Abstract
Arginine (Arg) is a conditionally-essential amino acid that is taken up by bovine mammary gland in excess of its output in milk. In this study we evaluated the effects of Arg concentration on the expression of casein and signaling pathway-related genes in mammary epithelial cells. The treatments (applied for 24 h) were designed to be devoid of Arg 0X (control; 0.00 mg/L), resemble the profile of Arg in casein (Arg 1X; 278.00 mg/L), be deficient [Arg 0.25X (69.50 mg/L) and Arg 0.5X (139.00 mg/L)], or be in excess of the amount in casein [Arg 2X (556.00 mg/L), Arg 4X (1,112 mg/L), and Arg 8X (2,224 mg/L)]. The expression of CSN1S, CSN3 and mTOR in the experimental groups was higher than those of the control group (P<0.05). Except for Arg 0.25X and Arg 8X (P>0.05), the expression of CSN1S2, CSN2 and JAK2 in other experimental groups was higher (P<0.05) than those in the control group. Except for Arg 8X (P>0.05), the expression of STAT5 in the other experimental groups was higher than those of the control (P<0.05). It also was observed that except for Arg 0.5X, the S6K expression was higher in other experimental groups than the control (P<0.05). In contrast, except for Arg 0.25X the other experimental groups resulted in lower 4EBP1 expression than the control (P<0.05). Among groups, the expression of CSN1S1, CSN1S2, CSN2, CSN3, JAK2, STAT5, mTOR and S6K gene was highest with Arg 2X (P<0.05); the reverse was true for 4EBP1 gene, with the lowest expression in this group (P<0.05). Taken together, Arg appears to play an important role in the transcriptional regulation of casein genes and mTOR-related genes in bovine mammary epithelial cells.
Introduction
There is growing recognition that besides their role as building blocks of proteins and polypeptides, some amino acids (AA) regulate key metabolic pathways that are necessary for maintenance, growth, reproduction, and immunity [1]. Arginine, as a AA, plays an important role in the control of cell division, wound healing, removal of ammonia from the body, protein synthesis, and the release of hormones [2], [3], [4]. Interestingly, reviews of the literature in dairy ruminants concluded that the uptake of arginine by the mammary gland greatly exceeded its output in milk [5], [6]. Except for its use during milk protein synthesis, those reviews concluded that the extra arginine might exert other unknown biological or metabolic functions in the mammary gland.
The availability of AA in the lactating cow is critical for mammary protein synthesis [7]. Previous work on AA metabolism in dairy cattle have focused on achieving an optimal balance in the diet to provide the required precursors for maintaining and enhancing milk protein synthesis [5], [8]. Recently, Rius et al. [9] investigated signaling pathways responsive to casein and starch infusion in primiparous mid-lactation Holstein cows. Results revealed that cell signaling molecules involved in the regulation of milk protein synthesis responded differently to the various nutritional stimuli. Subsequent work by Appuhamy et al. [10] revealed regulatory roles of essential AA other than arginine in mammary cell protein synthesis.
Research in monogastrics has underscored the importance of arginine in other biological functions, and recently the function and the metabolism of arginine in ruminant mammary gland was re-considered by Doepel and Lapierre [11]. Using abomasal infusions to increase arginine availability in Holstein cows, they evaluated if the supply of arginine relative to controls would alter milk and milk protein yield. Their results revealed that milk protein yield was increased by infusion of arginine compared with a control that included infusion of water. In their work, the Arg infusion had similar effects as other EAA infusion, this might be because Arg can be converted from other EAA in body. But the research also revealed that the range in the ratio of Arg uptake to output was between 2.52 to 2.12 and was still largely in excess of Arg secretion in milk protein [11]. Despite the positive result with arginine infusion, the functions and potential mechanisms of excess arginine uptake by mammary gland of the lactating cow remain unclear.
A compelling case was recently made for more in-depth studies of molecular regulation of lactation, particularly in livestock species [12], and specifically on the effect of mTOR in the regulation of milk protein synthesis [13]. Our previous work also demonstrated that arginine increases casein protein synthesis in bovine mammary epithelial cells [14], thus, we hypothesized that arginine might partly regulate milk protein synthesis through alterations in expression of genes related with the mTOR and STAT signaling pathways. The specific objective was to elucidate the effect of arginine on mRNA expression of casein, mTOR, and STAT genes in bovine mammary epithelial cells.
Materials and Methods
Animal and Mammary Tissue Sampling
Three multiparous healthy lactating Chinese Holstein cows at similar lactation stages (day 100±5 of lactation) were obtained from the Experimental Farm of Inner Mongolia Agricultural University. The cows were sacrificed within 1 h after milking. Immediately before exsanguination, milked mammary glands were split down the mid-line, and mammary tissue was excised from the center portion of each of the four quarters. Tissue samples from four quarters of the mammary glands of each cow were mixed and processed together. After removing obvious pieces of connective tissue and fat, the mammary tissues were cut into small pieces of 0.5 mm in thickness [15]. The tissue pieces from each cow were divided into 6 parts and placed in DMEM/F12 medium supplemented with the antibiotic-antimycotic mix (Sigma-A5955) as described by Chen et al. [14]. The study was approved by the Inner Mongolia Agricultural University Animal Care and Use Committee.
