Abstract
BACKGROUND AND PURPOSE
Genome-wide association studies have revealed multiple common variants associated with known risk factors for ischemic stroke (IS). However, their aggregate effect on risk is uncertain. We aimed to generate a multilocus genetic risk score (GRS) for IS based on genome-wide association studies data from clinical-based samples and to establish its external validity in prospective population-based cohorts.
METHODS
Three thousand five hundred forty-eight clinic-based IS cases and 6399 controls from the Wellcome Trust Case Control Consortium 2 were used for derivation of the GRS. Subjects from the METASTROKE consortium served as a replication sample. The validation sample consisted of 22 751 participants from the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium. We selected variants that had reached genome-wide significance in previous association studies on established risk factors for IS.
RESULTS
A combined GRS for atrial fibrillation, coronary artery disease, hypertension, and systolic blood pressure significantly associated with IS both in the case-control samples and in the prospective population-based studies. Subjects in the top quintile of the combined GRS had >2-fold increased risk of IS compared with subjects in the lowest quintile. Addition of the combined GRS to a simple model based on sex significantly improved the prediction of IS in the combined clinic-based samples but not in the population-based studies, and there was no significant improvement in net reclassification.
CONCLUSIONS
A multilocus GRS based on common variants for established cardiovascular risk factors was significantly associated with IS both in clinic-based samples and in the general population. However, the improvement in clinical risk prediction was found to be small.
Keywords: genetics, polymorphism, genetic, risk assessment, risk factors
Introduction
Stroke is the leading cause of permanent disability and the third most common cause of death in high-income nations.1,2 Approximately 80% of stroke cases are caused by ischemia, with large artery atherosclerosis and cardioembolism from atrial fibrillation (AF) being among the most common mechanisms.
Ischemic stroke (IS) is highly heritable.3,4 Probably reflecting some of the heritability, recent genome-wide association studies (GWAS) have identified common variants in several genomic regions that are associated with IS5 (L.L. Kilarski et al, unpublished data, 2013) or specific stroke subtypes such as large artery stroke.6,7 Furthermore, GWAS have identified multiple variants in multiple genomic regions that are associated with established risk factors for IS, including AF,8 coronary artery disease,9,10 and hypertension.11,12 Several of these variants were also found to be associated with IS risk typically through their association with specific stroke mechanisms, such as large artery disease6,13–15 or cardioembolism,16,17 although some loci were also found to be associated with IS as a whole5 (L.L. Kilarski et al, unpublished data, 2013). In general, the observed increase in risk associated with individual variants was found to be small usually in the range of 1.2 to 1.6. However, because most risk alleles are common in people of European ancestry, their effect on a population level is likely to be substantial. Current risk prediction models, based on conventional risk factors,18 perform reasonably well and are used to guide clinical decision making. Yet, efforts to add emerging factors into risk prediction scores continue19–21 because even incremental improvements in predictive performance might lead to clinically meaningful changes in risk classification. Combining the effects of genetic risk variants into multilocus genetic risk scores (GRSs) may aid in risk prediction. In fact, recent studies have demonstrated significant improvements in coronary risk prediction using multilocus GRS for coronary artery disease.22–25 Despite the increase in direct-to-consumer testing for genetic variants for stroke,26 the use of adding genetic variants to stroke risk prediction has not been evaluated systematically.
The purpose of this study therefore was to construct a multilocus GRS based on common variants previously shown to reach genome-wide significance for an association with known risk factors for stroke. Single nucleotide polymorphisms (SNPs) were identified on the basis of systematic literature and database review using predefined criteria. We hypothesized that a GRS derived from clinic-based case–control samples would replicate in independent clinic-based case–control samples and improve the ability to predict IS in population-based cohorts.
Methods
Study Sample
The clinic-based case–control sample for derivation of the GRS consisted of 3548 IS cases and 6399 controls from the Wellcome Trust Case Control Consortium 2.6The clinic-based replication sample included 3856 IS cases and 4069 controls from the METASTROKE consortium.15 The prospective population-based cohorts included 22 751 participants from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium.27 A detailed description of these cohorts together with information on the methods used for genotyping is provided in Table 1 and the online-only Data Supplement.
Table 1.
