Abstract
The methylation of cytosine to 5-methylcytosine (5-meC) is an important epigenetic DNA modification in many bacteria, plants, and mammals, but its relevance for important model organisms, including Caenorhabditis elegans and Drosophila melanogaster, is still equivocal. By reporting the presence of 5-meC in a broad variety of wild, laboratory, and industrial yeasts, a recent study also challenged the dogma about the absence of DNA methylation in yeast species. We would like to bring to attention that the protocol used for gas chromatography/mass spectrometry involved hydrolysis of the DNA preparations. As this process separates cytosine and 5-meC from the sugar phosphate backbone, this method is unable to distinguish DNA- from RNA-derived 5-meC. We employed an alternative LC–MS/MS protocol where by targeting 5-methyldeoxycytidine moieties after enzymatic digestion, only 5-meC specifically derived from DNA is quantified. This technique unambiguously identified cytosine DNA methylation in Arabidopsis thaliana (14.0% of cytosines methylated), Mus musculus (7.6%), and Escherichia coli (2.3%). Despite achieving a detection limit at 250 attomoles (corresponding to <0.00002 methylated cytosines per nonmethylated cytosine), we could not confirm any cytosine DNA methylation in laboratory and industrial strains of Saccharomyces cerevisiae, Schizosaccharomyces pombe, Saccharomyces boulardii, Saccharomyces paradoxus, or Pichia pastoris. The protocol however unequivocally confirmed DNA methylation in adult Drosophila melanogaster at a value (0.034%) that is up to 2 orders of magnitude below the detection limit of bisulphite sequencing. Thus, 5-meC is a rare DNA modification in drosophila but absent in yeast.
A covalently modified DNA base, 5-methylcytosine (5-meC), is widely found in bacteria, plants, and mammalian cells and is associated with the epigenetic regulation of gene expression.1 However, this DNA modification is not ubiquitous. 5-meC is thought to be absent in the DNA of many species, including popular laboratory model organisms such as Caenorhabditis elegans.2 Other organisms might possess low amounts of cytosine DNA methylation, including Neurospara crassa,3Dictyostelium discoidium,4Schistosoma mansoni,5 and Drosophila melanogaster,6 for some of which DNA methylation was initially thought to be absent. In several of these cases, the content of DNA methylation is at the edge of the detection limit of bisulphite sequencing approaches, currently the dominating technique for analysis of DNA methylation. Despite that bisulphite sequencing gives highly valuable information about the sequence context of methylated cytosine, it has limitations at the lower concentration range: incomplete bisulphite conversion of unmethylated cytosines and missalignment of sequencing reads in the mapping process in repetitive, telomeric, and GC-rich regions result in false positive rates of ∼0.5% and higher.7−10 For this reason, the existence and biological function of low methylation levels is still ambiguous and/or debated in some species.
Until recently, also DNA of budding yeast Saccharomyces cerevisiae has been considered to be free of 5-methylcytosine. Despite this modification had been described in the first studies that date back to the late 1970’s;11 later studies did not confirm DNA methylation of S. cerevisiae. Yeast DNA is not cut by methylation-dependent restriction endonucleases, and 5-meC was not found in DNA digests analyzed by HPLC/UV–VIS where a detection limit of 1 per 3100 to 6000 residues was achieved.12 Recently however, Tang et al. challenged this result. With the use of a GC–MS method, a much lower detection limit (6.4 fmol) compared to the HPLC protocols was achieved. This protocol detected 5-meC in DNA extracts of three budding yeast laboratory strains.13 Moreover, this study reported DNA methylation in several other yeast species as well, which included the medically important tropical yeast Saccharomyces boulardii and the popular laboratory model fission yeast (Schizosaccharomyces pombe). Indeed, the evidence for DNA methylation is debated in the latter species. In difference to S. cerevisiae, S. pombe possesses a cytosine methyltransferase homologue of the DNMT2 family, termed Pombe MethylTransferase 1 (Pmt1).14 Pmt1, however, appears to be specific for tRNA modification.15 The 5-meC contents reported by Tang et al. ranged from 0.036% for S. boulardii to 0.205% for S. pombe. For S. cerevisiae strains, values from 0.085% (strain W1588-4C) to 0.128% in the common laboratory yeast strain BY4741, a derivative of S288c, the parent of the yeast genome project,16 were reported. These results appeared paradigm-shifting, implying that a huge variety of yeast species, including S. cerevisiae, are capable of cytosine DNA methylation and would indicate that these important model organisms could be employed to analyze the function of low-level DNA methylation.