Tissue Incubation and Purification of Mammary Epithelial Cells
The tissue culture and the mammary epithelial cell isolation were carried out according to the methods described by Wu [16], O’Quinn et al. [15] and Xu et al. [17]. The method for isolation and purification of mammary cells is described in detail below. Firstly, collagenase digestion was used to isolate the mammary cells from tissue. An equal volume of collagenase (Type II, Gibco 17101-015) was added into 5 ml tube (the working solution is 0.5%) containing tissue slices, placed in a constant temperature shaker at 37°C for 1 h, and then filtered through 80 mesh prior to collecting cells by centrifugation (1,500 r/min for 5 min at room temperature). Cells were then added to 25-mL polypropylene flasks containing DMEM/F12 medium and placed in an incubator at 37°C. When cells reached 90% confluence, 0.25% Trypsin was added into the culture to purify epithelial cells from fibroblast according to their different sensitivity to Trypsin.
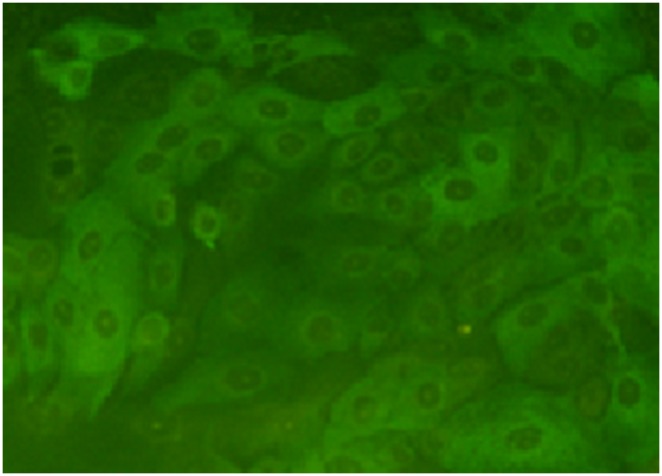
The isolated mammary epithelial cells were evaluated by morphology and immunofluorescence cell staining with fluoresceinisothiocyanate. The antibodies used in immunofluorescence were CK18 mouse monoclonal antibody (Sigma-Aldrich: SAB3300015-100UL) as first antibody and goat anti-mouse IgG conjugated FITC (Sigma-Aldrich: F5387) as secondary antibody following the manufacturer’s protocol. From figure 1 it can be seen that the purified cells had the characteristic cobblestone morphology of epithelial cells, and there were clear boundaries between cells but with close connections. Additionally, cell monolayer growth showed visible dome structures. From figure 2 it can be seen that the expression of CK-18 was positive in purified mammary epithelial cells. Lastly, the purified epithelial cells were mixed and incubated at 5×105/mL (counting by Cytorecon CYT-1000) on plastic dishes, and the 2nd generation cells were used for the arginine study.
Figure 1. Microscope image of F2 mammary epithelial Cells (×100).
Figure 2. Immunofluorescence of Cytokeratin 18 (CK-18) (×200).
Preparation of the Arginine-removal Basic Medium
As arginine exists in commercial cell medium, a medium without arginine had to be prepared taking into account the composition of DMEM/F12 (Gibco, Invitrogen, Catalog #11320082, Life Technologies Corporation, US). The basic medium devoid of arginine was prepared by combining the individual reagents (according to the composition of DMEM/F12 in ultrapure water). The basic medium was used in the transition process immediately before the arginine study. The AA profile of this arginine-removal basic medium was described in our previous work [14]; for the experimental treatments the arginine was added into the medium in accordance with the experimental design described below.
Experimental Design and Treatments
A single factorial experimental design varying the arginine levels was used to achieve the specific treatment groups. The target response that we focused on was casein production, thus, we attempted to develop media with combinations of AA closely resembling the profile in casein. The media containing arginine that most-closely resembled the profile of casein was defined as Arg 1X. The other treatment groups were made simply by varying the arginine concentration and without changing the concentration of other AA. The design included 7 groups, the same as our previous work [14]: Arg 0.25X (arginine at 69.50 mg/L), Arg 0.5X (arginine at 139.00 mg/L), Arg 1X (arginine at 278.00 mg/L), Arg 2X (arginine at 556.00 mg/L), Arg 4X (arginine at 1,112 mg/L), Arg 8X (arginine at 2, 224 mg/L) as experimental groups, and Arg 0X (arginine at 0.00 mg/L) was set as the control group (without arginine in the medium). Each treatment was run in triplicate, and a set of appropriate blanks (without cell incubation) was included.