Description of Case–Control and Population-Based Cohorts Used in the Analysis
| Age, y | Women, % |
No. of Cases |
No. of Controls |
No. of Incident Cases |
Total No. of Subjects |
Ancestry | Genotyping | |
|---|---|---|---|---|---|---|---|---|
| Clinic-based derivation samples | ||||||||
| WTCCC2-UK | 72.2±12.5 | 48.5 | 2374 | 5562 | … | 7936 | CEU | Illumina 660 (cases) |
| Illumina 1.2M-Duo (controls) | ||||||||
| WTCCC2-Munich | 66.6±12.9 | 42.1 | 1174 | 837 | … | 2011 | CEU | Illumina 660 (cases) |
| Illumina 550 (controls) | ||||||||
| Clinic-based replication samples | ||||||||
| ASGC | 72.9±13.2 | 45.5 | 1177 | 1244 | … | 2421 | CEU | Illumina 610 |
| MGH-GASROS | 66.6±14.5 | 42.9 | 767 | 395 | … | 1162 | CEU | Affymetrix 6.0 |
| BRAINS | 74.4±7.2 | 46.7 | 394 | 444 | … | 838 | CEU | Illumina 610 |
| GEOS | 41.0±6.9 | 41.1 | 448 | 498 | … | 946 | CEU | Illumina HumanOmni1 |
| ISGS/SWISS | 66.5±13.6 | 48.4 | 1070 | 1488 | … | 2558 | CEU | Illumina 650 (cases) |
| Illumina 550 (controls) | ||||||||
| Prospective population-based cohorts | ||||||||
| ARIC | 54.3±5.7 | 52.8 | … | … | 437 (4.6%) | 9349 | CEU | Affymetrix 6.0 |
| CHS | 72.3±5.4 | 60.9 | … | … | 453 (13.9%) | 3268 | CEU | Illumina 370 |
| FHS | 66.1±12.6 | 54.7 | … | … | 198 (4.5%) | 4371 | CEU | Affymetrix 550 |
| Rotterdam | 69.1±9.0 | 40.6 | … | … | 467 (8.1%) | 5763 | CEU | Illumina 550 |
ARIC indicates The Atherosclerosis Risk in Communities study; ASGC, Australian Stroke Genetics Collaborative; BRAINS, Bio-Repository of DNA in stroke; CEU, Caucasian European; CHS, Cardiovascular Health Study; FHS, Framingham Heart Study; GEOS, Genetics of Early Onset Stroke; ISGS, Ischemic Stroke Genetics Study; MGH-GASROS, Massachusetts General Hospital—Genes Associated with Stroke Risk and Outcomes Study; SWISS, Siblings With Ischemic Stroke Study; WTCCC2-UK, The Wellcome Trust Case Control Consortium II UK; and WTCCC2-Munich, The Wellcome Trust Case Control Consortium II Munich.
Selection of Genetic Variants
We selected SNPs from GWAS on modifiable risk factors for IS as defined by the American Heart Association stroke statistics and the American Heart Association/American Stroke Association guideline on primary prevention of stroke28,29 that were published before January 2012. This included GWAS on the following risk factors: hypertension, diastolic blood pressure, systolic blood pressure, smoking/tobacco use, type 1 diabetes mellitus, type 2 diabetes mellitus, low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, overweight and obesity, AF, hypertriglyceridemia, coronary artery disease, myocardial infarction, alcohol abuse, uric acid, elevated circulating urate levels, and hyperhomocysteinemia (Table I in the online-only Data Supplement).
The National Human Genome Research Institute GWAS catalogue30 was used as the primary source for published GWAS. We considered studies assessing >100 000 markers in the discovery stage and variants reaching a global P value of <1E–5.31,32 Findings from candidate gene studies were excluded as were studies based on somatic mutations (eg, in cancer cells) compared with naturally occurring mutations. We further searched PubMed using the following MeSH terms “risk factor” AND “GWAS” OR “genome wide” OR “genome wide association.” The resulting citations and abstracts were reviewed manually as were cited articles in the selected publications. Studies on non-Central European populations and studies where the effect allele was not reported were disregarded. A full list of the 521 SNPs included in the analysis is provided in Table I in the online-only Data Supplement.