These conclusions are challenged by the application of an improved technique. Employing a liquid chromatography selective reaction monitoring (LC-SRM) method that is insensitive to contamination with RNA-derived methylcytosine, we achieve a detection limit of 250 attomoles for methylcytidine (around 25 times more sensitive compared to the previous study13). We detect no cytosine DNA methylation in the different yeast species S. cerevisiae, S. boulardii, S. paradoxus, S. pombe, and P. pastoris. In contrast, the protocol unambiguously detected and accurately quantified DNA methylation in Mus musculus, Arabidopsis thaliana, Escherichia coli, and at a low but significant level in Drosophila melanogaster.
Results and Discussion
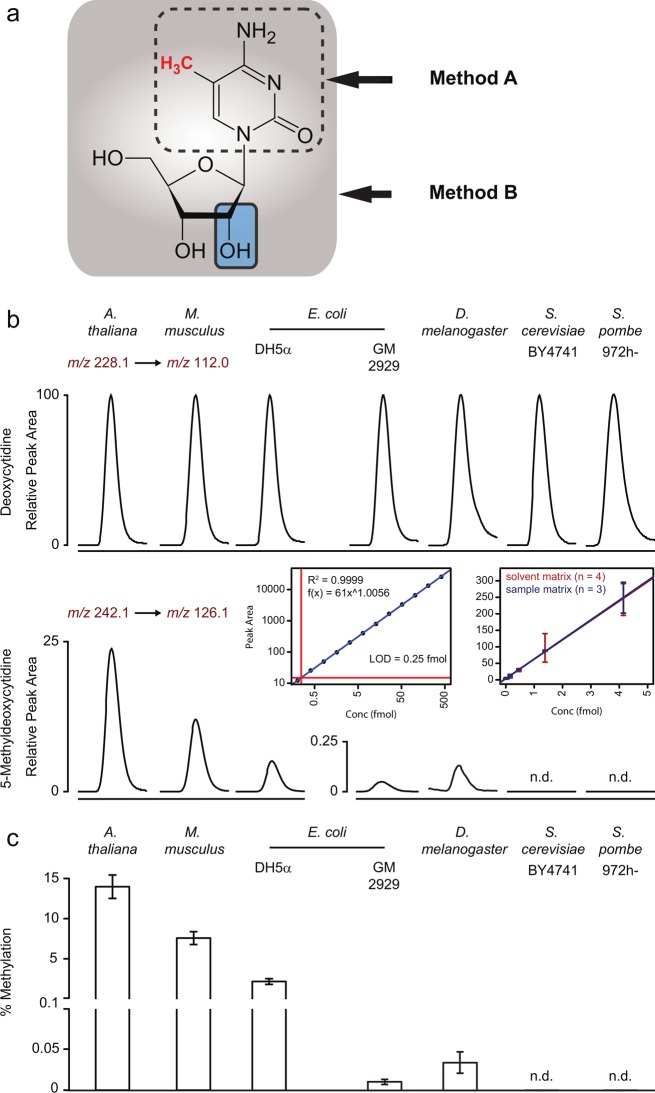
Similar to a variety of common LC and LC–MS protocols for the detection of 5-methylcytosine,13,17,18 Tang et al. analyzed the 5-meC content in yeast DNA upon hydrolysis. This procedure separates base and sugar phosphate and quantifies eventually the (free) methylated and unmethylated cytosine.13,17,18 Applying a similar approach that is based on liquid chromatography-tandem mass spectrometry (LC–MS/MS),17 we obtained 5-meC concentration values for S. cerevisiae DNA fully comparable to those reported by Tang et al. (data not shown), confirming the precision of the GC–MS technology. Surprisingly, however, this method revealed significant 5-meC content also in negative control DNA purified from the E. coli strain GM2929 (Marinus, CGSC no: 7080). This was unexpected, as this E. coli strain is deficient in bacterial DNA methyltransferases, including the dcm-6 allele and is used in laboratories around the world to produce nonmethylated DNA for cloning experiments.19,20 In the search for a potential source of the methylated cytosine in GM2929 DNA preparations, we considered copurified methylcytosine derived from RNA as its potential source. Cytosine methylation has been found as an abundant modification on different RNA species, especially on rRNA in prokaryotes and tRNA in eukaryotes.21−23 In order to setup a quantification method that restricts the detection of 5-meC to DNA-derived nucleosides, we used a protocol based on enzymatic release of nucleosides using the enzyme DNA Degradase Plus (ZymoResearch).24 The enzymatic digest preserves the base coupled to the deoxyribose (nucleoside, Figure 1a). This enables the separation of the DNA-derived deoxycytidine and the RNA derived (hydroxyl)-cytidine by mass spectrometry due to significant mass difference. Samples processed in this manner were analyzed using a state of the art HPLC (Agilent 1290 Infinity) coupled to a triple quadrupole mass spectrometer (Agilent 6460), set to quantify deoxyribose-cytosine (Figure 1b, upper panel) and deoxyribose-5-methylcytosine (5-methylcytidine, Figure 1b, middle panel).
Figure 1.