RNA Extraction from Mammary Cells
After culture for 24 hours the reactions were stopped by removing medium and collecting cells. Cells were then frozen in liquid nitrogen prior to storage at −80°C. The RNA was extracted and purified using a cellular RNA extraction kit (DP430 spin column kit, Tiangen Biotech, Beijing, China). The extraction steps were carried out following the manufacturer's protocol. RNA quantity and integrity were confirmed with a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, US).
Measurement of Target Gene Expression
RNA samples were reverse-transcribed using PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa Code: D6110A, TaKaRa Biotechnology (Dalian) Co., Ltd., Dalian, China), according to the manufacturer’s instructions. The real time-PCR was performed in a Bio-Rad IQ5 Real-Time PCR (Bio-Rad Laboratories, Inc. Hercules, CA, US), using SYBR Premix Ex Taq II kit (TaKaRa Code: DRR081A, TaKaRa Biotechnology (Dalian) Co., Ltd., Dalian, China). All reactions were run using the protocol below: 30 s at 95°C; 10 s at 95°C, 20 s at annealing temperature, and 30 s at 72°C for 40 cycles. The same conditions were performed on an equal amount of RNAase-free water as a negative control. The information of primers for target and internal reference gene were displayed in table 1 including the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as internal control gene. It is recognized that using a single ICG is not ideal [18] but GAPDH has been used previously as the sole ICG in studies of mammary cell gene expression.
Table 1. The information of real time PCR primers of target genes detected in mammary epithelial cell from dairy cattle by in vitro culture with different arginine levels.
| Items | Sequences of primers (5′ –3′) | Genebank ID | Production | Annealing temperature |
| CSN1S2 (αs2-casein) | F:AGCTCTCCACCAGTGAGGAA | NM_174528.2 | 150 bp | 56°C |
| R:GCAAGGCGAATTTCTGGTAA | ||||
| CSN3 (kappa-casein) | F:CCAGGAGCAAAACCAAGAAC | NM_174294 | 148 bp | 56°C |
| R:TGCAACTGGTTTCTGTTGGT | ||||
| JAK2 (Janus kinase 2) | F:ACAGGGGCTGGCGTTCA | XM_002689603.1 | 146 bp | 61°C |
| R:TATTGGTAACCAACAGCTCAAGG | ||||
| mTOR (Mechanistic target of rapamycin) | F:ATGCTGTCCCTGGTCCTTATG | XM_001788228.1 | 199 bp | 63°C |
| R:GGGTCAGAGAGTGGCCTTCAA | ||||
| STAT5 (Signal transducer and activatorof transcription 5) | F:AAGACCCAGACCAAGTTCGC | NM_001012673.1 | 203 bp | 64°C |
| R:AGCACCGTGGCAGTAGCAT | ||||
| S6K (Ribosomal protein S6 kinase) | F:GGACATGGCAGGGGTGTTT | NM_205816.1 | 162 bp | 58°C |
| R:GGTATTTGCTCCTGTTACTTTTCG | ||||
| 4EBP1 (4E binding protein 1) | F:GGCAGGCGGTGAAGAGTC | BC120290.1 | 177 bp | 60°C |
| R:CCTGGGCTGCGGGAT | ||||
| GAPDH (glyceraldehyde-3-phosphate dehydrogenase) | F:GGGTCATCATCTCTGCACCT | XM_001252479 | 177 bp | 59°C |
| R:GGTCATAAGTCCCTCCACGA |
Statistical Analysis
The statistical analysis was carried out by one-way analysis of variance (ANOVA) followed by the Duncan’s test for post-hoc multiple comparisons of treatment means using SAS software (SAS Inst., Cary, NC, USA). In addition, a regression analysis was conducted using SPSS (v16.0). The random effect in the ANOVA model was replicate and the fixed effect was level of arginine. Significance was declared at P <0.05.
Results
Effects of Arginine Level on the Expressions of Casein Genes
Figure 3A/B shows the relative expression of Casein alpha s2 (CSN1S2) and Casein kappa (CSN3). The relative expression of Casein alpha s1 (CSN1S1) and Casein beta (CSN2) has been published previously (Chen et al., 2013). Expression of CSN1S2 and CSN3 in the Arg 2X (P<0.05) (containing 556.00 mg/L arginine in culture medium) was higher than other treatments and the control. Except for CSN1S2 in the Arg 0.25X and the Arg 8X, all other Arg concentrations resulted in greater expression of these genes compared with the control group (P<0.05).
Figure 3. In vitro effect of arginine level on the expression of casein genes in mammary epithelial cells from lactating dairy cattle.
