hyperamplification. Immunofluorescence staining showed that stable HBx transfection resulted in centrosome aberrations with three or more centrosomes. The proportions of L-O2, L-O2-P, and L-O2-X cells with either multipolar spindles or dipolar/monopolar spindles were about 3.0%, 4.0%, and 93.0%, respectively, suggesting that HBx leads to genomic instability of host cells.
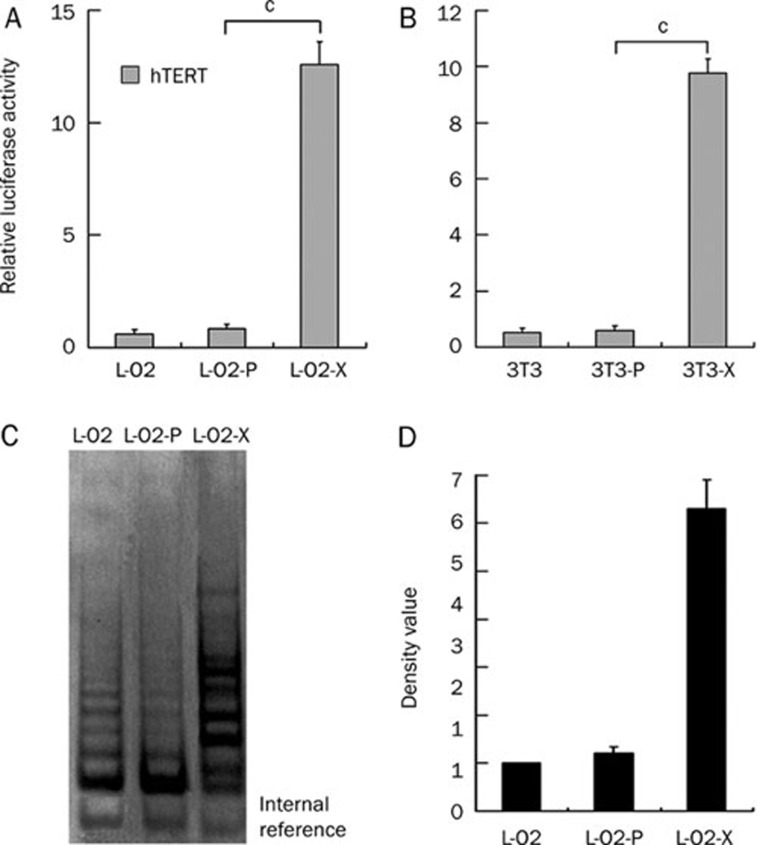
HBx enhances telomerase function
To further provide evidence of a malignant phenotype in the L-O2-X cells, we examined the transcriptional activity of hTERT using a luciferase reporter gene assay and TRAP assay. The data showed that the transcriptional activity of hTERT was remarkably increased in L-O2-X or 3T3-X cells relative to controls (P<0.01, Student's t test, Figure 7A and 7B). TRAP showed that telomerase activity of hTERT was significantly enhanced in L-O2-X cells compared with control cells (Figure 7C and 7D), suggesting that L-O2-X cells have a malignant phenotype.
Figure 7.
HBx activates telomerase function. (A and B) A luciferase reporter gene assay revealed that HBx was able to promote the transcriptional activities of hTERT in L-O2-X/3T3-X cells relative to L-O2-P/3T3-P cells (cP<0.01 vs L-O2-P or 3T3-P, Student's t test). (C and D) A TRAP assay confirmed telomerase activity; proteins were separated by electrophoresis and visualized after silver staining. Lane 1, L-O2 cells; lane 2, L-O2-P cells; lane 3, L-O2-X cells. The data suggest that L-O2-X cells have a malignant phenotype.
Discussion
HBV has an oncogenic role in the development of HCC17, 36; however, the mechanism underlying HBV-induced HCC remains unclear. It has been reported that HBx is preferentially maintained in HCC in the absence of other viral antigens25. However, the function of HBx in the transformation of human liver cells is unknown. Many studies have shown that HBx alone could not transform cells without other factors in transgenic mouse and rodent NIH/3T3 cells8, 32, 33, 34, 35. In the present study, we show that stable HBx transfection resulted in carcinogenesis using an immortalized human liver cell line L-O2 as a model. However, the reasons for the different results remain unclear. The different integration patterns of the HBx gene in different host cells may play an important role in the transformation, in addition to the role of transactivation by the HBx protein.
