Summary
The CRISPR-Cas system is a prokaryotic host defense system against genetic elements. The Type III-B CRISPR-Cas system of the bacterium Thermus thermophilus, the TtCmr complex, is composed of six different protein subunits (Cmr1-6) and one crRNA with a stoichiometry of Cmr112131445361:crRNA1. The TtCmr complex co-purifies with crRNA species of 40 and 46 nt, originating from a distinct subset of CRISPR loci and spacers. The TtCmr complex cleaves the target RNA at multiple sites with 6 nt intervals via a 5’ ruler mechanism. Electron microscopy revealed that the structure of TtCmr resembles a ‘sea worm’ and is composed of a Cmr2-3 heterodimer ‘tail’, a helical backbone of Cmr4 subunits capped by Cmr5 subunits, and a curled ‘head’ containing Cmr1 and Cmr6. Despite having a backbone of only four Cmr4 subunits and being both longer and narrower, the overall architecture of TtCmr resembles that of Type I Cascade complexes.
Introduction
The CRISPR-Cas (clustered regularly interspaced short palindromic repeat/CRISPR-associated proteins) system is a recently discovered, prokaryotic host defense system (found in most archaea and many bacteria) targeting mobile genetic elements such as phages and plasmids (Barrangou et al., 2007; Brouns et al., 2008; Horvath and Barrangou, 2010; Karginov and Hannon, 2010). Typically, CRISPR loci are composed of ~25- to ~38 bp direct DNA repeats, separated by unique spacer sequences of a similar length (Grissa et al., 2007), which are derived from previous encounters with invading elements. The cas genes are often located in close proximity to these CRISPR loci, and are used to categorize the system into one of the three major CRISPR-Cas (sub)types (Makarova et al., 2011). The exact mechanisms of CRISPR-mediated defense are distinct between the different CRISPR-Cas types and subtypes. In general, the CRISPR loci are transcribed and processed into small RNAs called CRISPR RNAs (crRNAs), each composed of a separate spacer sequence flanked by part(s) of the repeat sequence. The mature crRNAs are then loaded on either a protein complex (for Type I and Type III CRISPR-Cas systems), composed of several Cas proteins (Brouns et al., 2008; Hale et al., 2009), or a single Cas protein, Cas9 (Gasiunas et al., 2012; Jinek et al., 2012), in the case of a Type II CRISPR-Cas system. The complex is then guided towards its complementary target, resulting in base-pairing interactions between the crRNA and the invading genetic element and generally, followed by degradation of the invader. New spacers can be acquired in a still poorly-understood process called ‘adaptation’, in which a new spacer (derived from the invader) will be integrated in a directional fashion into the chromosomal CRISPR spacer array.
In type III systems, crRNA synthesis starts with the transcription of the CRISPR array. The resulting pre-crRNA is most likely first cleaved in the repeat sequence by one or more Cas6 proteins followed by further processing at the 3’ end, potentially involving other Cas proteins to generate the mature crRNA (Carte et al., 2008; Scholz et al., 2013). Consistent with other CRISPR systems, subtype III-A seems to target DNA invaders (Marraffini and Sontheimer, 2008). However, the subtype III-B is unique, in that it has been shown to target RNA rather than DNA. The effector complex of the subtype III-B system, the Cmr complex, binds crRNA, and cleaves a target RNA complementary to the bound crRNA (Hale et al., 2009). Comparison of different Cmr modules revealed that subtype III-B contains either six Cmr genes (Cmr-α module) and an associated csx1 gene, such as the Cmr complex from Pyrococcus furiosus (Hale et al., 2009), or seven Cmr genes (Cmr-β module), as has been shown in Sulfolobus species (Deng et al., 2013; Zhang et al., 2012). Surprisingly, the modus operandi of the Cmr complexes encoded by these Cmr modules appears to be markedly different. The Cmr-α complex of P. furiosus cuts target RNA molecules according to a ‘ruler mechanism’, cleaving the target 14 nucleotides (nt) upstream of the 3’ end of the basepaired crRNA (Hale et al., 2009). The Cmr-β complex of S. solfataricus on the other hand, has been shown to cleave substrate RNAs at UA dinucleotides (Zhang et al., 2012). The ribonuclease activities of a bacterial Cmr complex have not been addressed so far.
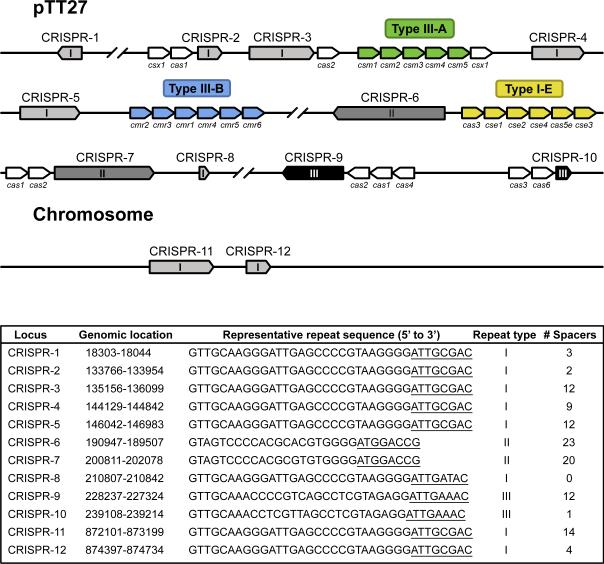
The thermophilic bacterium Thermus thermophilus HB8 has eleven CRISPR loci in total, two of which are located on the chromosome, while the other nine loci are found on a 260-kbp plasmid pTT27 (Agari et al., 2010) (Fig. 1). Here, the CRISPR locus 8 is not considered a genuine CRISPR because this locus consists of only one (type-I) repeat sequence and does not contain any spacers (Agari et al., 2010). These loci are classified into three types depending on the nucleotide sequence of the repeat (Agari et al., 2010). All 11 CRISPRs are unidirectionally expressed and processed to mature crRNAs, although the stability or processing can be different between crRNAs (Juranek et al., 2012). The 5’ ends of all the crRNAs predominantly retain eight nucleotides derived from repeat sequences (the 5’ handle), while their 3’ ends are variable depending on the repeat sequence type, suggesting that this strain has multiple crRNA processing systems (Juranek et al., 2012). More than 30 Cas proteins are encoded by pTT27, including subtypes I-E, III-A, and III-B (Agari et al., 2010; Juranek et al., 2012) (Fig 1.). Expression of CRISPR loci and most cas genes are up-regulated by infection with the myophage ΦYS40 (Agari et al., 2010). Furthermore, an operon containing the subtype I-E genes and one containing the subtype III-A genes are positively regulated by cAMP receptor protein (CRP) in a cAMP-dependent manner (Agari et al., 2010; Shinkai et al., 2007). Elucidation of the functional and structural mechanisms and roles of all CRISPR-Cas machineries in this strain will provide a systematic understanding of the host defense systems. In this study, we investigated the structure and function of a bacterial Cmr-α effector complex of the subtype III-B CRISPR-Cas system: the T. thermophilus Cmr complex (TtCmr).
Figure 1.