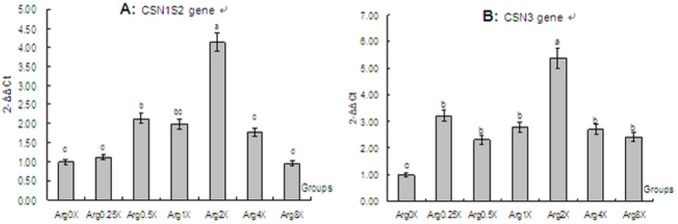
And: Panel A, CSN1S2 gene; and Panel B, CSN3 gene. Arg 0X, control without supplemental Arg (0.00 mg/L); Arg 0.25X, deficiency in casein (69.50 mg/L); Arg 0.5X, deficiency in casein (139.00 mg/L); Arg 1X, to match its concentration in casein (278.00 mg/L); Arg 2X, excess in casein (556.00 mg/L); Arg 4X, excess in casein (1,112 mg/L); and Arg 8X, excess in casein (2,224 mg/L). Different letters indicate significant difference between means (P<0.05).
The relative expression of CSN1S2and CSN3 increased from the Arg 0.25X to the Arg 2X; however, expression decreased when comparing the Arg 4X to the Arg 8X.
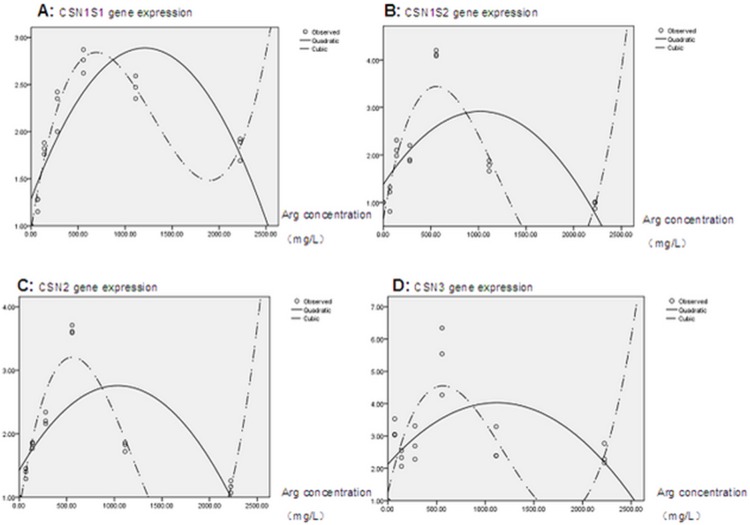
The quadratic and cubic regression analysis results for casein gene expression (see figure 4A–D, data of CSN1S1 and CSN2 were cited from the previous work by Chen et al. [14]) showed that, compared with a quadratic regression equation, a cubic regression equation had higher adjusted coefficient of determination (adjusted R2). The cubic equations for casein genes described below were used in this work to evaluate gene expression.
Figure 4. Cubic regression analysis of casein gene expression and arginine concentration.
Panel A, CSN1S1 (data cited from Chen et al. (2013)); Panel B, CSN1S2; Panel C, CSN2 (data cited from Chen et al. (2013)); and Panel D, CSN3 gene.
CSN1S1,
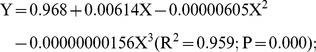 |
CSN1S2,
 |
CSN2,
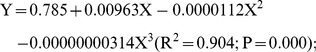 |
CSN3,
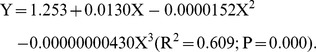 |
According to these equations, the calculated highest expression for casein genes was 2.37, 2.81, 2.66, and 3.76 for CSN1S1, CSN1S2, CSN2, CSN3, respectively; the corresponding arginine concentration was 434.44 mg/L, 372.36 mg/L, 371.784 mg/L, and 369.65 mg/L.
Effect of Arginine Level on the Expression of Signaling Pathway Related Genes
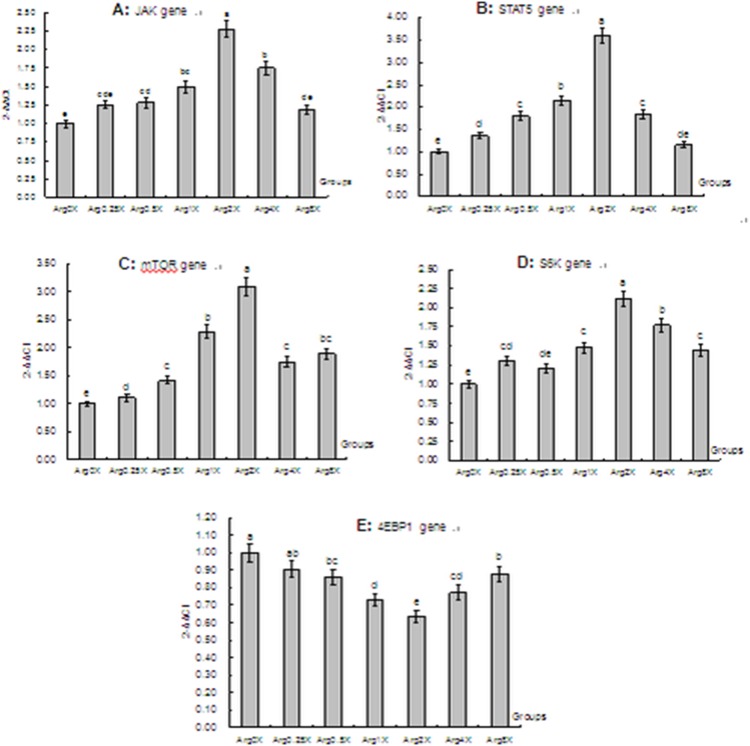
The results of the expression of signaling pathway related genes are presented in figure 5A–E. Similar to results of the casein genes, the relative expression of Janus kinase 2 (JAK2), Signal transducer and activator of transcription 5 (STAT5), Mechanistic target of rapamycin (mTOR), and Ribosomal protein S6 kinase (S6K) was highest in the Arg 2X (P<0.05) compared with other doses and the control group. In addition, the pattern of change in the expression of these genes was similar to the casein genes, with a gradual increase up to Arg 2X followed by a decrease as the concentration of arginine increases. The opposite response to Arg dose was observed for the expression of Eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1), with Arg 2X leading to the lowest expression among the groups (P<0.05); additionally, except for the Arg 0.25X, the other experimental groups led to lower 4EBP1 expression than the control group (P<0.05).