In the present study, to verify the effect of HBx on human liver cells, we chose human immortalized liver L-O2 cells and NIH/3T3 cells as models. The expression of AFP, a tumor-associated antigen, is silenced in normal adult hepatocytes, but reactivated in human HCC37, 38, 39. AFP may play a role in one of the fundamental steps in the progression of hepatoma. We performed stable HBx transfection in L-O2 cells and examined the expression of AFP in the engineered L-O2-X cells. Usually, a hallmark of transformed cells is the ability to form colonies in soft agar and induce tumor formation in animals—non-transformed cells fail to do so8, 32, 33, 34, 35. Interestingly, our findings showed that L-O2-X cells were able to form colonies in soft agar and tumors in nude mice (Figure 2), suggesting that the HBx gene alone may have strong oncogenic potential. Thus, we provide evidence that HBx contributes to hepatocarcinogenesis in vivo and in vitro.
Next, by flow cytometry analysis, we showed that stable HBx expression in L-O2 cells caused an acceleration in cell proliferation (Figure 3). Some investigators have reported that the HBx protein transactivates multiple DNA elements, including AP-1 and NF-κB18, 40. Indeed, our findings further support that NF-κB and AP-1 are targets of HBx (Figure 4). The activation of NF-κB and AP-1 may play a critical role in the functional switch of the viral protein HBx in cell apoptosis and transformation. A growing body of evidence has shown that HBx can affect many genes and signal pathways involved in cell function9, 11, 16. Tumorigenesis is a multistage process, which involves activation of cellular oncogenes and inactivation of tumor suppressor genes. Some studies have shown that c-Myc plays a crucial role in tumor development and that it is an inducible oncogene41. In addition, cancer often correlates with an enhanced level of PCNA, and PCNA levels can be used as a prognostic marker in some cases42, 43, 44. In our experiments we showed that c-Myc, PCNA, Bcl-2, and survivin were upregulated in L-O2-X cells (Figure 5), suggesting that our data are consistent with previous reports. Chromosomal instability is a characteristic feature of HCC. It plays an important role in liver carcinogenesis9, 45. Here, we explored centrosome production in L-O2-X cells by immunofluorescence analysis (Figure 6), revealing that HBx may dysregulate the normal processes during cell division, resulting in genomic instability. It has been reported that hTERT expression and telomerase activity are both detected in HCC samples but not in normal liver tissues46, 47. In our study, we found that HBx was able to activate hTERT (Figure 7). It has been reported that c-Myc plays a key role in the process of telomerase activation, and it induces telomerase activity and prolongs the lifespans of certain normal cells, such as human mammary epithelial cells (HMECS)48. c-Myc protein can bind the two c-Myc sites on the hTERT gene promoter to trigger hTERT gene expression and activate telomerase activity49, and cells can evade apoptosis via c-Myc and other oncogenes, leading to carcinogenesis. Our data provide more evidence that stable HBx transfection could result in a malignant phenotype because hTERT is activated in the engineered cells.
Accordingly, we suppose that the integration of HBx in the host genome may result in genetic instability. Furthermore, as a transcriptional co-activator, HBx exerts a pleiotropic effect on diverse cellular functions. The aberrant high expression of the HBx gene in normal cells may provide a strong influence on both cell cycle progression and growth advantage, which promotes cell transformation. HBx affects cellular fate by shifting the balance toward cell survival, probably causing loss of apoptotic functions or directly contributing to oncogenesis by gain of transforming functions. Therefore, HBx is not acutely oncogenic, but contributes to oncogenesis by stimulating proliferation of cells. Meanwhile, the transactivation by the HBx protein contributes to the development of HCC.
Our findings provide valuable evidence that stable HBx transfection in L-O2 cells is an ideal model, which is superior to the hepatoma cell model23 and mouse models, for further exploration of the mechanisms involved in hepatocarcinogenesis associated with HBx. Using our model, further studies may provide new insights into the mechanisms of pathogenesis and the contribution of HBx to hepatocarcinogenesis.
Author contribution
Prof Xiao-dong ZHANG and Li-hong YE designed the research; Wei-ying ZHANG performed the research; Na CAI helped to perform part of the research. Wei-ying ZHANG analyzed the data and wrote the paper. Prof Xiao-dong ZHANG revised the paper.
Acknowledgments
This project was supported by grants from the National Basic Research Program of China (973 Program, No 2007CB914802, No 2007CB914804, No 2009CB521702) and the National Natural Science Foundation (No 30670959).
