Abstract
Erk1/2 activation contributes to mouse ES cell pluripotency. We found a direct role of Erk1/2 in modulating chromatin features required for regulated developmental gene expression. Erk2 binds to specific DNA sequence motifs typically accessed by Jarid2 and PRC2. Negating Erk1/2 activation leads to increased nucleosome occupancy, and decreased occupancy of PRC2 and poised RNAPII at Erk2-PRC2 targeted developmental genes. Surprisingly, Erk2-PRC2 targeted genes are specifically devoid of TFIIH, known to phosphorylate RNA polymerase II (RNAPII) at Serine-5, giving rise to its initiated form. Erk2 interacts with and phosphorylates RNAPII at its Serine-5 residue, consistent with the presence of poised RNAPII as a function of Erk1/2 activation. These findings underscore a key role for Erk1/2 activation in promoting the primed status of developmental genes in mouse ES cells and suggest that the transcription complex at developmental genes is different than the complexes formed at other genes, offering alternative pathways of regulation.
INTRODUCTION
Pluripotent embryonic stem cells (ESCs) are widely used in the study of epigenetic mechanisms due to their unique properties in self-renewal and their ability to undergo multi-lineage differentiation in response to appropriate signaling cues (Ng and Surani, 2011; Young, 2011). ESCs generally have a characteristic epigenetic signature that reflects their broad developmental potential. One such signature is a ‘poised’ chromatin state present on developmental genes (Bernstein et al., 2006; Mikkelsen et al., 2007). This ‘poised’ state comprises a bivalent chromatin domain containing a histone modification associated with transcriptional activation, histone H3 trimethylated at lysine 4 (H3K4me3), along with another associated with transcriptional repression, H3K27me3 (for a review, see Voigt et al., 2013). The H3K4me3 mark is imposed by the SET1A/B and MLL family of Trithorax complexes, specifically by the MLL2 complex (Shilatifard, 2012), whereas H3K27me3 is catalyzed and propagated by the Polycomb Repressive Complex 2 (PRC2) (Margueron and Reinberg, 2011). Canonical PRC2 comprises four core polypeptides, Ezh2, Eed, Suz12 and RbAp46/48, with Ezh2 being the enzymatic subunit that catalyzes H3K27me2/3. This Ezh2 activity is regulated by its partner proteins, such as Jarid2 and Aebp2 (Margueron and Reinberg, 2011). PRC2 occupies a large cohort of developmental genes in undifferentiated ESCs (Bernstein et al., 2006; Boyer et al., 2006; Lee et al., 2006). Yet despite this prevalent deposition, the determinants responsible for directing PRC2 occupancy at specific loci are still a focus of active investigation.
In addition to bivalent chromatin marks, developmental genes also harbor a particular form of RNAPII that is preferentially phosphorylated on Ser-5 residues (RNAPIIS5P) (Brookes et al., 2012; Stock et al., 2007). In ESCs, this form of RNAPII is often referred to as the ‘poised’ RNAPII (Brookes and Pombo, 2012) although the exact transcriptional state has yet to be fully defined. These chromatin and transcriptional features are regarded as representing a transcriptionally poised state that is amenable to rapid gene induction. Although the enzymes that catalyze the chromatin modifications comprising this transcriptionally poised state are well understood, less is known about the upstream ‘signal(s)’ that confers this transcriptional competency and how specific chromatin signatures are established and reconfigured during cell fate transitions (Voigt et al., 2013).
Cell fate determination entails signaling pathways that often activate downstream transcription factors, directing their deposition to specific genomic loci to elicit changes in gene expression through local changes in chromatin architecture (Badeaux and Shi, 2013; Johnson and Dent, 2013). However, emerging evidence suggest that terminal signaling effectors themselves may also cooperate with chromatin regulators to induce changes in chromatin organization, highlighting a direct convergence of signaling effectors on chromatin (Dawson et al., 2009; Goke et al., 2013; Ho et al., 2011; Klein et al., 2013). Several signaling pathways that are crucial for ESC pluripotency have been identified. Amongst these, the cytokine leukemia inhibitory factor (LIF) and its downstream signaling effector, Stat3, have been shown to be critical for self-renewal (Niwa et al., 1998). Conversely, Fgf4 induced activation of Erk1/2 triggers the transition from self-renewal to lineage commitment (Kunath et al., 2007). It is interesting to note that although LIF primarily activates the Stat3 signaling pathway, it also induces the Erk1/2 signaling pathway in conjunction with Fgf4 (Niwa et al., 2009). Erk1/2 signaling is also implicated in modulating the transition between the pluripotency states of mouse ESCs and epiblast stem cells (EpiSCs) (Greber et al., 2010). Interestingly, whilst Erk1/2 signaling drives lineage commitment in mouse ESCs, it is requisite for the maintenance of self-renewal in human ESCs (Greber et al., 2011). A role of Erk2 in transcriptional regulation was also recently reported for human ESCs (Goke et al., 2013). These observations underscore the importance of Erk1/2 signaling in stem cells. Conceivably, the transition between different states of pluripotency and differentiation necessitates changes in the chromatin landscape to consolidate gene expression changes. However, little is known as to how Erk1/2 orchestrates this transition or the identity of the downstream effectors.
While the role of Erk1/2 signaling in lineage priming is a recognized feature of proper ESC differentiation, the functional significance of PRC2 in ESCs is unclear, stemming from an incongruity between its prevalent deposition on developmental genes, and the relatively mild defect in self-renewal exhibited by PRC2 mutant ESCs (Chamberlain et al., 2008; Leeb et al., 2010; Pasini et al., 2007; Shen et al., 2008). Instead, PRC2 mutant ESCs exhibit impaired differentiation potential in vitro, more consistent with the post-implantation defects observed in vivo (Faust et al., 1995; O’Carroll et al., 2001; Pasini et al., 2004). These genetic studies suggest that PRC2 may play a more important role during lineage commitment. Given the importance of Erk1/2 signaling in lineage priming, we set out to determine if a functional relationship between Erk1/2 activation and PRC2 might exist to bring about proper ESCs differentiation. In the studies presented herein, we analyzed the downstream effects of Erk1/2 signaling, showing that Erk2 directly impinges on chromatin, and serves to regulate both the deposition of PRC2 as well as the establishment of the poised RNAPII, specifically on developmental genes.