Figure 5. Effects of arginine level on the expressions of signaling pathways related genes in mammary epithelial cells from lactating dairy cattle in vitro.
Panel A, JAK2; Panel B, STAT5; Panel C, mTOR; Panel D, S6K; and Panel E, 4EBP1 gene. Arg 0X, control without supplemental Arg (0.00 mg/L); Arg 0.25X, deficiency in casein (69.50 mg/L); Arg 0.5X, deficiency in casein (139.00 mg/L); Arg 1X, to match its concentration in casein (278.00 mg/L); Arg 2X, excess in casein (556.00 mg/L); Arg 4X, excess in casein (1,112 mg/L); and Arg 8X, excess in casein (2,224 mg/L). Different letters indicate significant difference between means (P < 0.05).
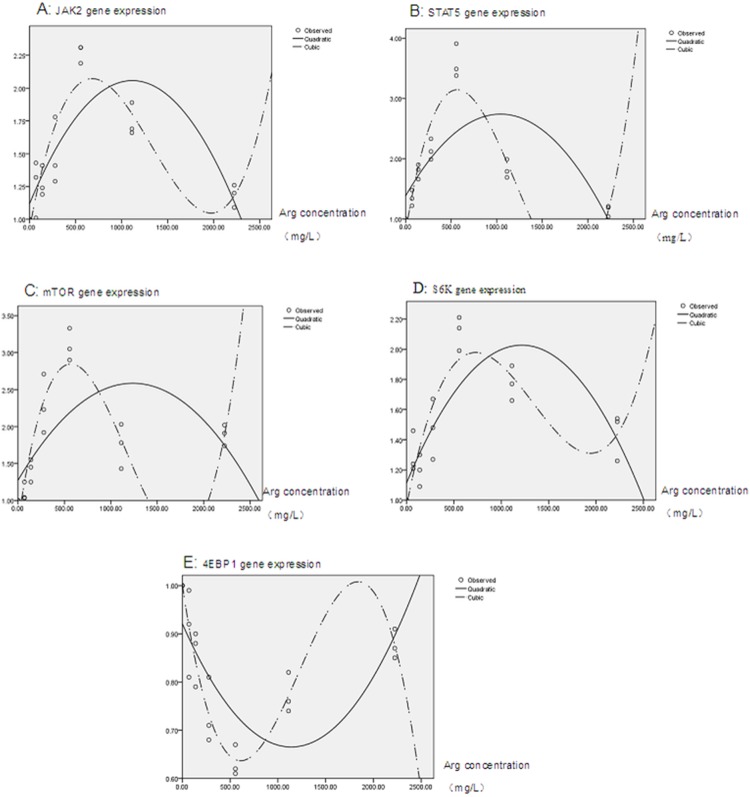
Quadratic and cubic regression analysis results (see figure 6A–E) of signaling pathway related gene showed that, a cubic regression equation (compared with the quadratic regression equation) with higher adjusted R2 provided a better fit to each set of observations. The cubic equations for signaling pathway related genes below were used in this work to evaluate gene expression.
Figure 6. Cubic regression analysis of signaling parthway related gene expression and arginine concentration.
Panel A, JAK2; Panel B, STAT5; Panel C, mTOR; Panel D, S6K; and Panel E, 4EBP1.