(A) Quantification methods involving hydrolysis of nucleic acids (method A) are unable to distinguish whether modified cytosine is of RNA or DNA origin. Structural representation of RNA-derived 5-methyl-cytidine and DNA-derived 5-methyl-deoxycytidine. (B) Quantification of deoxycytidine and 5me-deoxycytidine specifically derived from DNA. Top: Chromatograms corresponding to deoxycytidine, as measured by LC-SRM in DNA samples obtained from A. thaliana, M. musculus, E. coli (K12 derivates DH5alpha and the methyltransferase mutant GM2929), D. melanogaster (w118), as well as two yeast species (S. cerevisiae BY4741 and S. pombe 972h-). The chromatographic peak represents deoxycytidine. Bottom: The chromatographic peak represents 5me-deoxycytidine; the illustrated chromatogram is normalized to the corresponding deoxycytidine signal as shown in the top panel. Inset bottom panel: (left) calibration curve for 5me-deoxycytidine resulting in a linear correlation spanning over 3 orders of magnitude; limit of detection is estimated at 250 attomol; (right) external calibration (increasing amount of standard in solvent) and standard addition (increasing amount of standard spiked into 100 ng DNA digest prepared from S. cerevisiae BY4741). The calibration curve for 5me-deoxycytidine at the lower concentration limit excludes significant matrix effects on quantification and LOD. (C) Methylation levels in genomic DNA purified from six different species. The number of 5me-deoxycytidine is expressed as percentage of unmodified deoxycytidine for each species (n = 3 from biological triplicates, error bars = ± SD); n.d. = not detected.
This DNA specific method unambiguously detected deoxycytidine and 5-methyldeoxycytidine in purified standards over a concentration range of 3 orders of magnitude. 5-Methyldeoxycytidine and deoxycytidine were quantified by external calibration within the dynamic range of the analytical method (R2 = 0.9999 and R2 = 0.9965, respectively, Figure 1b). A limit of detection at 250 attomol per injection for 5-meC was obtained (Figure 1b, left inset). The absence of an inferring matrix effect of the DNA digest sample was confirmed by reproducing the limit of detection and linear range by standard addition of 5-meC to a fully processed sample of S. cerevisiae DNA (Figure 1b, right inset). This method clearly detected and quantified 5-meC in DNA samples of A. thaliana leaves (14.0% of deoxycytidines methylated), mouse liver (7.6%), and methylation competent E. coli K12 cells (strain DH5alpha 2.3%), confirming DNA methylation as expected (Figure 1, panels b and c). Taking into account the genome size and GC content, these values allowed an estimation about the number of methylated cytosine residues per genome (A. thaliana, 6 × 106; M. musculus, 9 × 107; E. coli, 5 × 104) (Table 1). Moreover, this protocol confirmed the expected decline in methylation in the DNA methyltransferases deficient (dcm-6) K12 derivate GM2929 (Figure 1b, Table 1). Compared to the methylation competent K12 strain DH5α, the content of methylated cytosines was reduced to 0.016% (equaling <500 modified cytosine bases per genome), which corresponds to a reduction in DNA methylation of 99.3%.