RESULTS
Loss of PRC2 components leads to impaired Erk1/2 activation
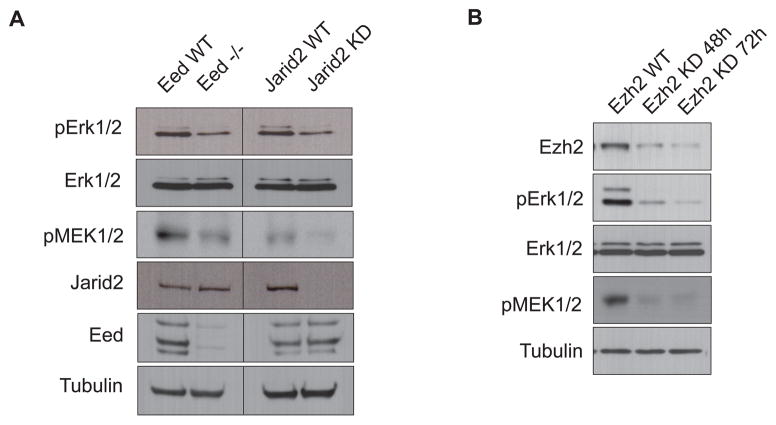
To address a possible functional relationship between Erk1/2 activation and PRC2-mediated repression, we first assessed the status of Erk1/2 activation in PRC2 mutant ESCs. Interestingly, knockdown of the PRC2 partner protein Jarid2 in ESCs led to a prominent reduction in phosphorylated Erk1/2 (pErk1/2) without an affect on Erk1/2 protein levels (Figure 1A). Moreover, an ESC line deficient in the core PRC2 subunit Eed (Eed−/−), also exhibited a similar reduction in pErk1/2 levels (Figure 1A). Assessment of the immediate upstream kinases, MEK1/2 in their activated phosphorylated forms (pMEK1/2) confirmed an impaired activation of the Fgf-MEK1/2-Erk1/2 pathway (Figure 1A). Notably, acute depletion of the PRC2 catalytic subunit Ezh2 by inducible shRNA knockdown provoked an even stronger reduction in pMEK1/2 and pErk1/2 (Figure 1B). Collectively, these data suggest that PRC2 mutant ESCs are defective at the level of MEK1/2-Erk1/2 activation that may relate to their aberrant differentiation.
Figure 1. Loss of PRC2 Components Abrogates Erk1/2 Activation.
(A) Western blot comparing phospho-Erk1/2 (pErk1/2) and phospho-MEK1/2 (pMEK1/2) levels in Eed null ESCs and in Jarid2 stable knockdown (KD) ESCs.
(B) Western blot comparing pErk1/2 and pMEK1/2 as a function of time of Ezh2 knockdown.
Reconfiguration of PRC2 occupancy on developmental genes upon Erk1/2 inactivation
To investigate if Erk1/2 activation and Polycomb-mediated repression are interdependent, we next examined the effects on PRC2 when Erk1/2 activation is thwarted through the use of a highly specific MEK1/2 inhibitor, PD0325901 (Ying et al., 2008). Strikingly, H3K27me3 was reduced on numerous developmentally regulated genes (Figure S1A). Importantly, pluripotency was preserved in these MEK1/2-inhibited ESCs, as evidenced by the up-regulation of stem cell markers such as Nanog, Tbx3 and Prdm14 and sustained repression of lineage specific genes (Figures S1B and S1C). Hence, the observed reduction in H3K27me3 does not account for transcriptional de-repression. This finding implicates Erk1/2 signaling in potentiating H3K27me3 deposition onto developmental genes and together with the data above, suggests an intricate relationship between Erk1/2 and Polycomb repression.
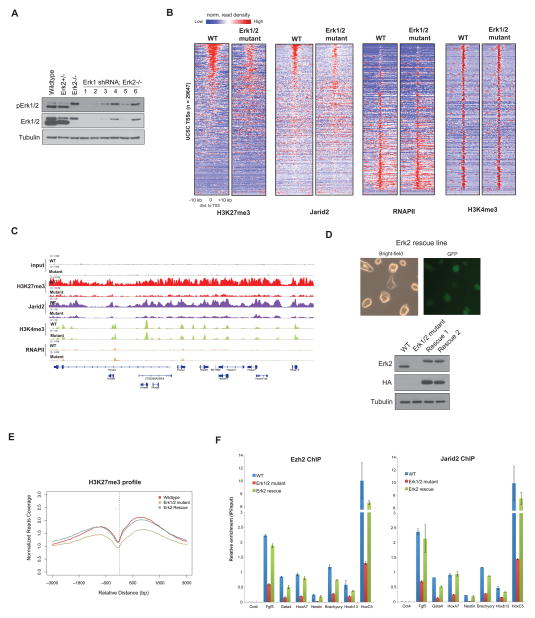
We next established ESCs that are deficient in both Erk1 and Erk2 to directly assess the role of these kinases in the context of Polycomb repression. We first obtained Erk2−/− ESCs (Kunath et al., 2007; Saba-El-Leil et al., 2003) and observed a compensatory up-regulation of phospho-Erk1 (Figure 2A), as reported previously (Kunath et al., 2007; Yao et al., 2003). We next depleted Erk1 using lentiviral based shRNA against Erk1 in the Erk2−/− ESCs, giving rise to Erk1/2 mutant ESCs (Figure 2A). As in the case of the MEK1/2-inhibited ESCs, we observed a similar up-regulation of pluripotency genes in the Erk1/2 mutant ESCs while many of the developmental genes remained repressed (Figure S2). Chromatin immunoprecipitation and sequencing (ChIP-seq) revealed that both H3K27me3 and Jarid2 exhibited a reduction that was focused primarily around the transcription start sites (TSS) of developmental genes in the Erk1/2 mutant cells (Figure 2B), essentially consistent with the results obtained using the MEK1/2 chemical inhibitor (Figure S3A). This effect appears to be specific for the Polycomb repressive machinery given that the pattern of H3K4me3 and H3K9me3 deposition remained largely unchanged on the developmental genes (Figures 2B, S3A and S3B). Examples of genes exhibiting this specific decrease in H3K27me3 and Jarid2 include the HoxA gene cluster, a classic target of PRC2 (Figure 2C). However, further cluster analysis revealed notable exceptions, with H3K27me3 levels being elevated at other loci (n=2221), that mainly encode cytokine signaling molecules (Figures S3C, S3D and Table S1). A significant reconfiguration of H3K27me3 and a decreased presence of PRC2 at developmental loci were demonstrated recently in mouse ESCs under ‘2i’ conditions that inhibit both MEK1/2 and GSK3 kinases (Marks et al., 2012). In our analysis using either the MEK1/2 inhibitor alone or Erk1/2 mutant cells, the specific loss of Erk1/2 activation resulted in this significant reconfiguration of H3K27me3 deposition. That some loci contained elevated H3K27me3 levels argues against a general loss in PRC2 enzymatic activity in accounting for the diminished H3K27me3 levels at the TSS. Moreover, the Erk1/2 mutant cells did not exhibit any loss in PRC2 components (Figure S3A), in accordance with the findings under ‘2i’ conditions (Marks et al., 2012). Notably, the reduction in H3K27me3 is not due to increased presence of H3K27me3 demethylases, Jmjd3 and Utx (Figure S3E). Taken together, our findings suggest that PRC2 targeting and/or its maintenance at target developmental genes has been compromised upon the specific loss of Erk1/2 in ESCs.