Weighting Scheme and Calculation of Risk Scores
SNPs were weighted by their estimated effect sizes (β) provided in the original reports (Table I in the online-only Data Supplement). For SNPs derived from quantitative trait studies, we arbitrarily used a uniform weight of 0.1, corresponding to an odds ratio (OR) of 1.105 (weak to moderate effect size) while accounting for directionality of effects. This was performed to account for the different scales and measures that were used in the original studies to measure the effects of SNPs on the respective traits.
Weighted multilocus GRSs (wGRSs) for individual risk factors (eg, hypertension) were calculated using the – score function implemented in PLINK33 and an additive model. Risk profile scores were derived by adding the number of risk alleles multiplied by the weight of the risk variant. SNPs with missing information were excluded from the model. Scores were expressed as the mean score per SNP in the set. The combined GRSs (cGRSs) were calculated by adding Z score transformed wGRS. Z scores were used to account for the variable numbers of risk alleles constituting the wGRS.
Statistical Analysis
For the case–control studies, risk profiles were analyzed by generalized linear models using logistic regression in R with phenotype (case or control) as the outcome variable and the risk profile score as the predictor variable. In the absence of additional information in the control cohorts, sex was used as the only covariate. To account for intrinsic genetic differences between cohorts, we added an indicator variable for recruiting site in all analyses of the merged derivation and replication sample. Step-wise logistic regression was performed using the stepAIC function in the MASS library in R. All normalized wGRSs were included in the analysis to discover the optimal set of wGRS for inclusion in the cGRS by exact Akaike information criterion. Odds ratios for the wGRS and cGRS are reported as an increase of risk per improvement of 1 SD of the respective GRS. The variance in case/control status explained by the score statistic was estimated as the difference in variance using Nagelkerke pseudo-R2 between a model including the cGRS, sex, and study site (full model) versus the covariates alone (reduced model). To evaluate the potential value of the cGRS in risk prediction, we compared the receiver operating characteristic curves of models with and without cGRS (lroc package for logistic regression in R). C-statistics were computed to assess the gain in predictive power of the cGRS and of the full versus reduced model.
For the prospective population-based cohorts, we used Cox regression models to evaluate the association of the cGRS with the incident IS and an R2 measure to estimate variance explained by the model. The statistical significance of change in the area under the receiver operating characteristic curve (AUC) between models was tested with the correlated C-index approach. C-statistics were meta-analyzed using the point estimates with SEs as input followed by inverse-variance meta-analysis. Significance of the full versus reduced model was evaluated using a method reported by Hanley and McNeil,34 which accounts for the correlation between both models. This correlation between the AUCs was obtained using the metacor package for R. The continuous net reclassification indices was calculated using published methods.35
Results
The clinic-based derivation sample included 3548 IS cases and 6399 controls from the Wellcome Trust Case Control Consortium 2 (WTCCC2; merged WTCCC2-UK and WTCCC2-Munich). Three thousand eight hundred fifty-six clinic-based cases and 4069 controls from METASTROKE served as a replication sample. A total of 22 751 participants from CHARGE (the Atherosclerosis Risk in Communities Study [ARIC], Cardiovascular Health Study [CHS], Framingham Heart Study [FHS], and Rotterdam) were included in the prospective cohort analyses. Median follow-up in the prospective cohorts was 14.4 years (interquartile range, 8.5). One thousand five hundred fifty-five (6.8%) incident cases of IS occurred during follow-up. Table 1 provides background characteristics for the clinic-based case–control samples and the population-based cohorts.