Schematic representation of CRISPR arrays and cas genes on the genome and plasmid pTT27 of T. thermophilus HB8. CRISPR arrays (1 to 12) are indicated in different grayscales, depending on the repeat type (I, II or III). Cas(-related) genes belonging to a particular CRISPR-Cas subtype are colored in green (subtype III-A), blue (subtype III-B) or yellow (subtype I-E). Additional cas genes are indicated in white. For each of these CRISPR arrays, the bottom panel summarized the genomic location, the representative repeat sequence, repeat type and the number of spacers. The 5’ handles, as found by our deep sequencing analysis, are underlined.
Results
Preparation and initial characterization of the Cmr complex from T. thermophilus
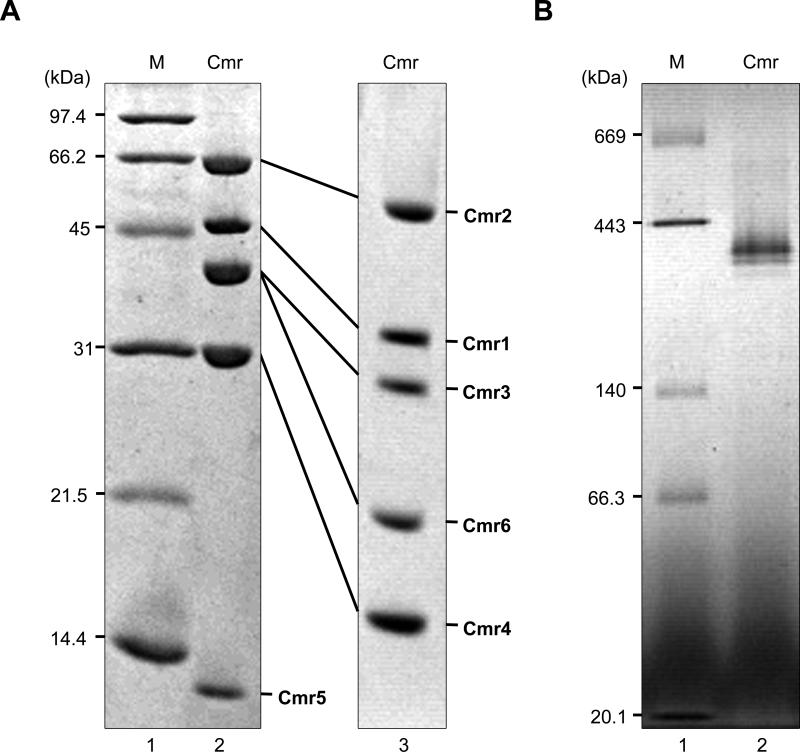
We constructed a recombinant T. thermophilus strain that produces the Cmr6 protein fused with a (His)6 tag at its C-terminus. The Cmr complex was purified to homogeneity by three different column chromatography steps, as described in the Supplemental experimental procedures. The purified protein complex was composed of six proteins (Fig. 2A). We confirmed by mass spectrometry based peptide sequencing that the proteins corresponded to TTHB162 (Cmr1), TTHB160 (Cmr2), TTHB161 (Cmr3), TTHB163 (Cmr4), TTHB164 (Cmr5), and TTHB165 (Cmr6) (data not shown), indicating that the T. thermophilus Cmr complex is composed of six subunits. According to the N-terminal amino acid sequence analysis, the N-terminal methionine residues of the Cmr4 and Cmr6 proteins in the complex were deleted. A blue native polyacrylamide gel electrophoresis (BN-PAGE) analysis of the Cmr complex revealed two bands: a ~350 kDa major band and a ~310 kDa minor band (Fig. 2B). These bands suggest multiple stoichiometries of the Cmr complex may co-occur (addressed below). The complex was subjected to the gel permeation chromatography (GPC)-high-performance liquid chromatography (HPLC) in the presence of 6 M guanidine-HCl. We identified the peaks shown in, using individually expressed recombinant Cmr proteins as standards (Fig. S1 and Fig. S2A-B). The UV spectrum of the peak at 44 min detected from the gel-filtration chromatogram was similar to that of a synthetic RNA (Fig. S2C), suggesting that this fraction is derived from crRNA. Transcription of T. thermophilus cmr genes increases after infection of ΦYS40 phage (Agari et al., 2010); however, yield and subunit composition of the Cmr complex from phage-infected cells was not significantly different from that of non-infected cells (data not shown).
Figure 2.
Isolation and characterization of the native TtCmr complex. (A) SDS-PAGE analysis of the T. thermophilus Cmr complex. The purified protein (2 μg) was analyzed on a 15% polyacrylamide gel (lane 2), or an 8% polyacrylamide gel containing 8 M urea (lane 3), and the gels were stained with Coomassie Brilliant Blue R-250. Each subunit is indicated. The Cmr6 has a (His)6-tag at its C-terminus. Lane 1, molecular-weight markers (Bio-Rad Laboratories, Inc.): phosphorylase b (97.4 kDa), serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21.5 kDa), lysozyme (14.4 kDa). (B) Blue native-PAGE analysis of the T. thermophilus Cmr complex. Two μg of the Cmr complex was analyzed on a 4 to 16% linear polyacrylamide gradient gel in the presence of 0.02% Coomassie Brilliant Blue G-250 (lane 2). Lane 1, molecular weight markers (Sekisui Medical, Co, Ltd.): thyroglobulin (669 kDa), ferritin (443 kDa), lactate dehydrogenase (140 kDa), bovine serum albumin (66.3 kDa), trypsin inhibitor (20.1 kDa). Protein concentration used was determined by the Bradford method (Bradford, 1976), using bovine serum albumin as a standard.
The crRNA content of the Cmr complex
One of the hallmarks of effector complexes from subtype III-B (and subtype III-A) CRISPR-Cas systems is the maturation of crRNAs into different sizes. While the exact sizes can vary slightly among different species (Hale et al., 2009; Zhang et al., 2012), and even within one species (Scholz et al., 2013), the population of complex-bound crRNA species is typically composed of two major classes, differing by 6 nt in size. The crRNAs were extracted from the purified T. thermophilus Cmr complex and analyzed by denaturing gel electrophoresis, which showed that the complex binds two major crRNA species of 40 and 46 nt, although low-abundant levels of crRNA species of other sizes could be observed as well (Fig. S3A). No substantial differences in abundance or size-distribution could be discerned when comparing the Cmr-bound crRNAs from either the uninfected or the phage-infected cells (data not shown). Successful polyadenylation of their 3’-ends by E. coli poly(A) polymerase, indicates that these crRNA have accessible, unphosphorylated 3’ ends (Fig. S3A). Furthermore, the 5’-phosphate-dependent ribonuclease Xrn-1 was not able to degrade the purified crRNAs prior to polynucleotide kinase (PNK) treatment, suggesting the presence of a 5’ hydroxyl group (Fig. S3B). In order to establish the exact nature of the Cmr-bound crRNAs, as well as to determine any changes in the crRNA population upon phage-infection, the crRNAs were prepared for high-throughput sequencing using 2×74 paired-end reads.