Figure 2. Erk1/2 Regulates PRC2 Occupancy on Developmental Genes in ESCs.
(A) Western blot for Erk1/2 and pErk1/2 in ESC lines developed toward the generation of Erk1/2 mutant ESCs. 1 – 6 denotes different shRNA constructs against Erk1.
(B) Heatmap representations of normalized read density of H3K27me3, H3K4me3, Jarid2 and total RNAPII corresponding to ~ 29000 annotated TSS (UCSC database) in wild type (WT) and Erk1/2 mutant ESCs. The normalized reads coverage against library size was calculated in the distance of ±10 kb to TSS with 200 bp bin. Heatmaps were ranked according to H3K27me3 enrichment in WT ESCs. Color scale represents normalized read density.
(C) Representative ChIP-seq tracks for H3K27me3, H3K4me3, Jarid2 and RNAPII on the HoxA gene cluster in WT and Erk1/2 mutant ESCs. The X-axis corresponds to genomic location, and the Y-axis to normalized ChIP-seq signal density.
(D) Generation of Erk2 rescue ESCs in the Erk1/2 mutant ESCs. Exogenous Erk2 is fused to an HA epitope tag and a downstream IRES-GFP. Western blots showing the expression of Erk2 (top panel) and HA-Erk2 (middle panel) in the respective cell lines.
(E) Average H3K27me3 profile for WT, Erk1/2 mutant and Erk2-rescue ESCs, centered on TSS. The average reads coverage of ~29000 TSS regions was calculated and normalized by total mapped reads. Histogram window size is ±3 kb with 10 bp bin size.
(F) ChIP-QPCR for Ezh2 and Jarid2 in WT, Erk1/2 mutant and Erk2-rescue ESCs. Results are presented as a percentage of input material. Bar represents mean of 2 replicates.
See also Figures S1, S2 and Table S1.
We next assessed if H3K27me3 could be restored upon stable expression of HA-tagged Erk2 in the Erk1/2 mutant ESCs (‘Erk2 rescue’ ESCs, Figure 2D). ChIP-seq analysis demonstrated that the genome-wide averaged signal of H3K27me3 was largely restored in the Erk2-rescue ESCs, at levels comparable to that in wild type ESCs (Figures 2E and S3F). Consistent with the restoration of H3K27me3, both Ezh2 and Jarid2 occupancies were similarly reconstituted in the Erk2-rescue cells (Figure 2F). Thus, activation of Erk1/2 signaling appears to impact PRC2 deposition on developmental promoters, either directly or indirectly. This interdependency between Erk1/2 signaling and PRC2 functioning suggests an unexpected feedback mechanism between these two processes.
Erk2 is recruited to a cohort of Polycomb target genes and alters nucleosome occupancy
To understand how Erk2 impacts PRC2 occupancy on developmental genes, we tested for the presence of Erk2 at the promoters of PRC2 target genes. ChIP-seq experiments demonstrate that Erk2 does indeed bind to developmental genes, as exemplified by the cases of the HoxA, HoxB gene clusters and Nkx2-2 (Figure 3A). We verified this Erk2 occupancy by comparing Erk2 ChIP-QPCR results between wild type versus Erk1/2 mutant ESCs (Figures S4A and S4B). Furthermore, inactivation of Erk1/2 using the MEK1/2 inhibitor (PD0325901 alone or ‘2i’) also led to an overall reduction in Erk2 occupancy (Figure 3B). We obtained approximately 2300 high confidence Erk2 enriched regions, of which 45% fall within 3 kb of the TSS (Table S2). Of these, ~90% overlap with H3K27me3 and Jarid2, corresponding to ~700 genes that contained Erk2-Jarid2-H3K27me3 within 3 kb of the TSS (Figures 3C, 3D and Table S3). Notably, the majority of the Erk2 enriched regions were depleted of H3K27me3 and deficient in Jarid2 levels in the Erk1/2 mutant cells (Figures 3C and S3C). Functional annotation of the Erk2 targets using GREAT (McLean et al., 2010) also showed strong association with lineage and cell commitment genes, typical of PRC2 regulation (Figure 3E). Examination of all Erk2 bound genomic sites revealed an overrepresentation of GA and GC rich DNA sequence motifs (Figure 3F), consistent with that previously reported for Jarid2 (Li et al., 2010; Peng et al., 2009). Further support for a direct binding of Erk2 to these repressive genomic regions was obtained using an in vitro gel shift assay with a representative double-stranded DNA oligomer containing an endogenously derived motif sequence (GA-rich) from the Pou3f2 locus along with recombinant Erk2, either wild type, or mutant in its DNA-binding (DBM) or kinase (KDM) domain (Hu et al., 2009). As shown in Figure 3G, wild type Erk2 was able to bind to the biotinylated DNA probe, whereas the DNA binding mutant was ineffectual. Erk2 binding was independent of its kinase activity (Figure 3G, left panel, lane 4) and largely unaffected by the presence of competing mutant oligomer harboring GA → GT mismatches (Figure 3G, lane 7). Moreover, competition experiments using probes, either wild type GA rich, or mutant GT rich, or GC rich, confirmed an affinity of Erk2 to GA motif sequences (Figure 3G, bottom right panel). Taken together, our data provides compelling evidence that Erk2 binds to specific repressive genomic loci in vivo, likely in a sequence specific manner.
Figure 3. Erk2 Binds to Polycomb-regulated developmental genes.
(A) ChIP-seq gene tracks showing occupancy of Erk2 on PRC2 target genes. The X-axis corresponds to genomic location, and the Y-axis corresponds to normalized ChIP-seq signal density.
(B) Average Erk2 profile for WT and MEK1/2 inhibited (PD0325901 and ‘2i’) ESCs, centered on TSS. The average reads coverage of ~29000 TSS regions was calculated and normalized by total mapped reads. Histogram window size is ±10 kb with 10 bp bin size.
(C) Heatmaps of H3K27me3 and Jarid2 normalized reads distribution on all 2296 Erk2 enriched regions. H3K27me3 and Jarid2 are present in the majority of Erk2 enriched regions in wild type (WT) ESCs, but are depleted in Erk1/2 mutant cells. Data matrix was constructed with Erk2 peak regions centered by peak summit with ±5 kb extension on both sides and a bin size of 50 bp, as described in Extended Experimental Procedures.
(D) Venn diagram of the overlap among target genes of Erk2, Jarid2 and H3K27me3 in WT ESCs. In this case, only respective target genes with at least one peak within 3 kb of the TSS were considered. A smaller list comprising Jarid2 and H3K27me3 target genes was generated by first annotating Jarid2 and H3K27me3 peaks to Erk2, as described in Extended Experimental Procedures.
(E) GO term analysis of all Erk2 targets.
(F) Motif analysis of all Erk2 enriched regions using MEME. Two distinct motifs were identified (GA- and GC-rich).