Weighted GRS for Individual Risk Factors
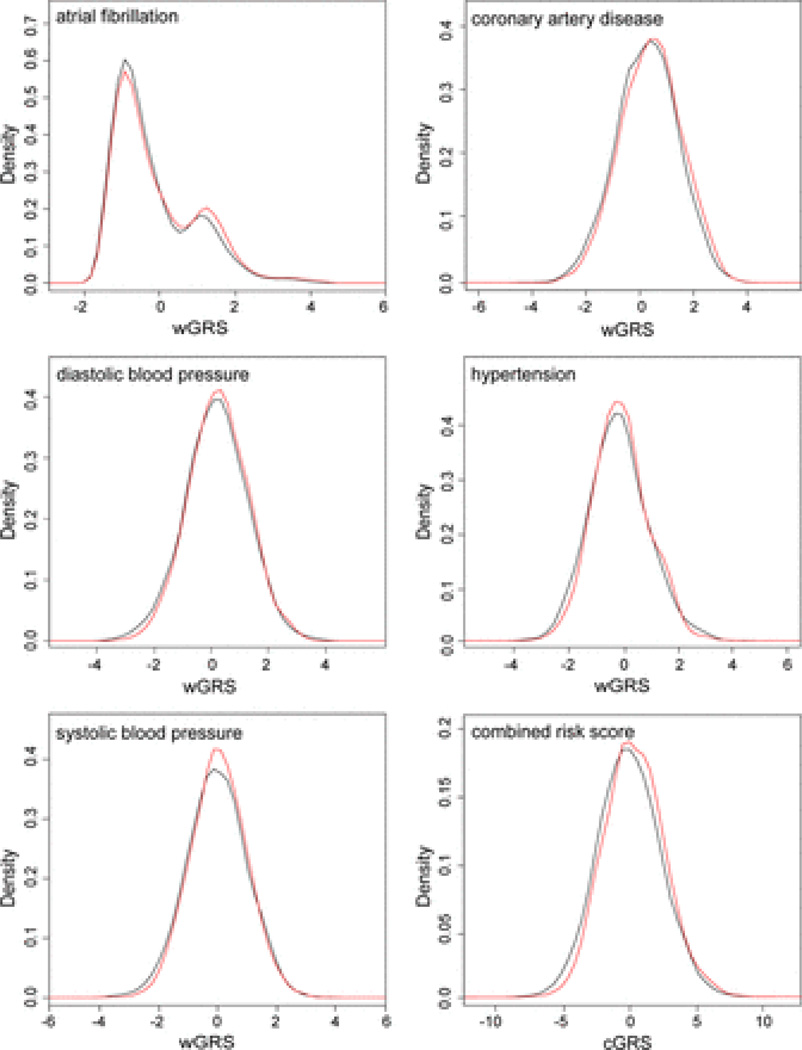
We first analyzed wGRS for individual risk factors (n=16) for IS in the merged clinic-based WTCCC2-UK and WTCCC2-Munich sample encompassing a total of 3548 cases and 6399 controls. Significant associations were found with wGRS for AF, coronary artery disease, diastolic blood pressure, hypertension, and systolic blood pressure (Table 2). Point estimates for OR in the merged derivation samples varied between 1.06 and 1.09 with similar effect sizes in the 2 derivation samples. Density plots for the wGRS are displayed in Figure 1. Analysis of the clinic-based replication and merged derivation and replication sample confirmed associations with the wGRS (Table II in the online-only Data Supplement) except for the wGRS for hypertension, which was not significant in the replication sample but reached significance in the merged derivation and replication sample.
Table 2.
Results for the wGRS for Individual Risk Factors in the Derivation Samples
| No. of Variants in wGRS |
WTCCC2-UK Derivation Sample |
WTCCC2-Munich Derivation Sample |
Merged Derivation Sample |
||||
|---|---|---|---|---|---|---|---|
| Risk Factor | Odds Ratio (95% CI) |
P Value | Odds Ratio (95% CI) |
P Value | Odds Ratio (95% CI) |
P Value | |
| Established RF28 | |||||||
| AF | 14 | 1.082 (1.031–1.134) | 1.16E-03* | 1.061 (0.970–1.160) | 1.99E-01 | 1.076 (1.032–1.122) | 6.19E-04* |
| BMI+weight | 95 | 1.007 (0.960–1.057) | 7.68E-01 | 1.000 (0.915–1.093) | 1.00 | 1.005 (0.963–1.050) | 8.09E-01 |
| CAD | 51 | 1.115 (1.062–1.170) | 1.04E-05* | 1.017 (0.930–1.111) | 7.20E-01 | 1.091 (1.046–1.139) | 5.84E-05* |
| DBP | 37 | 1.074 (1.024–1.127) | 3.50E-03* | 1.032 (0.944–1.128) | 4.93E-01 | 1.064 (1.020–1.110) | 4.21E-03* |
| HDL | 55 | 0.996 (0.949–1.045) | 8.64E-01 | 1.012 (0.926–1.107) | 7.89E-01 | 0.999 (0.957–1.042) | 9.54E-01 |
| HTN | 19 | 1.048 (0.999–1.099) | 5.39E-02 | 1.091 (0.998–1.194) | 5.59E-02 | 1.059 (1.014–1.105) | 8.25E-03* |
| LDL | 39 | 1.029 (0.981–1.080) | 2.37E-01 | 1.026 (0.938–1.121) | 5.81E-01 | 1.030 (0.987–1.074) | 1.75E-01 |
| MI | 12 | 1.064 (1.014–1.116) | 1.19E-02* | 0.977 (0.894–1.069) | 6.16E-01 | 1.043 (0.999–1.088) | 5.27E-02 |
| SBP | 29 | 1.077 (1.027–1.131) | 2.40E-03* | 1.037 (0.949–1.134) | 4.19E-01 | 1.069 (1.024–1.115) | 2.13E-03* |
| Smoking | 11 | 0.996 (0.950–1.045) | 8.80E-01 | 0.923 (0.845–1.010) | 8.03E-02 | 0.980 (0.939–1.022) | 3.47E-01 |
| T1D | 42 | 1.001 (0.954–1.050) | 9.73E-01 | 1.010 (0.924–1.104) | 8.27E-01 | 1.002 (0.961–1.045) | 9.20E-01 |