References
- Okuda K. Hepatocellular carcinoma. J Hepatol. 2000;32 1 Suppl:225–37. doi: 10.1016/s0168-8278(00)80428-6. [DOI] [PubMed] [Google Scholar]
- Feitelson MA, Duan LX. Hepatitis B virus X antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. Am J Pathol. 1997;150:1141–57. [PMC free article] [PubMed] [Google Scholar]
- Su Q, Schroder CH, Hofmann WJ, Otto G, Pichlmayr R, Bannasch P. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology. 1998;27:1109–20. doi: 10.1002/hep.510270428. [DOI] [PubMed] [Google Scholar]
- Ng RK, Lau CY, Lee SM, Tsui SK, Fung KP, Waye MM. cDNA microarray analysis of early gene expression profiles associated with hepatitis B virus X protein-mediated hepatocarcinogenesis. Biochem Biophys Res Commun. 2004;322:827–35. doi: 10.1016/j.bbrc.2004.07.188. [DOI] [PubMed] [Google Scholar]
- Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–20. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- Koike K, Moriya K, Iino S, Yotsuyanagi H, Endo Y, Miyamura T, et al. High-level expression of hepatitis B virus HBx gene and hepatocarcinogenesis in transgenic mice. Hepatology. 1994;19:810–9. [PubMed] [Google Scholar]
- Terradillos O, Billet O, Renard CA, Levy R, Molina T, Briand P, et al. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- Kim YC, Song KS, Yoon G, Nam MJ, Ryu WS. Activated ras oncogene collaborates with HBx gene of hepatitis B virus to transform cells by suppressing HBx-mediated apoptosis. Oncogene. 2001;20:16–23. doi: 10.1038/sj.onc.1203840. [DOI] [PubMed] [Google Scholar]
- Forgues M, Difilippantonio MJ, Linke SP, Ried T, Nagashima K, Feden J, et al. Involvement of Crm1 in hepatitis B virus X protein-induced aberrant centriole replication and abnormal mitotic spindles. Mol Cell Biol. 2003;23:5282–92. doi: 10.1128/MCB.23.15.5282-5292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C, Cho H, Kim SJ, Lee JH, Park SY, Chan GK. Mitotic aberration coupled with centrosome amplification is induced by hepatitis B virus X oncoprotein via the Ras-mitogen-activated protein/extracellular signal-regulated kinase-mitogen-activated protein pathway. Mol Cancer Res. 2004;2:159–69. [PubMed] [Google Scholar]
- Diao J, Khine AA, Sarangi F, Hsu E, Iorio C, Tibbles LA, et al. X protein of hepatitis B virus inhibits Fas-mediated apoptosis and is associated with up-regulation of the SAPK/JNK pathway. J Biol Chem. 2001;276:8328–40. doi: 10.1074/jbc.M006026200. [DOI] [PubMed] [Google Scholar]
- Fujii R, Zhu C, Wen Y, Marusawa H, Bailly-Maitre B, Matsuzawa S, et al. HBXIP, cellular target of hepatitis B virus oncoprotein, is a regulator of centrosome dynamics and cytokinesis. Cancer Res. 2006;66:9099–107. doi: 10.1158/0008-5472.CAN-06-1886. [DOI] [PubMed] [Google Scholar]
- Yoo YG, Oh SH, Park ES, Cho H, Lee N, Park H, et al. Hepatitis B virus X protein enhances transcriptional activity of hypoxia-inducible factor-1alpha through activation of mitogen-activated protein kinase pathway. J Biol Chem. 2003;278:39076–84. doi: 10.1074/jbc.M305101200. [DOI] [PubMed] [Google Scholar]
- Lee JO, Kwun HJ, Jung JK, Choi KH, Min DS, Jang KL. Hepatitis B virus X protein represses E-cadherin expression via activation of DNA methyltransferase 1. Oncogene. 2005;24:6617–25. doi: 10.1038/sj.onc.1208827. [DOI] [PubMed] [Google Scholar]
- Lara-Pezzi E, Gomez-Gaviro MV, Galvez BG, Mira E, Iniguez MA, Fresno M, et al. The hepatitis B virus X protein promotes tumor cell invasion by inducing membrane-type matrix metalloproteinase-1 and cyclooxygenase-2 expression. J Clin Invest. 2002;110:1831–8. doi: 10.1172/JCI200215887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung TW, Lee YC, Kim CH. Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERK and PI-3K/AKT pathways: involvement of invasive potential. FASEB J. 2004;18:1123–5. doi: 10.1096/fj.03-1429fje. [DOI] [PubMed] [Google Scholar]