(G) Left, Gel shift assay comparing recombinant Erk2, either wild type (WT), mutant in DNA-binding activity (DBM) or kinase activity (KDM) in the presence of probe containing a biotinylated GA-motif derived from the Pou3f2 locus. Top Right, Gel shift titrations and binding curve of WT- and KDM- Erk2. Increasing amounts of Erk2 proteins were used (0 – 10 μg) in the presence of the Pou3f2 GA-rich DNA probe. Bottom Right, Graph showing the effects of increasing amounts of different cold probes (0 – 250x) in affecting the binding between wild type Erk2 and the GA-rich DNA probe. ‘wt’ denotes wild type probe, while ‘mt’ denotes mutant GA -> GT probe.
See also Figures S3, S4, Table S2 and Table S3.
We considered the possibility that Erk2 binding to GA-rich regions might demarcate localized regions of chromatin accessibility. Thus, we performed histone H3 ChIP to directly assess nucleosome occupancy on select Erk2 binding sites in the case of (i) wild type, (ii) Erk1/2 mutant, (iii) two independently derived Erk2-rescue cell lines, and (iv) ESCs treated with either MEK1/2 inhibitor alone or (v) with both MEK1/2 and GSK3 inhibitors; ‘2i’. The results show a modest, but consistent increase in H3 occupancy on the Erk2 enriched regions tested in all the ESC lines wherein Erk1/2 was inactivated, relative to the cases of Erk2-rescue and wild type ESCs (Figure 4). On the other hand, H3 occupancy on pluripotency genes, represented by Oct4 and Sox2 promoters, was essentially similar, irrespective of Erk1/2 status (Figure 4). The addition of GSK3 inhibitor alone did not lead to any alterations in H3 occupancy on the Erk2 regions tested (Figure S5). These results suggest that Erk1/2 activation may be required for appropriate modulation of nucleosome occupancy in ESCs given that Erk2 binding corresponds to regions of histone eviction, and its loss promotes increased nucleosome density.
Figure 4. Erk1/2 regulates nucleosome occupancy.
Comparison of histone H3-ChIP QPCR for select Erk2-PRC2 bound developmental genes amongst wild type (WT) ESCs, two independent Erk2-rescue lines (rescue 1 and rescue 2), WT ESCs treated with MEK1/2 inhibitor (PD03), or with both MEK1/2 and GSK3 inhibitors (2i). Oct4 and Sox2 represent active genes, and the remaining represent Erk2-PRC2 developmental genes. Results are presented as a percentage of input material. Bar represents s.d of two replicates. (*) denotes p-value of <0.05, (#) denotes p-value <0.2 and (n.s) denotes not-significant. A two-tailed student t-test was performed.
See also Figure S5.
Activated Erk1/2 regulates establishment of poised RNAPII on Polycomb target genes
Interestingly, despite the apparent reduction in PRC2 occupancy when Erk1/2 signaling is impaired, many of these Erk2 targeted developmental genes remained repressed (Figures S2 and S6A). Recent studies detected the presence of a poised form of RNAPII(S5P) on PRC2 target promoters, that likely contributes to the facultative transcriptional state of these developmental genes in undifferentiated ESCs (Brookes et al., 2012; Stock et al., 2007). Thus, we next compared the status of RNAPII(S5P) in wild type versus Erk1/2 mutant cells by ChIP-seq. Erk1/2 mutant cells exhibited a significant reduction in RNAPII(S5P) occupancy on Erk2-PRC2 bound promoters such as the HoxA gene cluster and Cebpa, but not on active genes such as Nanog (Figure 5A). Importantly, genome-wide analysis confirmed that this reduction in RNAPII(S5P) was specific for Erk2-PRC2 bound promoters and not relevant in the case of highly active promoters (Figure 5B). Consistently, there were no detectable changes in RNAPII(S5P) levels in the case of the control group of silenced genes, as expected (Figure 5B). Moreover, the reduction in RNAPII(S5P) observed for Erk2-PRC2 genes cannot be attributed to a general reduction in RNAPII occupancy (Figures 5A and S6A). We also extended this analysis to include ESCs treated with MEK1/2 inhibitor, and observed a similar depletion of RNAPII(S5P) on select developmental genes (Figure S6B). Thus, abrogation of Erk1/2 leads to a specific reduction in RNAPII(S5P) on PRC2 target genes. That the absence of the well-recognized kinase Erk1/2, or impairment of its activity, leads to loss of the poised version of RNAPII at developmental loci, led us to investigate if Erk1/2 itself is responsible for the phosphorylation of Ser-5 on the CTD of RNAPII at developmental genes in ESCs. The results of in vitro kinase assays indicate that Erk2 is able to phosphorylate RNAPII on Ser-5 (Figure 5C). Moreover, Erk2 interacts with RNAPII in vivo as evidenced by endogenous immunoprecipitation experiments (Figure 5C, right panel).
Figure 5. Interplay Between Erk2 and TFIIH Determines Transcriptional States.
(A) Representative ChIP-seq gene tracks showing loss of RNAPII(S5P) on developmental genes (HoxA cluster and Cebpa), but not on the active gene, Nanog. ‘WT’ denotes wild type ESCs and ‘Mutant’ denotes Erk1/2 mutant ESCs. The gene tracks for H3K27me3, Jarid2 and RNAPII are also shown. The X-axis represents genomic locus and the Y-axis corresponds to normalized read density.
(B) Average normalized profiles of RNAPII(S5P), in wild type and Erk1/2 mutant cells, for different cohorts of genes as designated. Reads were centered at either ±500 bp or ±10 kb to the TSS. ‘High expressed’ and ‘silent’ genes represent the top 2000 most active and bottom 2000 least expressed genes in mouse ESCs respectively, as described in Extended Experimental Procedures. The ‘Erk2-PRC2’ group refers to the cohort of genes bound by Erk2 and H3K27me3, as classified in Figure 3D. A 10 bp bin was used in the plot generation.
(C) Left, In vitro kinase assay using Erk2 and RNAPII-CTD as substrate. Right, Endogenous co-immunoprecipitation using HA antibody and extracts of wild type ESCs (control) or ESCs stably expressing HA-tagged Erk2.
(D) Top, Ercc3 is only present on active genes (such as Klf2 and histone gene cluster) in ESCs, but not on Erk2-PRC2 bound developmental genes such as Gata6.
(E) Average profile of Ercc3 in wild type and Erk1/2 mutant ESCs, based on gene expression levels (similar to the classification used in Figure 5B). A 10 bp bin was used in the generation of the plots.
(F) ChIP-QPCR for RNAPII(S5P) comparing Ercc3-bound and Erk2-bound targets following Triptolide treatment (1 μM for 30 min). Bar represents s.d of two replicates.