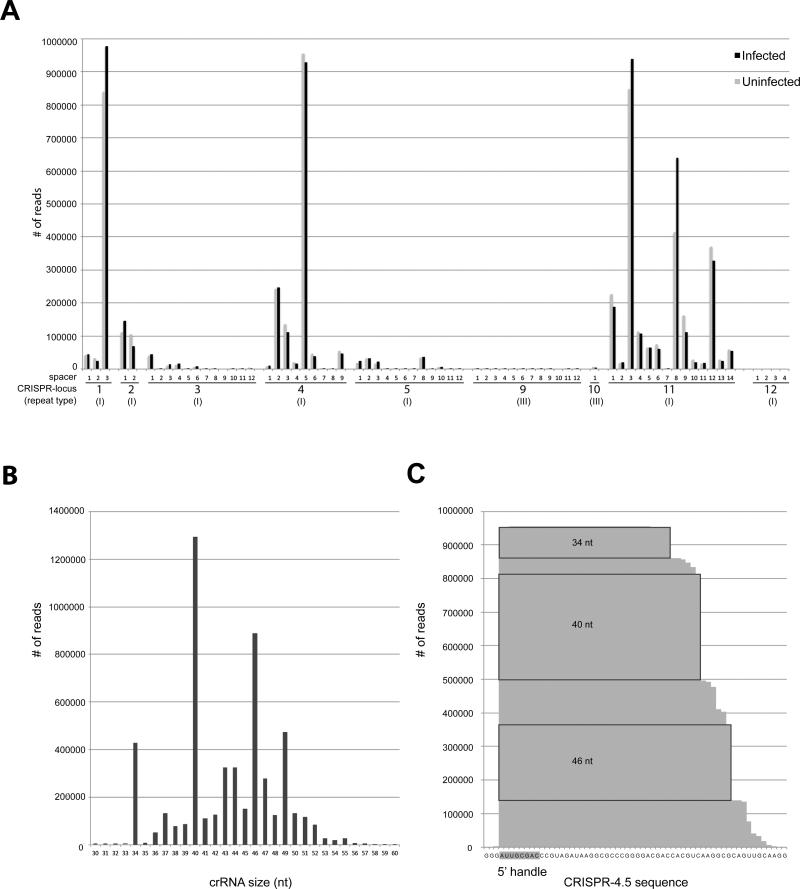
The sequencing reads were mapped to the genome of T. thermophilus, which revealed a considerable bias in the abundance of different crRNAs incorporated into the complex (Fig. 3A). This variation in abundance is evident when individual spacers within one CRISPR locus are considered and some CRISPR loci seem to be favored in contributing mature crRNAs for the Cmr complex when compared to others. A detailed overview of all the reads is given in Fig. S4. Out of the 11 total CRISPR loci, crRNAs that are destined for incorporation into the Cmr complex originated predominantly from only four of these loci (CRISPR-1,2,4 and 11). Although CRISPR-2 was slightly underrepresented in our dataset, this bias is in good agreement with the relatively high expression of these loci (Juranek et al., 2012). In accordance with the observed crRNA sizes on denaturing gels (Fig. S3A), the size-distribution resulting from the crRNA sequencing showed that the major population consists of lengths of 40 (24.3%) and 46 nt (16.7%) (Fig. 3B). This size-distribution is observed for the total crRNA population, and also when individual cases (such as CRISPR-4.5, spacer 5 from locus 4, the most abundant crRNAs in the dataset) are considered (Fig. 3C). The presence of crRNAs of other sizes within the complex are reminiscent of processing intermediates, suggesting that the final steps in crRNA maturation can occur when bound to the complex. crRNAs bound to the Cmr complex retain the last 8 nt of the type I repeat sequence at their 5’end (Agari et al., 2010). The observation that the majority of reads (81.2%) started with the sequence “ATTGCGAC” could suggest that this 5’ handle promotes binding to the complex. While reads originating from type-II repeats were almost completely absent in our dataset, a small number of reads originated from CRISPR-9 and -10 (containing type-III repeats). The last 8 nt of the repeats in these latter two CRISPR loci are very similar to those of type-I repeats (“ATTGAAAC”). These data indicate that the Cmr complex favors 40 and 46 nt crRNAs with type I(-like) repeats.
Figure 3.
Deep sequencing analysis of the crRNA-content of the TtCmr complex. (A) Distribution of crRNA reads over the 12 CRISPR loci present in the genome of T. thermophilus HB8. (B) Histogram illustrating the size-distribution of the mapped crRNA reads. (C) Deep-sequencing profile of the crRNA reads mapping to CRISPR-4.5. The 5’ handle, which are the first 8 nucleotides, derived from the upstream repeat sequence are indicated in gray. Abundant crRNA species of 34, 40 and 46 nt are indicated in boxes. An overview of all the mapped reads is given in Fig. S4.
Stoichiometry of the Cmr complex
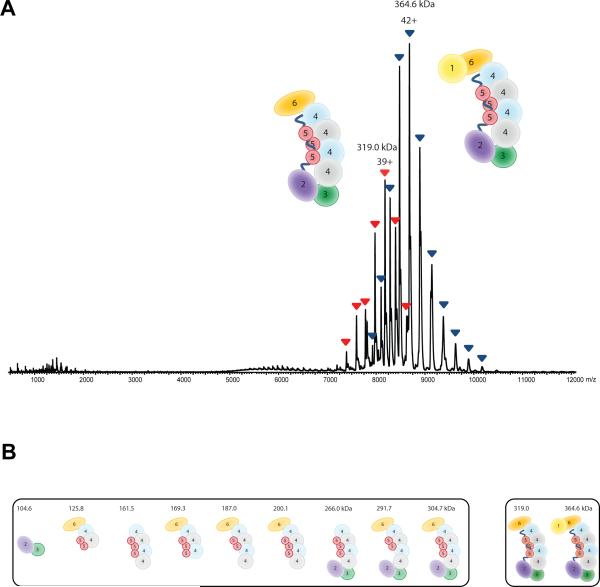
To further investigate the structural basis of the interaction between Cmr subunits and the stoichiometry of the complex, we determined the composition of the Cmr protein assembly using native mass spectrometry (Heck, 2008; van Duijn et al., 2012). Denaturing and tandem MS analyses provided accurate mass measurements for each subunit of Cmr. The measured masses of the individual Cmr subunits were consistent with their theoretical values (Table S1). Analysis of the intact assembly by native MS revealed the presence of two major components. From their well-resolved charge state distributions, we could determine masses of 364,610 ± 46 Da and 319,060 ± 40 Da (Fig. 4A), in good agreement with the data from native gel electrophoresis. Although we could measure the masses quite accurately, this did not provide direct information on the stoichiometry of the assembly. However, complexes composed of only one copy of each Cmr component (Cmr112131415161-crRNA1) would have a mass of only 246 kDa, indicating that some subunits are present in multiple copies.
Figure 4.
Subunit composition of the TtCmr complex. (A) Native nano-electrospray ionization mass spectrum of Cmr. Two major, well-resolved charge state distributions are present at high m/z values, corresponding to complexes of 365 kDa (blue) and 319 kDa (red). More exact masses are provided in Table S1. (B) Cmr (sub)complexes detected by native mass spectrometry. These (sub)complexes were detected and mass analyzed by reconstituting the Cmr complex lacking individual Cmr subunits. More accurate masses and assignments are given in Table S2.