Table 1. Content of 5me-Deoxycytidine (5-meC) in DNA Preparations of Ten Species and the Estimated Average Numbers of 5mdC per (Haploid) Genome of A. thaliana, M. musculus, D. melanogaster, E. coli, S. cerevisiae, S. boulardii, S. paradoxus, P. pastoris, and S. pombea.
| species | genome size MB | % methylation | Est. no. 5mdC per MB | Est. no. 5mdC/genome |
|---|---|---|---|---|
| A. thaliana | 119.7 | 14 ± 1.5 | 5.0 × 104 | 6.0 × 106 |
| M. musculus | 2800 | 7.6 ± 0.8 | 3.2 × 104 | 8.9 × 107 |
| D. melanogaster | 139.5 | 0.034 ± 0.013 | 1.4 × 102 | 2 × 104 |
| E. coli DH5α | 4.6 | 2.3 ± 0.1 | 1.2 × 104 | 5.4 × 104 |
| E. coli GM2929 (dcm-6) | 4.6 | 0.016 ± 0.002 | 81 | 3.8 × 102 |
| S. cerevisiae BY4741** | 12.5 | not detected (<0.002) | not detected (LOD = 8) | not detected (LOD = 96) |
| S. cerevisiae D27–310B** | 12.5 (b.f.) | not detected | not detected | not detected |
| S. cerevisiae AWRI 796** | 12.5 (b.f.) | not detected | not detected | not detected |
| S. cerevisiae SCHD0308 | 12.5 (b.f.) | not detected | not detected | not detected |
| S. cerevisiae SaflagerW-34/70 | 12.5 (b.f.) | not detected | not detected | not detected |
| S. boulardii CBS 5926 | 11.4 | not detected | not detected | not detected |
| S. paradoxus KPN3829 | 11.8 | not detected | not detected | not detected |
| S. pombe 972h- | 13.8 | not detected | not detected | not detected |
| P. pastoris SMD1168 | 9.4 | not detected | not detected | not detected |
Whereas, unmethylated cytidines were readily detected in all yeast species (Figure 1b, upper panel), the protocol applied on S. cerevisiae, S. boulardii, S. paradoxus, P. pastoris, and S. pombe DNA did not detect any evidence for the content of 5-methyldeoxycytidine (Figure 1b, lower panel; Table 1). Taking into account the limit of detection at 250 attomol for methyldeoxycytidine, its content in yeast would be lower than 0.00002 per deoxycytidine (Table 1). Considering the importance of budding yeast in basic research and industry, the analysis was conducted on five different S. cerevisiae yeasts. These included the laboratory strains BY4741 (in its prototrophic version, BY4741-pHLUM25), which had yielded the highest S. cerevisiae 5-meC content in the study by Tang et al.13 and the strain D273–10B that was used in the seminal study that claimed for the first time the absence of DNA methylation in S. cerevisiae.12 The measurements were conducted on DNA purified both from cells grown to stationary and exponential phase. Under both conditions DNA methylation remained undetectable. In addition, the absence of methylation was confirmed in industrial yeast. Analysis of the vine production strain AWRI 796, the lager/pilsner beer production yeast Saflager W-34/70, and the baking “dry-yeast” SCHD0308 (Ruf, Germany) revealed no evidence for DNA methylation either (Table 1).