(G) ChIP-QPCR for RNAPII(S5P) showing restoration of RNAPII(S5P) on developmental genes upon wild type (WT) Erk2 expression, but not in the case of the kinase domain mutant (KDM). Bar represents s.d of two replicates.
(H) ChIP-QPCR for Ezh2 showing restoration of Ezh2 on developmental genes upon wild type (WT) Erk2 expression but not in the case of kinase domain mutant (KDM). Bar represents s.d of two replicates.
See also Figure S6.
These findings are consistent with Erk2 (and likely Erk1) contributing to direct phosphorylation of RNAPII on Ser-5, specifically at targeted developmental genes. However, it is generally accepted that phosphorylation of Ser-5 (as well as Ser-7) on the CTD of RNAPII is largely mediated by the TFIIH complex (Akhtar et al., 2009; Komarnitsky et al., 2000; Lu et al., 1992). Therefore, we next assessed the presence of TFIIH in wild type versus Erk1/2 mutant ESCs using ChIP-seq for Ercc3, the largest subunit of the TFIIH complex (Drapkin et al., 1994; Schaeffer et al., 1993). Strikingly, Ercc3 showed an enrichment pattern inverse to that of Erk2 in the wild type case. Unlike Erk2, Ercc3 was largely absent on low/inactive promoters and specifically enriched on highly active promoters such as Klf2 and histone gene cluster 1 (Figures 5D, 5E and S6C). It should be emphasized that the decrease in RNAPII(S5P) occupancy in the Erk1/2 mutant cells was only observed on developmental genes, not on highly active genes (Figure 5B). Notably, in the Erk1/2 mutant ESCs, the highly active genes showed on average, an increased Ercc3 promoter signal (Figure 5E), mirroring the gain in RNAPII(S5P) (Figures 5B and S6A). This is in accordance with the well-documented role of TFIIH in promoting transcription initiation (Kim et al., 2000; Kumar et al., 1998). Importantly, inhibition of TFIIH activity by Triptolide (Titov et al., 2011) results in a selective decrease in RNAPII(S5P) signal on Ercc3-target genes such as Klf2 and Hist1h1a, but not on Erk2-bound developmental genes (Figures 5F and S6D). This finding thus provides additional support for a TFIIH-independent role of Erk2 in the establishment of RNAPII(S5P) on developmental genes. Finally, to ascertain that phosphorylation of RNAPII at Ser-5 is dependent on the kinase activity of Erk2, we performed rescue experiments to compare the ability of Erk2, either wild type or mutant in its kinase domain (KDM), to restore RNAPII(S5P) levels on developmental genes. Whereas expression of wild type Erk2 led to a significant increase in RNAPII(S5P) levels on developmental genes relative to Erk1/2 mutant cells, expression of Erk2-KDM was ineffectual (Figures 5G and S6E). Moreover, specific to the case of the wild type Erk2 rescue cells, the restoration of RNAPII(S5P) was accompanied by an increase in Ezh2 occupancy on the same developmental loci (Figure 5H). Taken together, our in vitro as well as in vivo data strongly indicate that activated Erk2 is likely a bona fide RNAPII CTD kinase in ESCs and through its kinase activity, is responsible for the establishment of the poised chromatin features on developmental genes.
Dynamics of Erk2 binding during ESC differentiation
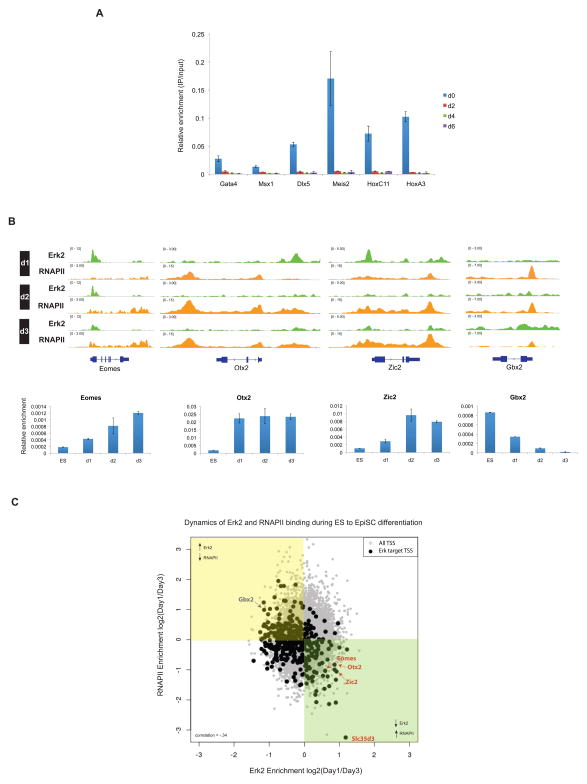
The results thus far have shown that Erk1/2 directly accesses PRC2 target genes, and impacts RNAPII(S5P) as well as PRC2 occupancy in ESCs. If PRC2 functions downstream of Erk1/2 activation to preclude gene activation in the ESC state, then transcription activation of development genes during differentiation should be accompanied by the loss of Erk2 binding and alleviation of PRC2-mediated repression. Indeed, Erk2 binding on developmental genes was reduced as a function of retinoic acid (RA)-mediated ESC differentiation, as early as day 2 of differentiation, and concomitant with transcription activation (Figures 6A and S7). We next followed the dynamics of Erk2 binding and RNAPII occupancy during early events of ESC conversion to EpiSC, whereby changes in gene expression occur more gradually. Developmental genes known to be upregulated in EpiSCs, relative to ESCs, e.g. Eomes, Otx2 and Zic2, exhibited the expected activation, along with decreased Erk2 binding (Figure 6B). On the other hand, Gbx2, whose expression is up-regulated in ESCs relative to EpiSCs, showed the reverse trend, with RNAPII decreasing and Erk2 occupancy increasing, as a function of differentiation (Figure 6B). Notably, an unbiased analysis of all Erk2 target genes supports a general inverse correlation between Erk2 occupancy and transcriptional activity during differentiation (Figure 6C).
Figure 6. Dynamics of Erk2 Binding During Differentiation.
(A) ChIP-QPCR for Erk2 showing loss of Erk2 binding on Erk2-PRC2 targets upon retinoic acid differentiation (day 0 to day 6). Results are presented as a percentage of input material. Bar represents s.d of two replicates.
(B) Top, ChIP-seq tracks showing the kinetics of Erk2 binding and RNAPII occupancy during mouse ES to Epiblast stem cell (EpiSC) differentiation (day 1 to day 3). Bottom, mRNA expression levels of the target genes specified, normalized to Gapdh. Bar represents s.d of two replicates.
(C) Scatter plot of Erk2 and RNAPII occupancy at Erk2 target genes during ES to EpiSC conversion (day 1 to day 3). X and Y-axes indicate the log2 ratio (day1/day3) of normalized ChIP-seq read density for Erk2 and RNAPII (total) mapping within ±3 kb of each TSS. Black dots correspond to the TSS of high confidence Erk2 target genes, and grey dots represent all other TSS. Correlation coefficient = −.34
See also Figure S7.