JAK2,
 |
STAT5,
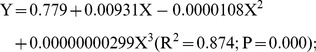 |
mTOR,
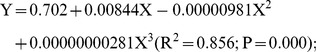 |
S6K,
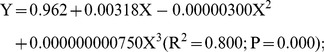 |
4EBP1,
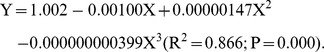 |
According to these equations, the calculated highest expression for signaling pathway related genes was 2.072, 3.131, 2.846, and 1.976 for JAK2, STAT5, mTOR, and S6K, respectively; the corresponding arginine concentration was 679.42 mg/L, 562.34 mg/L, 569.55 mg/L, and 729.64 mg/L. The minimum expression was 0.81 and the corresponding arginine concentration was 407.87 mg/L for 4EBP1 gene.
Discussion
A number of studies in monogastric animals have revealed that arginine plays several key functions including serving as precursor for the synthesis of nitric oxide and decreasing blood pressure [19], enhancing cell division [20], [21], and inducing the release of hormones and promoting protein and DNA synthesis [22]. Our preliminary in vitro studies showed that arginine increased the synthesis of casein in mammary epithelial cells from dairy cattle. Those results were substantiated by the fact that casein protein, including αs-casein and β-casein, in cultures with Arg 2X was the highest among the groups. For example, αs-casein and β-casein concentration due to Arg 2X increased by 120% and 96% compared with Arg 0X, by 64% and 19% compared with Arg 1X, by 141% and 24% compared with 4X, and by 152% and 53% compared with 8X [14]. In the present study, we attempted to find potential gene transcription mechanisms of Arg stimulation of casein synthesis.
In general, as a key previous step in protein expression, there is a correlation between gene transcription (e.g. mRNA expression) and protein expression [23]. We speculate that the observed stimulatory effects of Arg on casein expression (up to 2X the basal concentration) was associated at least in part with alterations, direct or indirect, with mRNA transcription of the casein genes. The results of casein gene expression in the current work confirmed our speculation that an “optimal” concentration of arginine in culture medium could increase casein gene expression in mammary epithelial cells.
From the multi-comparison ANOVA analysis we observed that the “optimal” concentration of arginine was 556.00 mg/L; further cubic regression analysis indicated that the “optimal” concentration of arginine for upregulation of casein gene expression varied depending on the specific casein gene (CSN1S1, CSN1S2, CSN2, and CSN3), but within a small range from 369.65 mg/L to 434.44 mg/L in culture medium. The stimulatory effects of Arg on casein expression were also associated with greater cell proliferation/growth as reported previously by our group [17].
Several transcription factors control expression of the major milk proteins, particularly the caseins [24]. The casein genes are all present on one chromosome (e.g., chromosome 6 for bovine and chromosome 5 for mouse) and in a cluster of single-copy genes [25]. Previous reports confirmed that casein gene transcription is mediated by Stat5, a member of a transcription factor family that transmits signals from cytokine and growth factor receptors to nuclear target genes [26]. While Stat5 is phosphorylated by JAK2 on a conserved tyrosine (Y694 or Y699, depending on species), resulting in its dimerization, translocation to the nucleus, and binding to a specific sequence [27].
Nutrients and hormones may modulate mammary protein synthesis through the mTOR signaling pathway [28]. In the bovine, STAT5 responds to prolactin and other lactogenic growth factors and its activity increases during lactation mostly due to phosphorylation [29]. Subsequently, Toerien et al. [30] showed that infusion of essential AA (EAA) plus glucose reduced phosphorylation of the IRS target eukaryotic initiation factor (eIF) 2 in mammary tissue and increased phosphorylation of the mTOR targets, ribosomal S6 kinase 1 (S6K1) and S6. Wu [1] concluded that functional AA such as arginine, cysteine, glutamine, leucine, act not only as cell signaling molecules but also as regulators of gene expression and the protein phosphorylation cascade. Increasing evidence suggests that arginine might directly activate anabolic cellular signaling pathways [31], [32].
The increase in expression of JAK2, STAT5, mTOR and S6K with the Arg 2X group (556.00 mg/L in culture medium) in this study might suggest a stimulation of protein synthesis [23], [28] as reported in previous work cited above. In the same fashion, the downregulation of 4EBP1 agreed with the negative role of 4EBP1 on protein translation [33], [34], a process that consequently stimulates protein synthesis. The cubic regression analysis revealed that the “optimal” concentrations of arginine varied depending on the specific gene (JAK2, STAT5, mTOR, and S6K), but within a range from 562.34 mg/L to 729.64 mg/L. Bauchart-Thevret et al. [35] reported that the arginine-dependent effect on cell survival and protein synthesis signaling in IPEC-J2 cells (a porcine intestinal epithelial cell line) was mediated by mTOR. The results discussed above might provide one of the explanations for the increase of casein gene expression when intracellular arginine is in an “optimal” concentration range, i.e. direct regulation of the mTOR signaling pathway by arginine.
The decrease in expression of the casein genes when arginine levels continually increased in culture medium was noteworthy. This response seems to agree to some extent with the results from Raggio et al. [36], i.e. the efficiency of transfer of absorbed AA into milk protein decreases markedly as protein supply increases. Clearly, further work needs to be conducted to clarify the nature of the response by mammary cells to increased AA supply.