The results obtained in budding yeast were representative for other yeast species. We measured DNA samples obtained from S. boulardii, a tropical yeast of medicinal use isolated from lychee and mangosteen fruit in 1923, S. paradoxus, a yeast species living on the bark of deciduous trees, the common laboratory models fission yeast S. pombe, and the methylotrophic yeast commonly used in protein production, P. pastoris. DNA methylation was not detected in any of these yeast species either (Table 1).
Finally, we applied this protocol to DNA obtained from D. melanogaster DNA. The fruit fly is a common model organism, but the role and existence of its DNA methylation is the subject of an ongoing debate.6,26,27 We could clearly and unambiguously detect Drosophila cytosine DNA methylation (Figure 1b). Revealing 0.034% of cytosines methylated, the total number of methylcytosine in the Drosophila genome is thus estimated to be in the range of 2 × 104 modified bases per genome (Table 1). This result thus confirms methylation in drosophila, reveals however a lower content as compared to estimations obtained with previous methods.6 The total content of methylated cytosines in Drosophila is thus comparable to those in the E. coli genome, albeit the latter genome is substantially smaller (Table 1). This value is several orders of magnitude above the detection limit of the LC–MS/MS method (Figure 1). It is however 10–100 fold below the error rate/detection limit of bisulphite sequencing. Bisulphite sequencing, at least in its current implementations, is thus not applicable for the analysis of Drosophila DNA methylation.
Conclusions
An LC-SRM method specific to DNA-derived nucleosides was used to assess cytosine DNA methylation in different model organisms. A detection limit of 250 attomoles for methyl-deoxycytidine, which corresponded to one modified base per 50000 deoxycytidines, was achieved. As expected, cytosine DNA methylation was detected and could be precisely quantified in A. thaliana, M. musculus, and E. coli DNA. Substantially lower, but unequivocal, evidence for DNA methylation was obtained in D. melanogaster DNA. However, DNA methylation was not detected in laboratory and industrial strains of S. cerevisiae, S. pombe, S. boulardii, S. paradoxus, and P. pastoris. In summary, these results reveal that methylcytosine quantification methods that are based on hydrolytic DNA cleavage are sensitive to copurification artifacts. Moreover, species such as Drosophila contain quantities of DNA methylation too low to be measured by bilsulphite sequencing. Importantly however, this study demonstrates that while a low amount of DNA methylation is detectable in D. melanogaster, yeast species do not possess cytosine DNA methylation.
Materials and Methods
Chemical and Reagents
Chemical standards were obtained from Sigma at a purity >99% [2′-deoxycytidine (dC, SD3897), cytidine (C, C122106), 5-methylcytidine (5mC, M4254)] and the 5-methyl-2′-deoxycytidine (5mdC, sc-278256, (purity >99%)) from Santa Cruz Biotechnology. UPLC-grade methanol, water, and formic acid were purchased from Greyhound. RNase A was purchased from Roche (10109169001).
Yeast and Bacteria Culture Conditions
The S. cerevisiae strains BY4741 pHLUM,25 D273–10B,28 and AWRI 796 (NCBI Taxon ID: 764097), as well as industrial yeast SCHD0308 and Saflager W-34/70, S. boulardii CBS 5926, S. paradoxus KPN3829, and P. pastoris SMD1168 were inoculated in 300 mL YPD [yeast extract (10 g/L), peptone (20 g/L), and 2% glucose] with 0.2 OD600 of yeast culture and incubated at 30 °C, 200 rpm. S. pombe strain 972h- was inoculated in 300 mL YED [yeast extract (5 g/L), 3% glucose]. Yeast cells were collected both at exponential (OD600 = 2) and stationary (OD600 > 8) growth phase. The E. coli K12 derivates strain DH5α [F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1] and GM2929 [F– ara-14 leuB6 thi-1 fhuA31 lacY1 tsx-78 galK2galT22glnV44hisG4rpsL136 (StrR) xyl-5 mtl-1 dam13::Tn9 (CamR) dcm-6 mcrB1 hsdR2 (rK– mK+) mcrA recF143] were grown as triplicates starting from a single colony each in 50 mL LB and incubated at 37 °C, 200 rpm until stationary growth phase. The number of bacterial and yeast cells were estimated on a CASY TT (Roche) cell counter. Yeast and bacterial cell pellets were centrifuged at 5000g for 3 min and washed with water before storage at −80 °C.