DISCUSSION
The presence and functional significance of poised RNAPII(S5P) on developmental genes have been a subject of debate. Although genome-wide RNAPII ChIP-seq studies have revealed a widespread presence of low levels of RNAPII at inactive promoters, including those bound by PRC2 (Guenther et al., 2007; Kim et al., 2005; Muse et al., 2007; Zeitlinger et al., 2007), many outstanding questions remain regarding the regulation of RNAPII recruitment to these loci, the identity of the CTD kinase(s) responsible for its Ser-5 phosphorylation, and the exact transcriptional configuration of this particular form of RNAPII, RNAPII(S5P). Recent studies have reassessed the transcriptional status of Polycomb repressed genes and found that Polycomb binding is surprisingly compatible with moderate gene expression (Brookes et al., 2012; Enderle et al., 2011; Kaneko et al., 2013) and hence, may facilitate the fine-tuning of gene expression, rather than enforce an obligatory silencing. This reflects a paradigm shift in how Polycomb proteins are widely perceived to regulate gene expression in mammals. Furthermore, the detection of stalled RNAPII on Polycomb targets (Chopra et al., 2009; Min et al., 2011; Stock et al., 2007) coupled with the detection of short RNAs emanating from these loci (Kanhere et al., 2010) imply an interplay between PRCs and the transcriptional apparatus (Brookes et al., 2012; Enderle et al., 2011; Min et al., 2011). Indeed, our results showing interplay between Erk1/2 activity and both RNAPII and PRC2 bear directly on this intricate regulation.
In this study, we describe the unexpected deposition of Erk2 on developmental promoters, as well as the corresponding decrease in RNAPII(S5P) upon Erk1/2 inactivation. Notably, the reduction in RNAPII(S5P) is specific to developmental genes and not active ones. These observations, along with those demonstrating the ability of Erk2 to interact with and phosphorylate RNAPII at the Ser-5 residue (Trigon et al., 1998), as also shown here, point to Erk2 being a bona fide Ser-5 RNAPII CTD kinase that acts primarily at developmental loci. This supposition is in accordance with earlier biochemical studies showing that Erk1/2 preferentially targets Ser-5 but not other serine residues on the CTD (Bonnet et al., 1999; Dubois et al., 1994; Trigon et al., 1998), as well as the implication of MAPKs in RNAPII phosphorylation during early development in vivo (Bellier et al., 1997), and during heat shock (Venetianer et al., 1995). Buttressing this role of Erk1/2 is the complementary, also unexpected finding that the presence of TFIIH at developmental genes is low and/or undetectable and that the levels of RNAPII(S5P) on these loci are unperturbed upon TFIIH inhibition by Triptolide, yet sensitive to Erk2 activation. This specific deficiency in TFIIH is in line with the reported lack of RNAPII(S7P) (also imposed by TFIIH) on Polycomb target genes (Brookes et al., 2012). Notably the lack of TFIIH on the developmental loci cannot be simply attributed to a lower level of transcription initiation at these loci relative to active genes (Figure S6F). Although we cannot completely rule out that a very low and/or transient occupancy of TFIIH may exist, our results clearly underscore a previously unrecognized role of Erk1/2 kinase in catalyzing RNAPII(S5P) at these developmental promoters. Future studies will address the interplay between these two CTD kinases and if certain roles may be subsumed and/or interchanged during transcription initiation.
The CTD of mammalian RNAPII contains 52 repeats of the heptapeptide, containing the consensus sequence, Tyr-Ser-Pro-Thr-Ser-Pro-Ser. Despite the similarity in sequence, previous studies in yeast have demonstrated that not every heptapeptide repeat in the CTD is functionally equivalent (Nonet et al., 1987; Scafe et al., 1990). We postulate that Erk-mediated phosphorylation of the CTD is specific to developmental genes and that this occurs at unique heptapeptide(s) within the CTD, perhaps distinct from that targeted by TFIIH (see model in Figure 7). In light of our previous reports demonstrating that the Ercc3 subunit of TFIIH is necessary for the establishment of an open complex (Kim et al., 2000), how then is transcription initiation achieved on developmental genes in its absence? Previous studies had demonstrated that the TFIIH requirement for transcription could be bypassed upon induced superhelicity of the DNA template (Goodrich and Tjian, 1994; Parvin and Sharp, 1993). Thus, it will be interesting to assess any changes in promoter DNA topology on these developmental genes as a function of Erk activity, and how that relates to transcriptional priming. Notably, Erk2 target genes are highly enriched for DNA Topoisomerase 2a (Top2a, an enzyme required to resolve torsional stress resulting from DNA supercoiling), despite a conspicuous lack of Ercc3 occupancy (data not shown). This corroborates our interpretation that distinct mechanisms may be in place to regulate transcription initiation on active versus poised genes. Collectively, based on our findings and those of Parvin and Sharp as well as Goodrich and Tjian, we propose that combinatorial utilization of different Ser-5 RNAPII CTD kinases and differential phosphorylation of different CTD repeats are necessary to activate or prime gene expression in ESCs. Promoters bound by Erk2 may be competent for transcription as evident by high levels of RNAPII(S5P) and Top2a, but robust transcription likely occurs only upon subsequent recruitment of the multi-subunit TFIIH complex, which fosters robust promoter melting and clearance through its different components (Compe and Egly, 2012). This is in good agreement with our data showing a progressive displacement of Erk2 on developmental genes in anticipation of transcription activation during differentiation. This proposed interplay between Erk and TFIIH may be analogous to the recent description of TFIIH in transcriptome amplification during B cell activation (Kouzine et al., 2013).
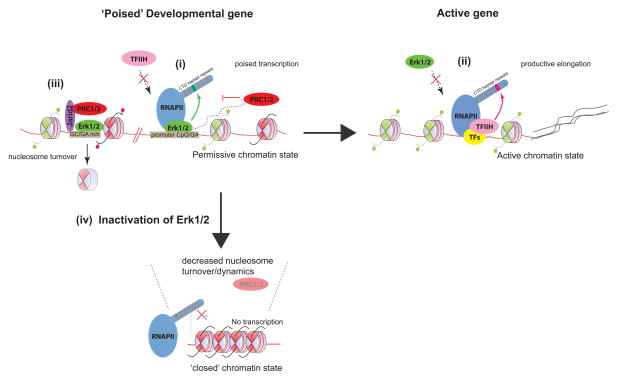
Figure 7. Model.