| T2D | 62 | 1.010 (0.962–1.059) | 6.94E-01 | 0.968 (0.886–1.059) | 4.78E-01 | 1.001 (0.960–1.044) | 9.63E-01 |
| Triglyceride levels | 49 | 0.995 (0.948–1.044) | 6.84E-01 | 1.039 (0.951–1.136) | 3.99E-01 | 1.005 (0.964–1.049) | 8.08E-01 |
| Less well-documented RF28 | |||||||
| Alcohol dependence | 26 | 1.025 (0.977–1.076) | 3.10E-01 | 0.885 (0.809–0.967) | 7.35E-03* | 0.991 (0.950–1.034) | 6.90E-01 |
| Homocysteine levels | 34 | 1.010 (0.963–1.060) | 6.73E-01 | 1.004 (0.919–1.010) | 9.28E-01 | 1.010 (0.968–1.053) | 6.54E-01 |
| Urate levels | 21 | 0.990 (0.944–1.039) | 8.23E-01 | 1.036 (0.948–1.133) | 4.33E-01 | 1.000 (0.958–1.044) | 9.98E-01 |
AF indicates atrial fibrillation; BMI, body mass index; CAD, coronary artery disease; CI, confidence interval; DBP, diastolic blood pressure; HDL, high-density lipoprotein; HTN, hypertension; LDL, low-density lipoprotein; MI, myocardial infarction; RF, risk factor; SBP, systolic blood pressure; wGRS, weighted genetic risk scores; WTCCC2-UK, The Wellcome Trust Case Control Consortium II UK; and WTCCC2-Munich, The Wellcome Trust Case Control Consortium II Munich.
Statistically significant risk factors at the P<0.05 level.
Figure 1.
Distribution of genetic risk score (GRS) in patients with ischemic stroke and controls in the merged derivation sample (WTCCC2-UK and WTCCC2-Munich). Shown are weighted GRSs (wGRSs) for atrial fibrillation, coronary artery disease, diastolic blood pressure, hypertension, and systolic blood pressure. Bottom right, The combined GRS (cGRS). Red line represents the distribution of the risk score in cases; and black line, the distribution of the risk score in controls. Note that the wGRS for AF is constituted by variants from 2 risk loci, which explains the bimodal distribution.
Combined Risk Score
We next tested combinations of the 16 wGRS for their association with IS. In step-wise logistic regression, a cGRS generated from wGRS for AF, coronary artery disease, hypertension, and systolic blood pressure encompassing a total of 113 variants (Table III in the online-only Data Supplement) showed the strongest association with IS in the merged WTCCC2-UK and WTCCC2-Munich sample (OR, 1.07 [95% confidence interval {CI}, 1.04–1.09]; P=5.78E–10; Table 3; Figure 1). The cGRS was found to replicate in the clinic-based replication sample (OR, 1.03 [95% CI, 1.01–1.04]; P=1.67E–03). Combining the derivation and replication samples and adding an indicator variable for recruiting site resulted in an OR similar to that found in the derivation sample (OR, 1.06 [95% CI, 1.04–1.08]; P=4.9E–07; Table 3).