DNA Extraction
A. thaliana DNA was extracted from 100 mg leaf tissue of Columbia 0 plants grown on a long day (16 h light, 8 h dark) for 3–4 weeks using a Plant Genomic DNA Miniprep Kit (GenElute G2N70–1KT). M. musculus DNA was extracted from 15 mg of liver tissue obtained from female mice of mixed C57BL/6/SV/129 background using the Genomic-tip 20/G kit (Qiagen). D. melanogaster DNA was extracted from a mixed population of 10 female and 10 male wt/w118 adult flies using a Gentra Puregene DNA purification kit (Qiagen). Yeast and bacterial DNA were extracted from 1.5 × 109 or 4.5 × 109 cells, respectively, using the Genomic-tip 20/G kit (Qiagen). DNA extracts were treated with RNase A at 37 °C for 45 min and DNA purification was performed according to the manufacturer’s instructions. Purified DNA was precipitated with isopropanol and washed with 70% ethanol and resuspended in 10 mM Tris·HCl, pH 8.0. DNA was quantified using dsDNA BR Assay Kit (Qubit) and quality controlled by gel electrophoresis.
Sample Preparation for LC–MS/MS
DNA samples were treated with DNA Degradase Plus (ZymoResearch, E2021) to obtain individual nucleosides. One microgram DNA was treated with 5U Degradase at 37 °C for 1 h, in a final volume of 25 μL and subsequently inactivated by adding 175 μL of 0.1% formic acid. The nucleobase standard solutions were prepared at a final concentration of 1 mg/mL in 50% methanol and mixed to obtain a standard mix with the following concentration: C, 800 ng/mL; 5mC, 8 ng/mL; dC, 80 μg/mL; and 5mdC, 80 ng/mL. Sixteen 1:2 serial dilutions were prepared for the external calibration curves. For the evaluation of a potential matrix effect on the calibration of the 5mdC, five 1:3 serial dilutions from a 5 ng/mL stock solution were prepared and 0.25 μL spiked in 25 μL of BY4741-pHLUM DNA digest.
Twenty microliters of the samples, diluted to contain approximately 100 ng of digested DNA, were injected onto a reverse phase ultraperformance liquid chromatography (UPLC) column (ZORBAX RRHD Eclipse Plus C18, 2.1 × 50 mm, 1.8 μm particle size, Agilent) using a 6 min isocratic run [water:methanol:formic acid (95:5:0.1) (100 μL/min)]. The eluent was directed to an electrospray ion source connected to a triple quadrupole mass spectrometer (Agilent 6460 QQQ), operating in positive mode. The following transitions were monitored for C m/z 244.1→112.0, 5mC m/z 258.2→126.1, dC m/z 228.1→112.0, and 5mdC m/z 242.1→126.1. Peak areas were extracted and integrated using MassHunter.
Acknowledgments
We thank Skirmantas Kriaucionis, Nick Kruger, and Rob Klose (all University of Oxford) for an inspiring discussion. We thank Josmar Langner for the E. coli strain GM2929, Nino Nikolovski for providing Arabidopsis thaliana leaf samples, Luca Pellegrinet for mouse liver samples, Nick Lowe and Johanna Rees for Drosophila melanogaster adult flies, Naresh Babu for yeast strain D273-10B, Elizabeth Bilsland for yeast strain AWRI 796, and Felix Krueger for help in the interpretation of the biSeq results. Work in the Ralser lab is funded by the Wellcome Trust (RG 093735/Z/10/Z) and the ERC (Starting Grant 260809). M.R. is a Wellcome Trust Research Career Development and Wellcome-Beit Prize fellow.