The two major complexes of 365 and 319 kDa, most likely represent the intact Cmr and Cmr subcomplex lacking Cmr1 respectively. The measured mass difference between the two assemblies is 45.5 kDa (mass of Cmr1 monomer is 44,479 Da). Collision induced dissociation experiments on mass-selected ions of the intact Cmr confirmed the facile dissociation of Cmr1, hinting at a more loosely attachment and a peripheral location of this subunit. Interestingly, besides the elimination of Cmr1, no other subunits were expelled from the intact complex under collision induced dissociation conditions. Selection and activation of the 319 kDa Cmr sub-complex did not result in substantial fragmentations, indicating a high gas-phase stability of the Cmr complex.
As the Cmr assembly could not be disrupted by tandem MS, we sought to further explore the Cmr structure using a combination of in-solution dissociation of the intact Cmr complex and, reconstitution of Cmr complexes lacking single Cmr proteins. From all these mass spectrometric data (Fig. 4B and Table S2) the stoichiometry of the complex was determined to be Cmr112131445361-crRNA1, i.e. containing multiple copies of Cmr4 and Cmr5. With this stoichiometry the expected mass would be 364,036 kDa, in good agreement with a measured mass of 364,610 kDa. As shown before (Jore et al., 2011; van Duijn et al., 2012) these types of mass spectrometric experiments can reveal a wide range of other structural details. For instance, we observed a stable heterodimer between Cmr2 and Cmr3, in agreement with earlier observations (Osawa et al., 2013; Shao et al., 2013). Having multiple copies present, the backbone of the Cmr complex is likely composed of Cmr4 and Cmr5, wrapped around the crRNA molecule.
Activity of the Cmr complex
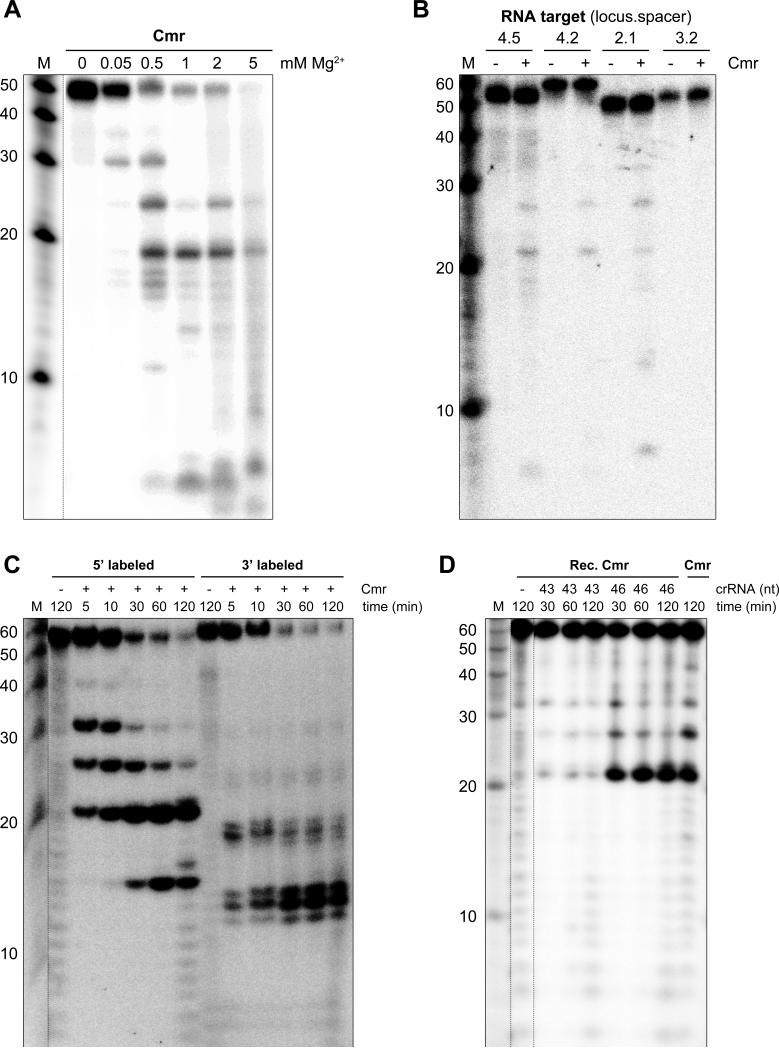
To evaluate the nucleolytic capabilities of the Cmr complex, an RNA substrate was generated by in-vitro transcription of a hybridized oligonucleotide pair in the presence of α-32P UTP. This resulted in a 50 nt internally labeled RNA substrate, complementary to one of the most abundant crRNAs found incorporated into the complex: CRISPR-4.5 (Fig. 3A). Ribonuclease activity was assayed by incubating the RNA substrate with the Cmr complex in buffers containing different concentrations of Mg2+ (Fig. 5A). The results showed that the complex harbors Mg2+-dependent endoribonuclease activity, although activity was also observed in the presence of Mn2+ (data not shown). The presence of multiple, distinct cleavage products indicates that the target RNA is cleaved at different sites. No activity was observed when complementary ssDNA or dsDNA substrates were used (Fig. S5).
Figure 5.
RNA-cleavage activity of the TtCmr complex. (A) An internally labeled RNA substrate complementary to CRISPR-4.5 was incubated with the endogenous Cmr complex for 1 h in the presence of different concentrations of Mg2+. Samples were analyzed by denaturing PAGE, followed by phosphorimaging. (B) Different internally labeled RNA targets (complementary to CRISPR-4.5, 4.2, 2.1 and 3.2) were tested in a similar assay (including 2 mM Mg2+), demonstrating the specificity of the Cmr complex. (C) Activity assay with the endogenous Cmr complex using a 5’ or 3’ labeled RNA substrate (CRISPR-4.5). (D) The 5’ labeled RNA substrate (CRISPR-4.5) was incubated with either the endogenous (“Cmr”) or the reconstituted Cmr complex (“Rec. Cmr”) for the indicated amount of time. The reconstituted complex was pre-loaded with the 43 or the 46 nt crRNA. Reconstituted Cmr complexes without crRNA (“-“) were included as a control. Noncontiguous lanes from the same gel are indicated with dotted lines. The results of a Cmr activity assay with complementary ssDNA and dsDNA substrates are depicted in Fig. S5.