Table 3.
Results for the cGRS* in the Clinic-Based Derivation and Replication Samples and in the Population-Based Sample
| Sample | Odds Ratio (95% CI) |
P Value | ΔR2, % |
ΔAUC, % (PValue) |
|---|---|---|---|---|
| Clinic-based derivation (WTCCC2-UK+WTCCC2-Munich; 3548 cases and 6399 controls) | 1.065 (1.044–1.087) | 5.78E-10 | 0.502 | 1.71 (1.5E-06) |
| Clinic-based replication (3856 cases and 4069 controls) | 1.026 (1.009–1.043) | 1.67E-03 | 0.041 | 0.19 (0.11) |
| Combined clinic-based derivation and replication (7404 cases and 10 468 controls) | 1.059 (1.036–1.083) | 4.87E-07 | 0.179 | 0.42 (1.8E-06) |
| Population-based (CHARGE; 1554 incident cases among 22 276 participants) | 1.027 (1.005–1.049) | 1.57E-02 | N/A | 0.11 (0.649) |
AUC indicates area under the receiver operating characteristic curve; cGRS, combined genetic risk score; CHARGE, Cohorts for Heart and Aging Research in Genomic Epidemiology; CI, confidence interval; N/A, not assessed; WTCCC2-UK, The Wellcome Trust Case Control Consortium II UK; and WTCCC2-Munich, The Wellcome Trust Case Control Consortium II Munich.
Generated from wGRS for atrial fibrillation, coronary artery disease, hypertension, and systolic blood pressure. The score included a total of 113 variants (for details see Table III in the online only Data Supplement).
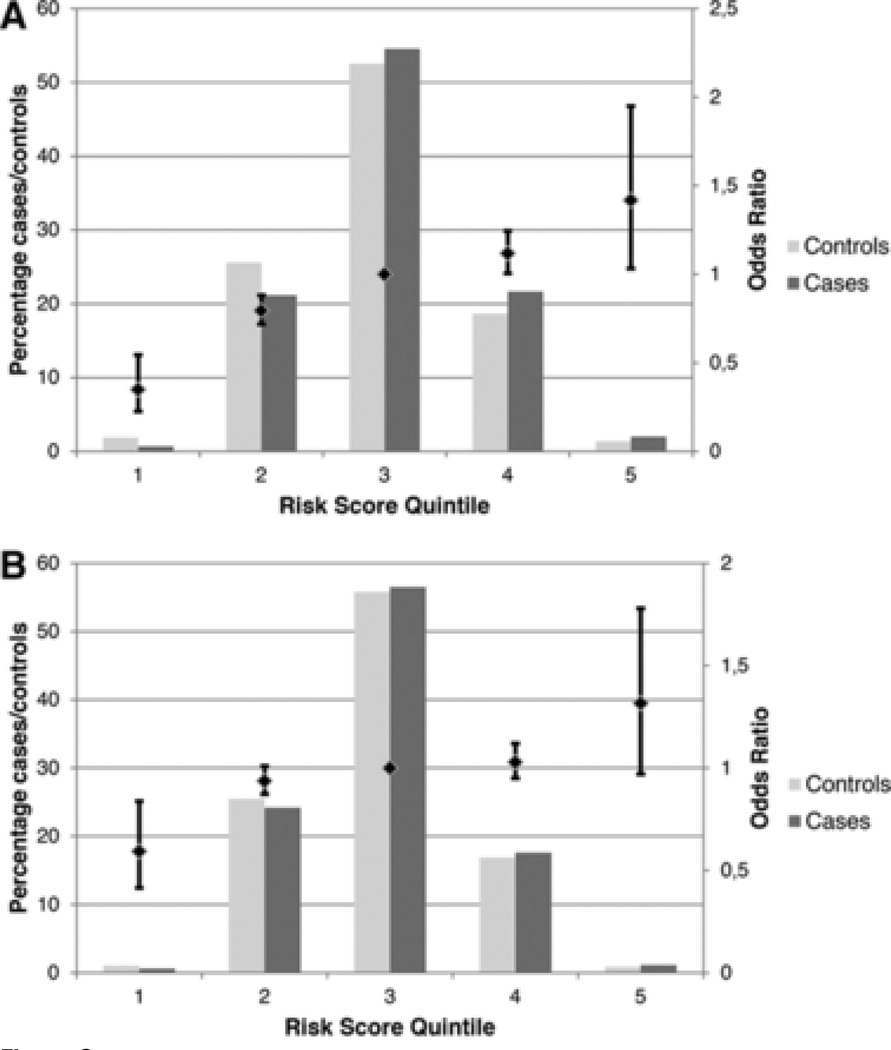
When we divided the cGRS in the derivation sample into quintiles, we found a linear increase in IS risk across quintiles (Cochran–Armitage test for linear trend across quintiles P=1E–13; Figure 2A). Compared with subjects in the third quintile (reference), subjects in the top quintile were estimated to have a 1.42× increased risk of IS (95% CI, 1.03–1.95), whereas subjects in the bottom quintile were estimated