Author Contributions
F.C. and M.M. established the LC-SRM method; F.C. prepared the DNA samples; F.C., M.M., and R.K. conducted the measurements; F.C., M.M., and M.R. analyzed the data; M.R. wrote the first manuscript draft; and all authors wrote the final manuscript.
The authors declare no competing financial interest.
References
- Law J. A.; Jacobsen S. E. Nat. Rev. Genet. 2010, 11, 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson V. J.; Johnson T. E.; Hammen R. F. Nucleic Acids Res. 1986, 14, 6711–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H.; Selker E. U. Nature 2001, 414, 277–283. [DOI] [PubMed] [Google Scholar]
- Katoh M.; Curk T.; Xu Q.; Zupan B.; Kuspa A.; Shaulsky G. Eukaryotic Cell 2006, 5, 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer K. K.; Rodríguez López C. M.; Chalmers I. W.; Munshi S. E.; Truscott M.; Heald J.; Wilkinson M. J.; Hoffmann K. F. Nat. Commun. 2011, 2, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H.; Leismann O.; Jeltsch A. EMBO J. 2000, 19, 6918–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke P. M.; Stirzaker C.; Song J.; Grunau C.; Melki J. R.; Clark S. J. Methods 2002, 27, 101–107. [DOI] [PubMed] [Google Scholar]
- Sievers S.; Fritzsch C.; Kuhnen C.; Müller O. Anticancer Res. 2008, 28, 2055–2060. [PubMed] [Google Scholar]
- Grunau C.; Clark S. J.; Rosenthal A. Nucleic Acids Res. 2001, 29, E65–E70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J.; Shoemaker R.; Xie B.; Gore A.; LeProust E. M.; Antosiewicz-Bourget J.; Egli D.; Maherali N.; Park I.-H.; Yu J.; Daley G. Q.; Eggan K.; Hochedlinger K.; Thomson J.; Wang W.; Gao Y.; Zhang K. Nat. Biotechnol. 2009, 27, 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S.; Kenny C.; Berger L.; Pratt K. J. Bacteriol. 1978, 135, 1156–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proffitt J. H.; Davie J. R.; Swinton D.; Hattman S. Mol. Cell. Biol. 1984, 4, 985–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.; Gao X.-D.; Wang Y.; Yuan B.-F.; Feng Y.-Q. Anal. Chem. 2012, 84, 7249–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C. R.; Bartlett R.; Nurse P.; Bird a P. Nucleic Acids Res. 1995, 23, 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.; Müller S.; Nellen W.; Jurkowski T. P.; Jeltsch A.; Ehrenhofer-Murray A. E. Nucleic Acids Res. 2012, 40, 11648–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B.; Davies A.; Cost G. J.; Caputo E.; Li J.; Hieter P.; Boeke J. D. Yeast 1998, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Rocha M. S.; Castro R.; Rivera I.; Kok R. M.; Smulders Y. M.; Jakobs C.; de Almeida I. T.; Blom H. J. Clin. Chem. Lab. Med. 2010, 48, 1793–1798. [DOI] [PubMed] [Google Scholar]
- Yamagata Y.; Szabó P.; Szüts D.; Bacquet C.; Arànyi T.; Páldi A. Epigenetics 2012, 7, 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer B. R.; Marinus M. G. Gene 1994, 143, 1–12. [DOI] [PubMed] [Google Scholar]
- Marinus M. G.; Carraway M.; Frey A. Z.; Brown L.; Arraj J. A. Mol. Gen. Genet. 1983, 192, 288–289. [DOI] [PubMed] [Google Scholar]
- Nau F. Biochimie 1976, 58, 629–645. [DOI] [PubMed] [Google Scholar]