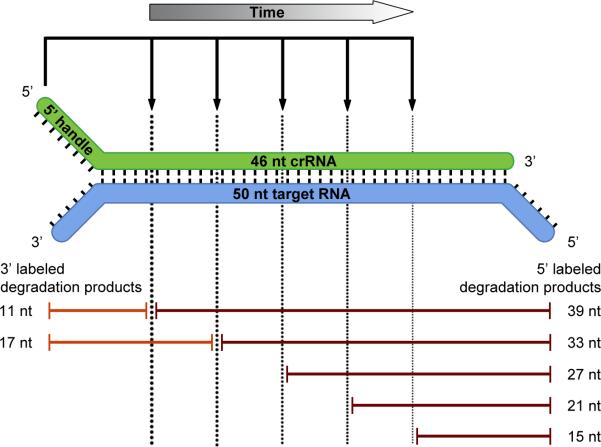
In order to determine the specificity of the complex, we tested several RNA substrates complementary to Cmr-associated crRNAs that were found in either high (CRISPR-4.5), moderate (CRISPR-4.2) or low (CRISPR-2.1) levels, as well as an RNA substrate complementary to a crRNA for which almost no reads were found (CRISPR-3.2) in our deep sequencing dataset (Fig. 3A). The results showed that the activity is specific for RNAs that are complementary to the Cmr-bound crRNAs and that this particular activity cannot be attributed to any potential contaminants present in the purified Cmr-complex sample (Fig. 5B). Regardless of the RNA substrate, sizes of the cleavage products were found to be identical (21, 27 and 33 nt), arguing against a sequence-dependent cleavage mechanism like the Cmr complex in S. solfataricus (Zhang et al., 2012). To study the activity in more detail, a similar experiment was performed using a 3’ or 5’ labeled RNA substrate (CRISPR-4.5). While the 5’ labeled substrate was degraded to 39, 33, 27, 21 and 15 nt products, a single 12 nt degradation product was observed with the 3’ labeled substrate (please note that the 3’ labeling reaction added one additional nucleotide at the 3’ end of the RNA substrate). These results suggest that the substrate is initially cleaved at its 3’ end, followed by additional cleavages processing towards its 5’ end (Fig. 5C). These findings are more in line with the ‘ruler mechanism’ of P. furiosus (Hale et al., 2009). However, the heterogeneity in the sizes of the Cmr-bound crRNAs (Fig. 3B) makes it difficult to draw firm conclusions about this possibility. Therefore, the Cmr complexes were reconstituted (using individually expressed and purified Cmr subunits from E. coli) and loaded with either a 43 or a 46 nt crRNA, which are 3’-truncated versions of the same crRNA (CRISPR-4.5). These reconstituted complexes (“Rec. Cmr”) were subsequently tested in an activity assay using a synthetic, 5’ labeled RNA substrate, and the activity of these complexes was compared to the endogenous Cmr complex (“Cmr”) (Fig. 5D). Surprisingly, the results showed that regardless of the 3’ end of the Cmr-bound crRNA(s), the RNA substrate was still cleaved at similar positions (Fig. 5D), excluding a ruler mechanism based on the 3’ end of the crRNA. Instead, these results imply either that the Cmr complex harbors multiple active sites, or that the RNA substrate is threaded through the complex towards a single active site. By analogy to Type I-E Cascade (Jore et al., 2011; Wiedenheft et al., 2011a), the 5’ handle of the crRNA is most likely recognized by one of the Cmr subunits, which in turn fixes the position of the RNA substrate within the complex. Therefore, we favor a scenario where the Cmr complex contains multiple active sites, with Cmr4 being the most likely candidate for providing the endoribonuclease activity. Based on this model, the substrate will be cleaved at fixed distances (6 nt intervals) measured from the 5’ end of the crRNA: a 5’ ruler mechanism (Fig. 6). Since the Cmr complex harbors just four Cmr4 subunits and we observed five differently sized degradation products, another Cmr subunit (possibly Cmr1 or Cmr6) might provide the remaining cleavage activity.
Figure 6.
Model for the cleavage activity of the TtCmr complex. Schematic representation of the cleavage activity deduced from the activity assays presented in Fig. 5. Sizes of the different 3’ and 5’ labeled cleavage products, that were observed in these assays, are indicated. Note that over time, the Cmr complex cleaves the RNA substrates from the 3’ end, cutting every 6 nt progressing towards the 5’ end. Cmr4 is the proposed subunit responsible for this catalytic activity, while another Cmr subunit might be responsible for the remaining cleavage.
Three-dimensional structure of the Cmr complex
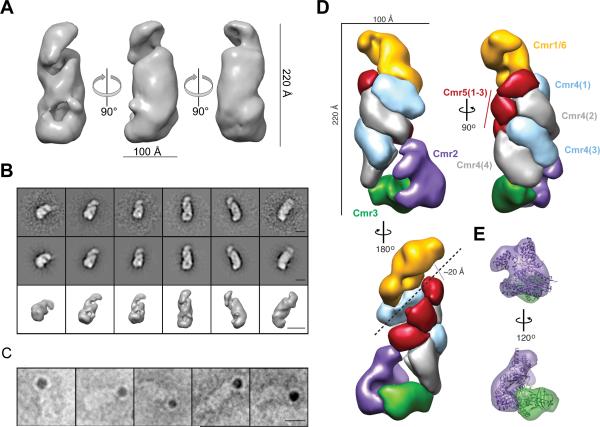
We gained insight into the three-dimensional structure of the Cmr complex containing crRNA using electron microscopy (EM) and single particle analysis of negatively stained particles (Fig. S6A). We reconstructed a three-dimensional (3D) map of TtCmr at ~26 Å resolution (using the 0.5 FSC criterion) from two-dimensional (2D) class averages using the common-line method followed by projection matching refinement of 9,000 individual particles images in EMAN and EMAN2 (Fig. 7A, Fig. S6B). Reprojections of the map matched with high fidelity to reference-free 2D class averages, indicating good agreement between our reconstruction and experimental data (Fig. 7B). The reconstruction of Cmr bound to crRNA resembled a ‘sea worm’ with overall dimensions of 90 × 100 × 220 Å, compatible with the expected molecular mass of the fully intact complex (~365 kDa). NTA-nanogold particle labeling of a His6 C-terminally tagged Cmr6 within the reconstituted complex, showed Cmr6 is located at either the head or the tail of the structure (Fig. 7C). Then, we collected a larger, higher-resolution data set using the automated molecular microscopy system LEGINON (Fig. S7A-C). Using the first reconstruction as a starting model, we obtained a reconstruction of the complex at ~22 Å resolution (using the 0.5 FSC criterion) from ~45,000 particles, showing clear additional structural features that could be segmented easily and automatically (Fig. 7D). The ‘sea worm’ is composed of a repeating helical backbone of four identical subunits with the size and shape of Cmr4 that are capped by three smaller, globular densities with the size and shape of Cmr5 juxtaposed with a ~20 Å channel or groove running the length of the complex. This channel could easily accommodate an A-form RNA duplex in a hypothetical target-bound state. The distance between adjacent Cmr4 subunits within this backbone is ~25 Å consistent with a length of ~6 to 7 bp of A-form RNA, closely mirroring the size of our observed cleavage products and suggesting these subunits as the catalytic centers. The Cmr2-Cmr3 subcomplex crystal structure from P. furiosus (Osawa et al., 2013; Shao et al., 2013) fits into the ‘tail’ with high fidelity (Fig. 7E). A curled ‘head’ containing Cmr1 and Cmr6 adjoins the other side of the proposed RNA-binding groove and positions these subunits on the opposite side of Cmr2-Cmr3.
Figure 7.