- Chan DW, Ng IO. Knock-down of hepatitis B virus X protein reduces the tumorigenicity of hepatocellular carcinoma cells. J Pathol. 2006;208:372–80. doi: 10.1002/path.1901. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang H, Ye L. Effects of hepatitis B virus X protein on the development of liver cancer. J Lab Clin Med. 2006;147:58–66. doi: 10.1016/j.lab.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Chen WL, Louie SG, Yen TS, Ou JH. Hepatitis B virus promotes hepatocarcinogenesis in transgenic mice. Hepatology. 2007;45:16–21. doi: 10.1002/hep.21445. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dong N, Zhang H, You J, Wang H, Ye L. Effects of hepatitis B virus X protein on human telomerase reverse transcriptase expression and activity in hepatoma cells. J Lab Clin Med. 2005;145:98–104. doi: 10.1016/j.lab.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dong N, Yin L, Cai N, Ma H, You J, et al. Hepatitis B virus X protein upregulates survivin expression in hepatoma tissues. J Med Virol. 2005;77:374–81. doi: 10.1002/jmv.20466. [DOI] [PubMed] [Google Scholar]
- Ye L, Dong N, Wang Q, Xu Z, Cai N, Wang H, et al. Progressive changes in hepatoma cells stably transfected with hepatitis B virus X gene. Intervirology. 2008;51:50–8. doi: 10.1159/000120289. [DOI] [PubMed] [Google Scholar]
- Zhang WY, Xu FQ, Shan CL, Xiang R, Ye LH, Zhang XD. Gene expression profiles of human liver cells mediated by hepatitis B virus X protein. Acta Pharmacol Sin. 2009;30:424–34. doi: 10.1038/aps.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Xing Z, Luan Z, Wu T, Wu X, Hu G. A specific splicing variant of SVH, a novel human armadillo repeat protein, is up-regulated in hepatocellular carcinomas. Cancer Res. 2003;63:3775–82. [PubMed] [Google Scholar]
- Hwang GY, Lin CY, Huang LM, Wang YH, Wang JC, Hsu CT, et al. Detection of the hepatitis B virus X protein (HBx) antigen and anti-HBx antibodies in cases of human hepatocellular carcinoma. J Clin Microbiol. 2003;41:5598–603. doi: 10.1128/JCM.41.12.5598-5603.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang NH, Chen YL, Wang XQ, Li XJ, Yin FZ, Wang XZ. Construction of IL-2 gene-modified human hepatocyte and its cultivation with microcarrier. World J Gastroenterol. 2003;9:79–83. doi: 10.3748/wjg.v9.i1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Wu T, Xu L, Liu A, Ji Y, Hu G. Upstream binding factor up-regulated in hepatocellular carcinoma is related to the survival and cisplatin-sensitivity of cancer cells. FASEB J. 2002;16:293–301. doi: 10.1096/fj.01-0687com. [DOI] [PubMed] [Google Scholar]
- Zhang H, Shan CL, Li N, Zhang X, Zhang XZ, Xu FQ, et al. Identification of a natural mutant of HBV X protein truncated 27 amino acids at the COOH terminal and its effect on liver cell proliferation. Acta Pharmacol Sin. 2008;29:473–80. doi: 10.1111/j.1745-7254.2008.00764.x. [DOI] [PubMed] [Google Scholar]
- Kim M, Kim JH, Jang HR, Kim HM, Lee CW, Noh SM, et al. LRRC3B, encoding a leucine-rich repeat-containing protein, is a putative tumor suppressor gene in gastric cancer. Cancer Res. 2008;68:7147–55. doi: 10.1158/0008-5472.CAN-08-0667. [DOI] [PubMed] [Google Scholar]
- Wang FZ, Sha L, Zhang WY, Wu LY, Qiao L, Li N, et al. Involvement of hepatitis B X-interacting protein (HBXIP) in proliferation regulation of cells. Acta Pharmacol Sin. 2007;28:431–8. doi: 10.1111/j.1745-7254.2007.00531.x. [DOI] [PubMed] [Google Scholar]
- Wang FZ, Sha L, Ye LH, Zhang XD. Promotion of cell proliferation by HBXIP via upregulation of human telomerase reverse transcriptase in human mesenchymal stem cells. Acta Pharmacol Sin. 2008;29:83–9. doi: 10.1111/j.1745-7254.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- Wei W, Huang W, Pan Y, Zhu F, Wu J. Functional switch of viral protein HBx on cell apoptosis, transformation, and tumorigenesis in association with oncoprotein Ras. Cancer Lett. 2006;244:119–28. doi: 10.1016/j.canlet.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Seifer M, Hohne M, Schaefer S, Gerlich WH. In vitro tumorigenicity of hepatitis B virus DNA and HBx protein. J Hepatol. 1991;13 Suppl 4:S61–5. doi: 10.1016/0168-8278(91)90026-8. [DOI] [PubMed] [Google Scholar]