- Sharma S.; Yang J.; Watzinger P.; Kötter P.; Entian K.-D. Nucleic Acids Res. 2013, 41, 9062–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y.; Lyko F.; Helm M. Nucleic Acids Res. 2010, 38, 1415–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T.; Kim K.-P.; Fan G.; Faull K. F. Anal. Biochem. 2011, 412, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulleder M.; Capuano F.; Pir P.; Christen S.; Sauer U.; Oliver S. G.; Ralser M. Nat. Biotechnol. 2012, 30, 1176–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz G.; Guzzardo P. M.; Olova N.; Fantappié M. R.; Rampp M.; Schaefer M.; Reik W.; Hannon G. J.; Lyko F. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 8627–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalke S.; Nickel O.; Walluscheck D.; Hortig F.; Onorati M. C.; Reuter G. Nat. Genet. 2009, 41, 696–702. [DOI] [PubMed] [Google Scholar]
- Sherman F. Genetics 1963, 48, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R.; Jorgensen P.; Moran U.; Weber G.; Springer M. Nucleic Acids Res. 2010, 38, D750–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D.; Trivedi U.; Thomson M.; Oliver F.; Kumar S.; Blaxter M. L. Genome Res. 2009, 19, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matecic M.; Stuart S.; Holmes S. G. Genetics 2002, 162, 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M.; Patterson N.; Endrizzi M.; Birren B.; Lander E. S. Nature 2003, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- Khatri I.; Akhtar A.; Kaur K.; Tomar R.; Prasad G. S.; Ramya T. N. C.; Subramanian S. Gut Pathog. 2013, 5, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K.; Lin Y.-C.; Tiels P.; Van Hecke A.; Glinka S.; Weber-Lehmann J.; Rouzé P.; Van de Peer Y.; Callewaert N. Nat. Biotechnol. 2009, 27, 561–566. [DOI] [PubMed] [Google Scholar]
- Wood V.; Gwilliam R.; Rajandream M.-A.; Lyne M.; Lyne R.; Stewart A.; Sgouros J.; Peat N.; Hayles J.; Baker S.; Basham D.; Bowman S.; Brooks K.; Brown D.; Brown S.; Chillingworth T.; Churcher C.; Collins M.; Connor R.; Cronin A.; Davis P.; Feltwell T.; Fraser A.; Gentles S.; Goble A.; Hamlin N.; Harris D.; Hidalgo J.; Hodgson G.; Holroyd S.; Hornsby T.; Howarth S.; Huckle E. J.; Hunt S.; Jagels K.; James K.; Jones L.; Jones M.; Leather S.; McDonald S.; McLean J.; Mooney P.; Moule S.; Mungall K.; Murphy L.; Niblett D.; Odell C.; Oliver K.; O’Neil S.; Pearson D.; Quail M. A.; Rabbinowitsch E.; Rutherford K.; Rutter S.; Saunders D.; Seeger K.; Sharp S.; Skelton J.; Simmonds M.; Squares R.; Squares S.; Stevens K.; Taylor K.; Taylor R. G.; Tivey A.; Walsh S.; Warren T.; Whitehead S.; Woodward J.; Volckaert G.; Aert R.; Robben J.; Grymonprez B.; Weltjens I.; Vanstreels E.; Rieger M.; Schäfer M.; Müller-Auer S.; Gabel C.; Fuchs M.; Düsterhöft A.; Fritzc C.; Holzer E.; Moestl D.; Hilbert H.; Borzym K.; Langer I.; Beck A.; Lehrach H.; Reinhardt R.; Pohl T. M.; Eger P.; Zimmermann W.; Wedler H.; Wambutt R.; Purnelle B.; Goffeau A.; Cadieu E.; Dréano S.; Gloux S.; Lelaure V.; Mottier S.; Galibert F.; Aves S. J.; Xiang Z.; Hunt C.; Moore K.; Hurst S. M.; Lucas M.; Rochet M.; Gaillardin C.; Tallada V. A.; Garzon A.; Thode G.; Daga R. R.; Cruzado L.; Jimenez J.; Sánchez M.; del Rey F.; Benito J.; Domínguez A.; Revuelta J. L.; Moreno S.; Armstrong J.; Forsburg S. L.; Cerutti L.; Lowe T.; McCombie W. R.; Paulsen I.; Potashkin J.; Shpakovski G. V; Ussery D.; Barrell B. G.; Nurse P.; Cerrutti L. Nature 2002, 415, 871–880. [DOI] [PubMed] [Google Scholar]