Molecular architecture of the T. thermophilus Cmr complex. (A) Surface representation of the 3D reconstruction at 26 Å resolution from negatively stained Cmr complexes. The contour level is ~1.5σ. (B) Representative reference-free 2D class averages (upper row), corresponding reprojections (middle row), and surface representation (lower row) of Cmr complexes. The scale bars represent 100 Å. (C) Representative raw particle images of Cmr complexes containing Cmr6-His6 and labeled with NTA-nanogold particles. The scale bars represent 100 Å. (D) Segmentation of the TtCmr complex reconstruction at 22 Å resolution highlighting the ‘sea worm’ architecture. Segmented regions are colored and labeled as: Cmr1/Cmr6 (orange), Cmr2 (purple), Cmr3 (green), Cmr4 (alternating light blue and grey), and Cmr5 (red). (E) The crystal structure of the homologous Cmr2-Cmr3 subcomplex from P. furiosus has been docked into the tail and is color-coded and labeled as in (D).
Discussion
In this study, we have analyzed the structure and activity of a bacterial Type III-B CRISPR-Cas effector complex: the Cmr complex from T. thermophilus. Both in terms of structure and activity, the TtCmr complex shows interesting differences compared to its previously studied archaeal homologues (Hale et al., 2012; Hale et al., 2009; Zhang et al., 2012).
crRNAs of different sizes were found to bind the TtCmr complex, although the major population consisted of 40 and 46 nt lengths, 1 nt smaller than previously reported for the P. furiosis Cmr complex (Hale et al., 2009). Interestingly, with the exception of the abundant 49 nt crRNA species, the size-distribution showed a trend for crRNAs that differ 6 nt in size: 34 nt, 40 nt and 46 nt (Fig. 3B). The maturation of these crRNAs is presumably initiated by Cas6-mediated single-site cleavage in the repeat sequence of the pre-crRNA, generating a “1x intermediate” crRNA that contains the last 8 nt of the repeat sequence at its 5’ end. The 3’end is subsequently shortened by an unknown ribonuclease (other than Cas6) to produce the mature crRNA (Carte et al., 2008; Hale et al., 2009; Wang et al., 2011). We confirmed that TTHB231 protein (NCBI accession number YP_145470) from T. thermophilus HB8 has Cas6-like activity, as incubation of a pre-cRNA with TTHB231 yielded cleavage products of approximately 70 to 80 nt, which are the expected sizes of the 1x intermediate crRNAs (data not shown). The occurrence of Cmr-bound crRNAs of other sizes supports the possibility that this step in crRNA maturation occurs when bound to the complex. In addition, a recent study (Hatoum-Aslan et al., 2013) has revealed that the 3’ trimming of crRNAs in a Type III-A (Csm) complex is catalyzed by a trans-acting factor not present in the complex; most likely, the same is true for Type III-B (Cmr) complexes.
The population of crRNAs that was found to bind the Cmr complex shows a remarkable bias for particular spacers, the majority of which consisted of CRISPR-1.3, CRISPR-4.5 and CRISPR-11.3, and to a lesser extent CRISPR-11.8 and CRISPR-11.12. As opposed to the upregulation of the cmr genes (Agari et al., 2010), the overall crRNA abundance profile did not change upon ΦYS40 phage infection (Fig. 3A), indicating that this observed bias is not regulated by infection of this particular phage. The abundance profile of Cmr-bound crRNAs corresponds well with the cellular expression patterns found for these RNAs (Juranek et al., 2012). Since crRNA maturation starts with the transcription of a long pre-crRNA transcript (Brouns et al., 2008; Pougach et al., 2010), the differences in abundance of the individual crRNAs can have different explanations. Firstly, Cas6 (and/or enzymes responsible for 3’ end trimming) could favor the maturation of particular crRNAs over others. Secondly, particular crRNAs might bind the Cmr complex more efficiently than others, potentially promoting their stability. In addition, differences in the stability of the crRNA itself might influence its abundance (Zhang et al., 2012). Lastly, internal promoters in repeat and/or spacer sequences could contribute to differences in crRNA abundance (Deng et al., 2012). Many of these explanations are related to the secondary structure and/or sequence of the repeat and spacer sequences. However, we did not observe a clear correlation between the thermodynamic stability of in-silico predicted crRNA structures and their abundance within the Cmr complex (data not shown). We found that protospacers corresponding to the CRISPR-11.1, -11.3, and -11.4 were located in the intergenic regions of the chromosomal DNA of this strain. Further studies are necessary to address the possibility for the Cmr complex to target self RNAs.
Recently, the structures of several CRISPR-Cas complexes have been elucidated. Cascade, the Type I-E complex of E. coli is composed of Cse1, Cse2, Cas7, Cas5e (Cas5), and Cas6e proteins and forms a 405-kDa complex with a stoichiometry of 1:2:6:1:1 that binds a 61 nt crRNA (Jore et al., 2011). The negative stain EM and small angle x-ray scattering structures of this complex show an unusual seahorse shape (Jore et al., 2011). A subsequent cryo-EM analysis resolved the location of the individual subunits. Moreover, structures with and without target have revealed a major conformational change, proposed to create a docking site for the Cas3 nuclease (Wiedenheft et al., 2011a). The negative stain EM structure of the Pseudomonas Type I-F Csy complex, a functional homolog of the E. coli Cascade, composed of Csy1, -2, -3, and -4 proteins and a stoichiometry of 1:1:6:1 with a 60 nt crRNA has revealed a crescent-shaped particle, the overall morphology of which is similar to that of the E. coli Cascade (Wiedenheft et al., 2011b). The Sulfolobus Type I-A Cascade complex is composed of Csa2, Cas5a, Cas6, and Csa5 proteins and crRNA, forms a helical filament, resembling the E. coli Cascade-like arch-shaped particle (Lintner et al., 2011). The Bacillus halodurans Type I-C complex, also reveals a Cascade-like architecture, with a helical backbone of Cas7 proteins (Nam et al., 2012).
On the other hand, the Sulfolobus Cmr complex is composed of Cmr1, -2, -3, -4, -5, -6, and -7 proteins with a proposed stoichiometry of 1:1:1:1:1:1:6, part of which forms a ‘crab claw’ structure unlike the Cascade complex (Zhang et al., 2012). The overall structure of the T. thermophilus Cmr complex reveals no obvious similarity to the ‘crab claw’ structure of the Sulfolobus Cmr complex. Interestingly, it does have a repeating backbone that resembles the helical Cas7 backbone and architecture of the E. coli Cascade complex (Fig. S7G-H). It might be that the crRNA is covered by the Cmr4 backbone of the T. thermophilus Cmr complex like the E. coli Cascade-crRNA complex (Wiedenheft et al., 2011a). The Cascade and Cmr complexes are classified into different types of the CRISPR-Cas systems, i.e. types I and III, respectively (Makarova et al., 2011). However, their architectural similarities suggest common ancestry of at least some of their core subunits (Reeks et al., 2013). Despite these similarities, Cascade(-like) complexes should be regarded as surveillance complexes, as nuclease activity (Cas3) is only recruited upon successful recognition of the protospacer, while Type II and Type III-B complexes have intrinsic nuclease activities, and should therefore be regarded as effector complexes.