In the presence of basal Erk1/2 activity, lineage specific promoters exist in a permissive chromatin state. Erk2 can access and bind to underlying sequences, including specific DNA motifs that are rich in GA dinucleotides, and/or to promoter CpG islands. (i) At promoters, binding of Erk2 may potentially antagonize TFIIH recruitment. Through its own kinase activity, Erk2 phosphorylates Ser-5 in a particular RNAPII CTD heptad repeat, thus establishing a stalled/poised form of RNAPII that permits regulated transcription of promoter derived ncRNAs or nascent transcripts (represented as black dashed lines). This event may trigger the subsequent recruitment of PRC1/2 complexes to buffer/fine-tune gene expression. (ii) Upon gene activation, Erk2 is displaced, and TFIIH must be recruited, alongside other transcription factors (TFs) and cofactors to promote robust gene activation (mRNA represented by black solid lines). TFIIH may target a CTD heptad repeat distinct from that of Erk2. (iii) Erk2 also binds to non-promoter regions, and may help potentiate Jarid2/PRC2 targeting and/or spreading through an initial wave of nucleosome eviction. (iv) In the absence of Erk1/2, developmental promoters may assume a more ‘inert’ chromatin state, with increased nucleosome stability and reduced propensity for permissive transcription. PRC2 may thereby be rendered dispensable on these developmental genes.
As shown here, the loss of Erk1/2 signaling also results in alterations in chromatin accessibility and a profound impact on PRC2 deposition. Negligible changes were observed for H3K4me3 and H3K9me3 depositions on developmental genes. This is in line with the dynamic and plastic nature of the Polycomb repressive machinery, the recruitment of which is attuned to environmental perturbations (Prezioso and Orlando, 2011). We speculate that Erk1/2-induced changes in chromatin accessibility may be key to potentiating PRC2 recruitment. Mechanistically, the increased nucleosome occupancy at Erk2 bound regions in the absence of Erk1/2 activation may occlude underlying DNA elements involved in PRC2 recruitment, thereby precluding focal deposition of H3K27me3. Interestingly, motif analysis of all Erk2 binding sites revealed a conspicuous enrichment for GA dinucleotides. This GA-rich polypurine motif was also previously described for Jarid2 (Peng et al., 2009) as well as the non-coding RNA (ncRNA), HOTAIR (Chu et al., 2011). It is noteworthy that a substantial proportion of Erk2 also resides in intergenic and intronic regions, apart from promoters. These observations raise a tantalizing possibility that Erk2 binding sites may demarcate genomic regions for Polycomb nucleation and subsequent spreading, perhaps analogous to that of Drosophila Polycomb Responsive Element (PRE). Our observation that Erk1/2 activation can regulate nucleosome disruption is of particular relevance given that PREs are typically depleted of nucleosomes (Mohd-Sarip et al., 2006). As such, it is conceivable that Erk1/2 may be part of a larger protein ensemble that is targeted to these putative Polycomb binding sites, and through local nucleosome turnover, promotes subsequent PRC2 recruitment (see model in Figure 7). Further investigation is needed, on a genome-wide level, to precisely define how changes in global nucleosome occupancy impacts PRC2 recruitment, and its relationship to RNAPII stalling (Gilchrist et al., 2010).
In addition to the aforementioned GA motif, other DNA elements including transcription factor binding sites, as well as promoter unmethylated CpG islands have been implicated in PRC2 recruitment (for a review see (Simon and Kingston, 2013)). At least in ESCs, the latter have been shown to play a causal role in PRC2 recruitment (Lynch et al., 2012; Mendenhall et al., 2010) and notably, in silico analysis reveals that increasing density of promoter CpG islands apparently correlates with the extent of H3K27me3 deposition (Orlando et al., 2012). Thus, an additional interesting hypothesis is that multiple CpG islands may potentiate the generation of nascent primary transcripts and/or ncRNAs that serve to recruit and/or facilitate PRC2 binding (see model in Figure 7). Indeed, a previous study determined that short ncRNAs are transcribed from PRC2 target loci (Kanhere et al., 2010). Although it remains to be formally tested if these CpG-related ncRNAs indeed direct PRC2 deposition, it nevertheless highlights a potential link between promoter effect transcription and PRC2 recruitment in cis. In this regard, our in vivo rescue experiments (Figures 5G and 5H) showing the concomitant restoration of both RNAPII(S5P) and PRC2 upon Erk activation is largely supportive of this hypothesis, and points toward a modus operandi wherein PRC2 recruitment occurs in a hierarchical manner downstream of transcriptional activity (see also (Kaneko et al., 2013)).
It was previously reported that Jarid2 deficient ESCs fail to efficiently establish the poised RNAPII on PRC2 targets (Landeira et al., 2010). That Jarid2/PRC2 deficient cells are compromised in Erk1/2 activation as shown here, may account, at least in part, for the reduced levels of RNAPII(S5P) on developmental genes. However, it remains unclear how loss of Jarid2/PRC2 impacts Erk1/2 activation. This may occur indirectly through the upregulation of Prdm14, a known target of PRC2, implicated in the attenuation of the Fgf-Erk pathway (Grabole et al., 2013; Yamaji et al., 2013) and/or through mis-regulation of other signaling cascades, such as the LIF/Stat3 pathway that also contribute towards Erk1/2 activation in ESCs (Niwa et al., 2009).
In summary, our data demonstrates how Erk1/2 induced effects on RNAPII poising and chromatin architecture may potentiate changes in PRC2 occupancy in ESCs, and emphasize the importance of signal-induced chromatin remodeling as a key epigenetic priming event that underlies the transcriptional competent state of developmental promoters in ESCs.
Experimental Procedures
Mouse ESCs maintenance and differentiation
All ESCs were grown in standard ESC medium containing LIF, unless otherwise stated. Erk2−/− ESCs were a kind gift from Dr. Sylvain Meloche. Erk1/2 mutant ESCs were maintained in the presence of 1 μg/ml puromycin and Erk2-rescue ESCs in 150 μg/ml hygromycin and 1 μg/ml puromycin. For ‘2i’ ESCs, 1 μM MEK1/2 inhibitor (PD0325901) and 3 μM GSK3 inhibitor (CHIR99021) were used. See also Extended Experimental Procedures.
Cloning, Transfections and shRNA knockdown
pLKO-based Erk1 lentiviral shRNA (TRCN0000023184) or control GFP shRNA (Thermo Scientific) plasmids were used to produce the lentiviruses. For generation of Erk2-rescue ESCs, Erk2 cDNA was PCR-amplified from ESCs and cloned into pCAGIG (GFP-marker) or pCAGIH (Hygromycin-marker) expression vectors. Site directed mutagenesis was carried out to obtain the KDM construct. See Extended Experimental Procedure for details on primer sequences.
EMSA
Each binding reaction was carried out with 1 ng of biotinylated dsDNA probe and 1 μg of Erk2 recombinant protein in 25 μl reaction buffer (50 mM Hepes, pH 7.9, 250 mM KCl, 2.5 mM EDTA, 25% glycerol, 5 μg BSA and 10 ng dI/dC). Reactions were carried out for 1 h at room temperature, separated on a 6% native TGE polyacrylamide gel at 4°C, and visualized using the LightShift EMSA kit (Pierce/Thermo).