Taken together, results of this study demonstrate for the first time that arginine plays an important role in the transcriptional regulation of casein genes and signaling pathway related genes in bovine mammary epithelial cells. Through its stimulatory effects on the expression of CSN1S1, CSN2 (in our previous work by Chen et al. [14]), CSN1S2, CSN3, JAK2, STAT5, mTOR and S6K, arginine supplementation at a concentration range from 369.65 mg/L to 729.64 mg/L in culture medium could stimulate casein synthesis in vitro. Additional experiments should be conducted to investigate the regulatory mechanism of arginine on casein expression in mammary gland of lactating cow.
Acknowledgments
The authors would like to give special thanks to Inner Mongolia Agricultural University for the experimental animal.
Funding Statement
The present work was funded by The Open Research Fund Projects of the State Key Laboratory of Animal Nutrition through project no. 2004DA125184F1106, and by National Basic Research Program of China (973 Program, http://www.dairy973.com/) through project no. 2011CB100803. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wu GY (2009) Amino acids: metabolism, functions, and nutrition. Amino Acids 37, 1–17. [DOI] [PubMed] [Google Scholar]
- 2. Wu G, Bazer FW, Davis TA, Jaeger LA, Johnson GA, et al. (2007) Important roles for the arginine family of amino acids in swine nutrition and production. Livest Sci 112, 8–22. [Google Scholar]
- 3. Mateo RD, Wu G, Moon HK, Carroll JA, Kim SW (2008) Effects of dietary arginine supplementation during gestation and lactation on the performance of lactating primiparous sows and nursing piglets. J Animal Sci 86, 827–835. [DOI] [PubMed] [Google Scholar]
- 4. Rhoads JM, Wu G (2009) Glutamine, arginine, and leucine signaling in the intestine. Amino Acids 7, 111–122. [DOI] [PubMed] [Google Scholar]
- 5. Bequette BJ, Backwell FRC, Crompton LA (1998) Current concepts of amino acid and protein metabolism in the mammary gland of the lactating ruminant. J Dairy Sci 81, 2540–2559. [DOI] [PubMed] [Google Scholar]
- 6. Lapierre H, Lobley GE, Doepel L, Raggio G, Rulquin H, et al. (2012) Triennial Lactation Symposium: Mammary metabolism of amino acids in dairy cows. J Animal Sci 90, 1708–1721. [DOI] [PubMed] [Google Scholar]
- 7.Bionaz M, Hurley W, Loor J (2012) Milk protein synthesis in the lactating mammary gland: Insights from transcriptomics analyses. In: Hurley WH, editor. Milk Protein, Chapter 11. InTech North America, New York, USA. 285–324. Available: http://dx.doi.org/10.5772/46054. Accessed 2012 Sep 12. [Google Scholar]
- 8. Hanigan MD, Crompton LA, Bequette BJ, Mills JA, France J (2002) Modelling mammary metabolism in the dairy cow to predict milk constituent yield, with emphasis on amino acid metabolism and milk protein production: model evaluation. J Theor Biol 217, 311–330. [DOI] [PubMed] [Google Scholar]
- 9. Rius AG, Appuhamy JA, Cyriac J, Kirovski D, Becvar O, et al. (2010) Regulation of protein synthesis in mammary glands of lactating dairy cows by starch and amino acids. J Dairy Sci 93, 3114–3127. [DOI] [PubMed] [Google Scholar]
- 10. Appuhamy JADRN, Bell AL, Nayananjalie WAD, Escobar J, Hanigan MD (2011) Essential amino acids regulate both initiation and elongation of mRNA translation independent of insulin in MAC-T cells and bovine mammary tissue slices. J Nutr 141, 1209–1215. [DOI] [PubMed] [Google Scholar]
- 11. Doepel L, Lapierre H (2011) Deletion of arginine from an abomasal infusion of amino acids does not decrease milk protein yield in Holstein cows. J Dairy Sci 94, 864–873. [DOI] [PubMed] [Google Scholar]
- 12. Loor JJ, Cohick WS (2009) ASAS centennial paper: Lactation biology for the twenty - first century. J Animal Sci 87, 813–824. [DOI] [PubMed] [Google Scholar]
- 13. Bionaz M, Loor JJ (2011) Gene networks driving bovine mammary protein synthesis during the lactation cycle. Bioinf Biol Insights 5, 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen LM, Li ZT, Wang MZ, Wang HR (2013) Preliminary report of arginine on synthesis and gene expression of casein in bovine mammary epithelial cell. IRJAS 3, 17–23. [Google Scholar]
- 15. O’Quinn PR, Knabe DA, Wu G (2002) Arginine catabolism in lactating porcine mammary tissue. J Animal Sci 80, 467–474. [DOI] [PubMed] [Google Scholar]