In this study, we have demonstrated that the TtCmr complex harbors Mg2+-dependent endoribonuclease activity (Fig. 5A), which is specific for RNA targets complementary to the crRNAs that reside in the complex (Fig. 5B). The Cmr2 subunit (NCBI accession number YP_145399) of the T. thermophilus Cmr complex does not have the N-terminal HD nuclease domain present in the Cmr2 proteins of the Pyrococcus and Sulfolobus Cmr complexes. This is consistent with the observation that the N-terminal HD domain is not involved in cleavage of target RNA (Cocozaki et al., 2012). Instead of a single cleavage site 14 nt upstream of the 3’ end of the crRNA observed in P. furiosus (Hale et al., 2009), the TtCmr complex has been demonstrated to cleave the target at multiple positions, spaced 6 nt apart (Fig. 5 and Fig. 6). Our experiments with the endogenous and reconstituted Cmr complexes with variable 3’ crRNA-ends showed that the degradation products are not a result of a 3’ ruler mechanism (Fig. 5D). Rather, our results demonstrated that cleavage is initiated at the 3’ end of the RNA substrate, followed by sequential endonucleolytic activity every six nucleotides towards the 5’ end (Fig. 5C), in other words, a 5’ ruler mechanism (Fig. 6). Cmr may use multiple cleavage sites to increase the cleavage efficiency and catalytic turnover, and possibly to improve the chance of interference in the case of targets with partial mismatches, secondary structure and/or associated proteins. Thus, this mechanism might render the Cmr complex more effective.
Although this study does not directly identify the ribonuclease of the complex, our data strongly suggest that Cmr4 fulfills this role. The main evidence for this is the observation that the Cmr complex probably harbors multiple active sites. If it is assumed that these sites are harbored in the same subunit, only Cmr4 and Cmr5 remain likely candidates. Cmr5 has previously been shown to be dispensable for activity, while Cmr4 was not (Hale et al., 2009). In addition, the sequential 6 nt cleavages corresponds well with the observed distance between two adjacent Cmr4 subunits. Interestingly, the 39, 33, 27 and 21 nt degradation products seem to be generated rather quickly when compared to the 15 nt degradation product (Fig. 5C). We propose that these early products are generated by Cmr4, while another Cmr subunit would be responsible for the 15 nt product. Generating and testing catalytically-dead mutants of Cmr4 (and other subunits) will therefore be an interesting challenge for future experiments.
Experimental Procedures
Purification of the Cmr complex
T. thermophilus HB8 cells producing the (His)6-tagged Cmr complex, constructed and cultivated as described in the Supplemental experimental procedures. The Cmr complex was isolated using a nickel resin column, followed by anion exchange and gel-filtration chromatography steps as described in detail in the Supplemental experimental procedures.
Expression and purification of recombinant Cmr proteins
The cmr genes were each amplified by genomic PCR and cloned under the control of the pET vector (Merck). The Cmr proteins were each expressed in the E. coli BL21(DE3) strain or its derivative with no tag, except for Cmr6, which has a (His)6 tag at its N-terminal followed by a tobacco etch virus (TEV) protease-recognition site (Fang et al., 2007). We confirmed the N-terminal amino acid sequences of the purified proteins. The N-terminal methionine of the Cmr4 protein was deleted. The Cmr6 protein has G-H residues at its N-terminal, which are derived from the TEV protease recognition site. The detailed experimental procedures are described in the Supplemental experimental procedures, and the SDS-PAGE analysis of the purified proteins is shown in Fig. S1.
Native mass spectrometry
Cmr was buffer-exchanged to 0.175 M ammonium acetate (pH 8.0) at 4°C, using five sequential steps on a centrifugal filter with a cut-off of 10 kDa (Sartorius). The complex was sprayed at a concentration of 1 μM from borosilicate glass capillaries. An LCT electrospray time-of-flight or modified quadrupole time-of-flight instruments (both from Waters, United Kingdom) adjusted for optimal performance in high mass detection was used (Tahallah et al., 2001; van den Heuvel et al., 2006). Exact mass measurements of the individual Cmr proteins were acquired under denaturing conditions (1 and 10 % formic acid). Reconstituted subcomplexes were heated at 65°C prior to buffer exchange (performed at 40°C). Instrument settings were as follows: needle voltage ~1.3 kV, cone voltage ~175 V, source pressure 9 mbar. Xenon was used as the collision gas for tandem mass spectrometric analysis at a pressure of 2 × 10−2 mbar. The collision voltage was varied between 10 to 200 V.
Deep sequencing of the bioinformatic analysis of the Cmr-bound crRNAs
Cmr-bound crRNAs were purified by phenol-chloroform isoamyl alcohol (PCI) extraction, as in the Supplemental experimental procedures. The strand-specific RNA-seq library was prepared according to the directional mRNA-seq library preparation protocol provided by Illumina. This protocol involves utilizing the Small RNA sample prep kit and the mRNA-seq library prep kit. crRNAs were treated with a phosphatase and kinase prior to ligation of RNA adapters to the 5’ and 3’ ends. The sequential ligation use of different adapters allowed for subsequent orientation of the sequencing reads. The ligated RNAs were then reverse transcribed and amplified by PCR. The library was sequenced with an Illumina Genome Analyzer IIx (Plateforme de Séquençage à Haut Débit Imagif, Gif-sur-Yvette, France). A total of 6,479,050 overlapping forward and reversed 74 nt Illumina reads were obtained and aligned against its mate using blast. Non-overlapping pairs were removed (774,457). Mapping of the adapter-stripped reads to the genome T. thermophilus HB8 was performed with Bowtie2 using the default settings (Langmead and Salzberg, 2012). Reads containing any insertions, deletions or mismatches regarding to the reference genome were discarded. As a result, 5,704,593 (92.74%) of the remaining reads could be mapped against the reference genome. Visualization was performed using Excel and Matplotlib (Hunter, 2007).
Reconstitution of the Cmr complex
Synthetic 43 and 46 nt crRNAs corresponding to CRISPR4.5 were purchased from Integrated DNA Technologies (IDT), the sequences of which are listed in Table S3. Cmr complexes were reconstituted by pre-heating the individual Cmr subunits (Cmr1-6) to 65°C, after which they were added together in a Cmr112131445361-crRNA1 molar ratio, followed by incubation for 10 min in at 65°C under gentle agitation.
RNA activity assays
RNA substrates for the activity assays were prepared by hybridizing two complementary DNA oligos, containing a SP6 promoter in front of the desired RNA sequence. A full description of all the oligos is given in Table S3. Internally, radiolabeled RNA was obtained by in-vitro transcription of the hybrized oligos with SP6 RNA polymerase in the presence of 32P α-UTP, followed by denaturing gel purification. The 50 nt target RNA sequence complementary to CRISPR-4.5 was purchased from IDT. This particular RNA was either 5’ or 3’ labeled using T4 polynucleotide kinase and 32P γ-ATP or T4 RNA ligase 1 and 32P pCp respectively, followed by denaturing gel purification. Target RNA substrates were incubated at 65°C with 5 nM endogenous Cmr complex or ~63 nM reconstituted Cmr complex in RNA degradation buffer (20 mM Tris-HCl, pH 8, 150 mM NaCl, 10 mM DTT, 1 mM ATP and 2 mM MgCl2). Reactions were stopped by transferring the tubes to ice followed by the addition of formamide RNA loading buffer. Samples and a 5’ 32P labeled RNA marker (Decade Markers, Ambion) were separated on a 20% polyacrylamide denaturing gel, containing 7 M urea and visualized by phosphorimaging.