Triptolide treatment
Triptolide was obtained from InvivoGen (catalog # ant-tpl). ESCs were treated at the stated concentrations for 30 min.
ChIP-PCR and ChIP seq
ChIP assays were performed as described previously (Gao et al., 2012). ChIP-westerns were also performed for each antibody used. For ChIP-seq, up to 30 ng of immunoprecipitated DNA was end-repaired, A-tailed, and ligated to custom barcode adapters with T4 ligase. Libraries were sequenced on Illumina HiSeq. A custom barcoding system was employed. The GEO accession number is SRP028688. See Extended Experimental Procedures for details on antibodies, Q-PCR primer sequences and computational analysis.
Supplementary Material
HIGHLIGHTS.
Erk1/2 activity regulates the targeting of PRC2-Jarid2 in mouse ESCs.
Erk2 binds to a cohort of PRC2-targeted developmental genes.
Erk2 phosphorylates RNAPII at its Serine 5 residue at developmental genes.
TFIIH is present on active genes but absent on Erk2-PRC2 developmental genes.
Acknowledgments
We thank the Genome Technology Center at NYU for help with sequencing, Theodoros Savvidis for technical support, Jinsook Son and Dr. Shuzo Kaneko for reagents, and Drs. Lynne Vales, Philipp Voigt and Roberto Bonasio for comments on the manuscript. We thank Drs. Yang Shi and Kristian Helin for kind gifts of the Jmjd3 and Utx antibodies, respectively. This work was supported by grants from the National Institute of Health (GM-64844 and R37-37120), NYSTEM (C028105) and the Howard Hughes Medical Institute (to D.R.). W-W. Tee is supported by a Druckenmiller post-doctoral fellowship from the New York Stem Cell Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Molecular cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nature reviews Molecular cell biology. 2013;14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellier S, Dubois MF, Nishida E, Almouzni G, Bensaude O. Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Molecular and cellular biology. 1997;17:1434–1440. doi: 10.1128/mcb.17.3.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bonnet F, Vigneron M, Bensaude O, Dubois MF. Transcription-independent phosphorylation of the RNA polymerase II C-terminal domain (CTD) involves ERK kinases (MEK1/2) Nucleic acids research. 1999;27:4399–4404. doi: 10.1093/nar/27.22.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell stem cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, Pombo A. Code breaking: the RNAPII modification code in pluripotency. Cell cycle. 2012;11:1267–1268. doi: 10.4161/cc.19990. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Hong JW, Levine M. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Current biology: CB. 2009;19:688–693. doi: 10.1016/j.cub.2009.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Molecular cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nature reviews Molecular cell biology. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- Dubois MF, Nguyen VT, Dahmus ME, Pages G, Pouyssegur J, Bensaude O. Enhanced phosphorylation of the C-terminal domain of RNA polymerase II upon serum stimulation of quiescent cells: possible involvement of MAP kinases. The EMBO journal. 1994;13:4787–4797. doi: 10.1002/j.1460-2075.1994.tb06804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome research. 2011;21:216–226. doi: 10.1101/gr.114348.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121:273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Molecular cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goke J, Chan YS, Yan J, Vingron M, Ng HH. Genome-wide kinase-chromatin interactions reveal the regulatory network of ERK signaling in human embryonic stem cells. Molecular cell. 2013;50:844–855. doi: 10.1016/j.molcel.2013.04.030. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Grabole N, Tischler J, Hackett JA, Kim S, Tang F, Leitch HG, Magnusdottir E, Surani MA. EMBO reports. 2013. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B, Coulon P, Zhang M, Moritz S, Frank S, Muller-Molina AJ, Arauzo-Bravo MJ, Han DW, Pape HC, Scholer HR. FGF signalling inhibits neural induction in human embryonic stem cells. The EMBO journal. 2011;30:4874–4884. doi: 10.1038/emboj.2011.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P, et al. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell stem cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nature cell biology. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Dent SY. Chromatin: receiver and quarterback for cellular signals. Cell. 2013;152:685–689. doi: 10.1016/j.cell.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nature structural & molecular biology. 2013 doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Molecular cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- Klein AM, Zaganjor E, Cobb MH. Chromatin-tethered MAPKs. Current opinion in cell biology. 2013;25:272–277. doi: 10.1016/j.ceb.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & development. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon KR, Bandle R, Nelson S, Nakahashi H, Awasthi P, Feigenbaum L, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–999. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KP, Akoulitchev S, Reinberg D. Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9767–9772. doi: 10.1073/pnas.95.17.9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nature cell biology. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes & development. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes & development. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- Lynch MD, Smith AJ, De Gobbi M, Flenley M, Hughes JR, Vernimmen D, Ayyub H, Sharpe JA, Sloane-Stanley JA, Sutherland L, et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. The EMBO journal. 2012;31:317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nature biotechnology. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS genetics. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes & development. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. Architecture of a polycomb nucleoprotein complex. Molecular cell. 2006;24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nature genetics. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nature cell biology. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes & development. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Nonet M, Sweetser D, Young RA. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Molecular and cellular biology. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Guenther MG, Frampton GM, Young RA. CpG island structure and trithorax/polycomb chromatin domains in human cells. Genomics. 2012;100:320–326. doi: 10.1016/j.ygeno.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Molecular and cellular biology. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. The EMBO journal. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezioso C, Orlando V. Polycomb proteins in mammalian cell differentiation and plasticity. FEBS letters. 2011;585:2067–2077. doi: 10.1016/j.febslet.2011.04.062. [DOI] [PubMed] [Google Scholar]
- Saba-El-Leil MK, Vella FD, Vernay B, Voisin L, Chen L, Labrecque N, Ang SL, Meloche S. An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO reports. 2003;4:964–968. doi: 10.1038/sj.embor.embor939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafe C, Chao D, Lopes J, Hirsch JP, Henry S, Young RA. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JH, Chambon P, Egly JM. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annual review of biochemistry. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Molecular cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nature cell biology. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nature chemical biology. 2011;7:182–188. doi: 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. The Journal of biological chemistry. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- Venetianer A, Dubois MF, Nguyen VT, Bellier S, Seo SJ, Bensaude O. Phosphorylation state of the RNA polymerase II C-terminal domain (CTD) in heat-shocked cells. Possible involvement of the stress-activated mitogen-activated protein (MAP) kinases. European journal of biochemistry/FEBS. 1995;233:83–92. doi: 10.1111/j.1432-1033.1995.083_1.x. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell stem cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Yao Y, Li W, Wu J, Germann UA, Su MS, Kuida K, Boucher DM. Extracellular signal-regulated kinase 2 is necessary for mesoderm differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12759–12764. doi: 10.1073/pnas.2134254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature genetics. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.