- Tarn C, Bilodeau ML, Hullinger RL, Andrisani OM. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J Biol Chem. 1999;274:2327–36. doi: 10.1074/jbc.274.4.2327. [DOI] [PubMed] [Google Scholar]
- Gottlob K, Pagano S, Levrero M, Graessmann A. Hepatitis B virus X protein transcription activation domains are neither required nor sufficient for cell transformation. Cancer Res. 1998;58:3566–70. [PubMed] [Google Scholar]
- Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118–29. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- Nakabayashi H, Hashimoto T, Miyao Y, Tjong KK, Chan J, Tamaoki T. A position-dependent silencer plays a major role in repressing alpha-fetoprotein expression in human hepatoma. Mol Cell Biol. 1991;11:5885–93. doi: 10.1128/mcb.11.12.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Ma XL, Qi TG, Liu XD, Meng YS, Guan GJ. Downregulation of alpha-fetoprotein siRNA inhibits proliferation of SMMC-7721 cells. World J Gastroenterol. 2005;11:6053–5. doi: 10.3748/wjg.v11.i38.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MS, Ma QL, Chen Q, Liu XH, Li PF, Du GG, et al. Alpha-fetoprotein triggers hepatoma cells escaping from immune surveillance through altering the expression of Fas/FasL and tumor necrosis factor related apoptosis-inducing ligand and its receptor of lymphocytes and liver cancer cells. World J Gastroenterol. 2005;11:2564–9. doi: 10.3748/wjg.v11.i17.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil R, Sirma H, Giannini C, Kremsdorf D, Bessia C, Dargemont C, et al. Direct association and nuclear import of the hepatitis B virus X protein with the NF-kappaB inhibitor IkappaBalpha. Mol Cell Biol. 1999;19:6345–54. doi: 10.1128/mcb.19.9.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang H, Li Z, Wu Q, Lathia JD, McLendon RE, et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS ONE. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzewska J, Guzinska-Ustymowicz K, Lebelt A, Zalewski B, Kemona A. Evaluation of proliferating markers Ki-67, PCNA in gastric cancers. Rocz Akad Med Bialymst. 2004;49 Suppl 1:64–6. [PubMed] [Google Scholar]
- Morell-Quadreny L, Clar-Blanch F, Fenollosa-Enterna B, Perez-Bacete M, Martinez-Lorente A, Llombart-Bosch A. Proliferating cell nuclear antigen (PCNA) as a prognostic factor in renal cell carcinoma. Anticancer Res. 1998;18:677–82. [PubMed] [Google Scholar]
- Naryzhny SN. Proliferating cell nuclear antigen: a proteomics view. Cell Mol Life Sci. 2008;65:3789–808. doi: 10.1007/s00018-008-8305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593–604. doi: 10.1038/sj.onc.1205434. [DOI] [PubMed] [Google Scholar]
- Kotoula V, Hytiroglou P, Pyrpasopoulou A, Saxena R, Thung SN, Papadimitriou CS. Expression of human telomerase reverse transcriptase in regenerative and precancerous lesions of cirrhotic livers. Liver. 2002;22:57–69. doi: 10.1046/j.0106-9543.2001.01594.x. [DOI] [PubMed] [Google Scholar]
- Harada K, Yasoshima M, Ozaki S, Sanzen T, Nakanuma Y. PCR and in situ hybridization studies of telomerase subunits in human non-neoplastic livers. J Pathol. 2001;193:210–7. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH786>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Wang J, Xie LY, Allan S, Beach D, Hannon GJ. Myc activates telomerase. Genes Dev. 1998;12:1769–74. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa K, Maehara K, Isobe K. Mechanism for the reduction of telomerase expression during muscle cell differentiation. J Biol Chem. 2001;276:22016–23. doi: 10.1074/jbc.M011181200. [DOI] [PubMed] [Google Scholar]