- 16. Wu G (1996) An important role for pentose cycle in the synthesis of citrulline and proline from glutamine in porcine enterocytes. Arch Biochem Biophys 336, 224–230. [DOI] [PubMed] [Google Scholar]
- 17. Xu BL, Wang MZ, Zhang XF, Ha S, Wang CY, et al. (2012) Arginine levels affect growth and CSN3 gene expression of dairy cows mammary epithelial cells in vitro . Chin J Animal Nutri 24, 852–858 (in Chinese with English abstract). [Google Scholar]
- 18. Bionaz M, Loor JJ (2007) Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol Genomics 29, 312–319. [DOI] [PubMed] [Google Scholar]
- 19. Gamboa A, Shibao C, Diedrich A, Paranjape SY, Farley G, et al. (2008) Excessive nitric oxide function and blood pressure regulation in patients with autonomic failure. Hypertension 51, 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rhoads JM, Chen W, Gookin J, Wu GY, Fu Q, et al. (2004) Arginine stimulates intestinal cell migration through a focal adhesion kinase dependent mechanism. Gut 53, 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jobgen WS, Fried SK, Fu WJ, Meininger CJ, Wu G (2006) Regulatory role for the arginine-nitric oxide pathway in energy-substrate metabolism. J Nutri Biochem 17, 571–588. [DOI] [PubMed] [Google Scholar]
- 22. Mateo RD, Wu G, Bazer FW, Park JC, Shinzato I, et al. (2007) Dietary l-arginine supplementation enhances the reproductive performance of gilts. J Nutri 137, 652–656. [DOI] [PubMed] [Google Scholar]
- 23. Guo Y, Xiao P, Lei S, Deng F, Xiao GG, et al. (2008) How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim Biophys Sin 40, 426–436. [DOI] [PubMed] [Google Scholar]
- 24. Rosen JM, Wyszomierski SL, Hadsell D (1999) Regulation of milk protein gene expression. Annu Rev Nutri 19, 407–436. [DOI] [PubMed] [Google Scholar]
- 25. Rijnkels M (2002) Multispecies comparison of the casein gene loci and evolution of casein gene family. J Mammary Gland Biol Neoplasia 7, 327–345. [DOI] [PubMed] [Google Scholar]
- 26. Levy DE, Darnell Jr JE (2002) STATs: transcriptional control and biological impact. Nature Review 3, 651–662. [DOI] [PubMed] [Google Scholar]
- 27. Watson CJ, Burdon TG (1996) Prolaction signal transduction mechanisms in the mammary gland: the role of the Jak/Stat pathway. Rev Reprod 1, 1–5. [DOI] [PubMed] [Google Scholar]
- 28. Burgos SA, Dai M, Cant JP (2010) Nutrient availability and lactogenic hormones regulate mammary protein synthesis through the mammalian target of rapamycin signaling pathway. J Dairy Sci 93, 153–161. [DOI] [PubMed] [Google Scholar]
- 29. Yang J, Kennelly JJ, Baracos VE (2000) Physiological levels of Stat5 DNA binding activity and protein in bovine mammary gland. J Animal Sci 78, 3126–3134. [DOI] [PubMed] [Google Scholar]
- 30. Toerien CA, Trout DR, Cant JP (2010) Nutritional stimulation of milk protein yield of cows is associated with changes in phosphorylation of mammary eukaryotic initiation factor 2 and ribosomal s6 kinase 1. J Nutri 140, 285–292. [DOI] [PubMed] [Google Scholar]
- 31. Corl BA, Odle J, Niu X, Moeser AJ, Gatlin LA, et al. (2008) Arginine activates intestinal p70S6K and protein synthesis in piglet rotavirus enteritis. J Nutri 138, 24–29. [DOI] [PubMed] [Google Scholar]
- 32. Yao K, Yin YL, Chu W, Liu Z, Deng D, et al. (2008) Dietary arginine supplementation increases mTOR signaling activity in skeletal muscle of neonatal pigs. J Nutri 138, 867–872. [DOI] [PubMed] [Google Scholar]
- 33. Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, et al. (1999) Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev 13, 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yanagiya A, Suyama E, Adachi H, Svitkin YV, Aza-Blanc P, et al. (2012) Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol Cell 46, 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bauchart-Thevret C, Cui L, Wu GY, Burrin DG (2010) Arginine-induced stimulation of protein synthesis and survival in IPEC-J2 cells is mediated by mTOR but not nitric oxide. Am J Physiol Endocrinol Metab 299, E899–E909. [DOI] [PubMed] [Google Scholar]
- 36. Raggio G, Pacheco D, Berthiaume R, Lobley GE, Pellerin D, et al. (2004) Effect of level of metabolizable protein on splanchnic flux of amino acids in lactating dairy cows. J Dairy Sci 87, 3461–3472. [DOI] [PubMed] [Google Scholar]