Single particle electron microscopy and analysis
A few microliters of 0.05 to 0.1 mg ml−1 purified protein solution was applied onto a carbon-coated grid and negatively stained with 2% uranyl acetate. We examined the sample grids with a JEM-2100 electron microscope (JEOL) with a LaB6 gun operated at an accelerating voltage of 200 kV. Images were recorded on a slow-scan charge-coupled device (SSCCD) camera (MegaScan; Gatan, Pleasanton, CA) at a final magnification of 65,000 and at defocus settings of 8,900 to 27,000 Å. The magnification was calibrated from catalase crystals. For single-particle analysis of the negatively stained particles, the EMAN (http://blake.bcm.edu/emanwiki/EMAN1) and EMAN2 (http://blake.bcm.edu/emanwiki/EMAN2) software suites were used for the following analysis. The contrast transfer function (CTF) was estimated using ‘e2ctf.py’. The phase reversal due to the CTF was corrected with ‘applyctf’. We manually picked particles from 105 EM images by using ‘e2boxer.py’. The samples might contain complexes lacking the Cmr1 subunit, but these complexes appeared to be less abundant (Fig. 2 and 4). We selected particles with consistent dimensions, most of which represent the full Cmr complex. After a low-pass filter was applied, the set of particles were iteratively aligned with ‘cenalignint’. Three-dimensional maps were constructed from the aligned images by using ‘startnrclasses’ and ‘startAny’. The structure was refined by ‘refine’. The three-dimensional map obtained was used as a starting model for higher-resolution refinement, with no filter applied. The total number of molecular images included in the three-dimensional reconstruction was 8,976, after a few dozen iterations starting from 9,105 molecules. The Fourier shell correlation (FSC) was calculated between two volumes, each generated from half of the data set. The resolution was taken to be the spatial frequency at which the FSC drops below 0.5. Full CTF correction produced a map with the identical structural features although the map appeared to be at lower resolution. We also examined TtCmr complexes with automated electron microscopy and carried out segmentation of the 3D reconstruction (Figs. 7D, S7), the details of which are described in the Supplemental experimental procedures.
Gold labeling
The Cmr complex with a C-terminally His6-tagged Cmr6 was incubated at 4°C overnight with 250 nM nickel-nitrilotriacetic acid (Ni2+-NTA) nanogold particles (5 nm; Nanoprobes) (Hainfeld et al., 1999). We examined the samples with the JEOL electron microscope as described above.
Supplementary Material
Highlights.
The Cmr complex binds crRNAs (40 and 46 nt) from a subset of CRISPR spacers.
The Cmr complex cleaves RNA with a 5’-based ruler mechanism at multiple sites.
The Stoichiometry of the complex is Cmr112131445361crRNA1.
The Cmr complex reveals a ‘sea worm’ shape with similarities to Cascade.
Acknowledgments
We thank Aimi Osaki for construction of the recombinant T. thermophilus strain, Kayoko Matsumoto and Toshi Arima for purification of the Cmr proteins, Drs. Kwang Kim and Seiki Kuramitsu for identification of the Cmr subunits by the MS/MS analysis, and Aaron Jansen for his help with the activity assays. E. Nogales, H.-W. Wang, P. Grob and T. Houweling are acknowledged for assistance with EM and image-processing. This work was supported by a Grant-in-Aid for Scientific Research (C), 25440013, from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to A.S.), and by an ALW grant (820.02.003, to J.vdO) from the Netherlands Organization for Scientific Research (NWO). EvD, AB and AJRH were supported by the Netherlands Proteomics Centre. D.W.T is a National Science Foundation Graduate Research Fellow, and J.A.D. is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The EM-derived density maps for the first structure and from the larger dataset have been deposited in the EMDB under accession numbers EMD-5719 and EMD-2418, respectively. The deep sequencing dataset of the Cmr-bound crRNAs are deposited in the SRA database under accession number SRP029745.
References
- Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J. Mol. Biol. 2010;395:270–281. doi: 10.1016/j.jmb.2009.10.057. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocozaki AI, Ramia NF, Shao Y, Hale CR, Terns RM, Terns MP, Li H. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure. 2012;20:545–553. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- Deng L, Kenchappa CS, Peng X, She Q, Garrett RA. Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012;40:2470–2480. doi: 10.1093/nar/gkr1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Jia KZ, Tang YL, Ma DY, Yu M, Hua ZC. An improved strategy for high-level production of TEV protease in Escherichia coli and its purification and characterization. Protein Expr. Purif. 2007;51:102–109. doi: 10.1016/j.pep.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainfeld JF, Liu W, Halsey CM, Freimuth P, Powell RD. Ni-NTA-gold clusters target His-tagged proteins. J. Struct. Biol. 1999;127:185–198. doi: 10.1006/jsbi.1999.4149. [DOI] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, 3rd, Graveley BR, Terns RM, Terns MP. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Samai P, Maniv I, Jiang W, Marraffini LA. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck AJ. Native mass spectrometry: a bridge between interactomics and structural biology. Nat. Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering. 2007;9:90–95. [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Juranek S, Eban T, Altuvia Y, Brown M, Morozov P, Tuschl T, Margalit H. A genome-wide view of the expression and processing patterns of Thermus thermophilus HB8 CRISPR RNAs. RNA. 2012;18:783–794. doi: 10.1261/rna.031468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J. Biol. Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, Ke A. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Inanaga H, Numata T. Crystal Structure of the Cmr2-Cmr3 Subcomplex in the CRISPR-Cas RNA Silencing Effector Complex. J. Mol. Biol. 2013 doi: 10.1016/j.jmb.2013.03.042. [DOI] [PubMed] [Google Scholar]
- Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, Severinov K. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 2010;77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeks J, Naismith JH, White MF. CRISPR interference: a structural perspective. Biochem. J. 2013;453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz I, Lange SJ, Hein S, Hess WR, Backofen R. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS One. 2013;8:e56470. doi: 10.1371/journal.pone.0056470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Cocozaki AI, Ramia NF, Terns RM, Terns MP, Li H. Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA silencing complex. Structure. 2013;21:376–384. doi: 10.1016/j.str.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai A, Kira S, Nakagawa N, Kashihara A, Kuramitsu S, Yokoyama S. Transcription activation mediated by a cyclic AMP receptor protein from Thermus thermophilus HB8. J. Bacteriol. 2007;189:3891–3901. doi: 10.1128/JB.01739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahallah N, Pinkse M, Maier CS, Heck AJ. The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun. Mass Spectrom. 2001;15:596–601. doi: 10.1002/rcm.275. [DOI] [PubMed] [Google Scholar]
- van den Heuvel RH, van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJ, Langridge D, van der Oost J, Hoyes J, Heck AJ. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal. Chem. 2006;78:7473–7483. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- van Duijn E, Barbu IM, Barendregt A, Jore MM, Wiedenheft B, Lundgren M, Westra ER, Brouns SJ, Doudna JA, van der Oost J, Heck AJ. Native tandem and ion mobility mass spectrometry highlight structural and modular similarities in clustered-regularly-interspaced shot-palindromic-repeats (CRISPR)-associated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol. Cell. Proteomics. 2012;11:1430–1441. doi: 10.1074/mcp.M112.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011a;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, Doudna JA. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. U S A. 2011b;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.