to have a 0.35-fold risk of IS (95% CI, 0.26–0.55). Similar results were obtained in the combined clinic-based derivation and replication sample (Figure 2B), although effect sizes were smaller (bottom quintile, 0.59 [95% CI, 0.41–0.84]; top quintile, 1.32 [95% CI, 0.97–1.78]).
Figure 2.
Odds ratios for risk categories defined using the combined genetic risk score. A, Clinic-based derivation sample. B, Merged clinic-based derivation and replication sample. The primary (left) y axis displays the percentage of cases and controls in each quintile, the secondary (right) y axis displays the odds ratio (OR) associated with each quintile where the middle (third) quintile serves as a reference.
Improvement of Prediction Quality
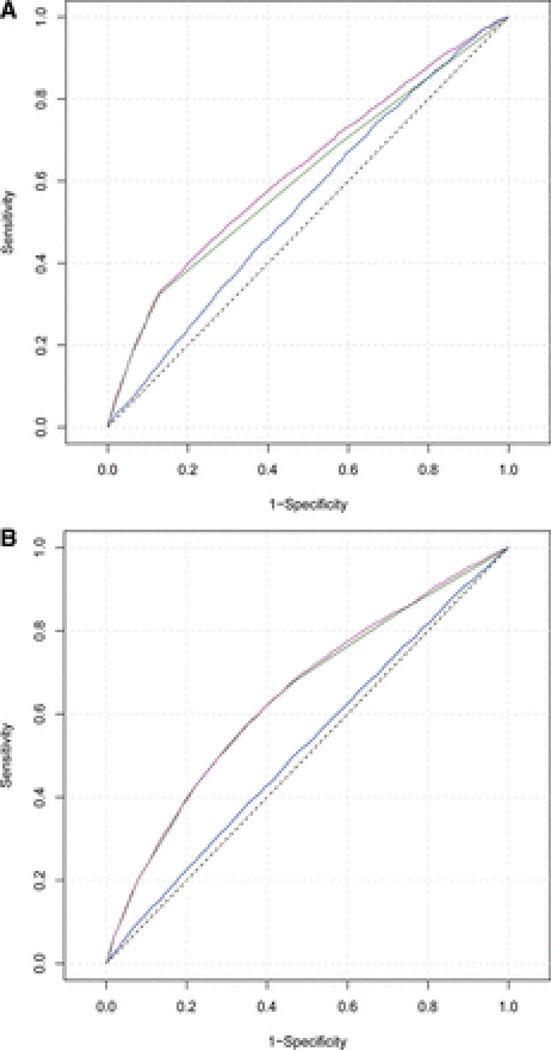
To determine the predictive value of the cGRS, we calculated the improvement in prediction quality of the cGRS compared with a model that included sex and study site (reduced model). The predictive strength, that is, the proportion to which the model explains variation in the data set, was 8.06% when using the cGRS in addition to all covariates (full model) and 7.56% when using only the covariates (reduced model). Hence, the cGRS resulted in an increase of 0.5% in predictive power (Table 2). This difference in explained variance was lower in the replication samples alone (0.041%) and was 0.179% in the combined clinic-based derivation and replication sample. The improvement in the c-statistic (AUC) for the cGRS was significant both in the clinic-based derivation sample (AUCfull model=62.75%, AUCreduced model=61.04%, ΔAUC=1.71%; P=1.5E–06) and in the combined derivation and replication samples (AUCfull model=64.51%, AUCreduced model=64.09%, ΔAUC=0.42%; P=1.8E–06; Table 3; Figure 3A and 3B).
Figure 3.
Receiver operating characteristic curves for models predicting a diagnosis of ischemic stroke in the derivation sample (A) and in the merged clinic-based derivation and replication sample (B). The reduced model of only covariates is shown in green, the full model including the combined genetic risk score (cGRS) in purple. The blue line represents the cGRS without covariates. The black dashed diagonal line represents a random prediction.
Validation of the cGRS in Prospective Population-Based Cohorts
To ascertain the validity and predictive power of the cGRS in stroke-free individuals, we meta-analyzed data from the prospective population-based ARIC, CHS, FHS, and Rotterdam cohorts. The pooled hazard ratio of the cGRS for incident IS was 1.03 (95% CI, 1.01–1.05; P=0.016) with no significant heterogeneity between studies (I2=19.9%; Figure I in the online-only Data Supplement).
AUC analyses in individual cohorts revealed largely variable c-statistics (AUCfull model=57.6%–72.2%), with no significant improvements in risk prediction in a combined meta-analysis (ΔAUC=0.11%; P=0.649; Table 3). The improvement of R2 with the full model compared with the reduced model ranged between 0.1% and 0.3% in individual cohorts.
We also assessed net reclassification results for the population-based cohorts. The continuous boundary-less net reclassification indices in individual cohorts ranged between −0.0101 and 0.1108, with no significant improvement in total reclassification.
Discussion
We found that a multilocus GRS composed of 113 common variants predicted IS risk. The cGRS was derived from clinic-based samples, replicated in independent samples, and validated using incident cases from prospective population-based cohorts. Subjects in the top quintile of the cGRS had >2-fold increased risk of IS when compared with subjects in the lowest quintile. However, the improvement in risk prediction by adding the cGRS to a simple model with sex and study site alone was small and not significant in the prospective validation cohort.
Considering >500 variants that had been shown previously to be associated with known risk factors for IS, we found weighted GRS for AF, coronary artery disease, hypertension, and systolic and diastolic blood pressure to show the strongest association with IS in the clinic-based derivation sample. These risk factors match with risk factors showing the strongest predictive value in conventional risk prediction models for stroke18,36 and with recent studies that have shown associations of individual risk alleles for AF, coronary artery disease, and hypertension with IS as a whole or with specific stroke subtypes.5,13,15,16Together, these findings suggest that multiple genetic variants at multiple chromosomal loci influence IS risk, possibly via known risk factors for IS. We did not examine our cGRS in relation to established risk scores such as the Framingham Risk Score for Stroke because information on risk factors was incomplete in the case–control samples and because one would not expect to see significant improvements over and above a score that contains actual information on the presence of these risk factors. It has been suggested that GRS may be most useful earlier in life, that is, before phenotypic variation in the risk factors incorporated into conventional risk prediction scores typically manifest.22However, our results suggest that the gain in predictive power by adding the GRS to information on sex alone is limited.
Our findings agree with studies in other conditions, including breast cancer,37diabetes mellitus,38,39 coronary artery disease,23,40 or multiple sclerosis,41 that found limited improvement in risk prediction with GRS based on GWAS discoveries. Our approach differs from most studies, in which we also considered variants that have to date not been associated with the clinical phenotype itself but instead reached genome-wide significance for association with known risk factors for the phenotype of interest, in this case IS. This enabled us to consider a much larger number of variants than were included in previous efforts to generate GRS. The validity of this approach is demonstrated by our finding that wGRS for systolic blood pressure, diastolic blood pressure, and hypertension all significantly associated with IS in the clinic-based case–control samples. Nevertheless, the improvement in risk prediction was small.
The limited use of our specific GRS for IS and of GRS in many other conditions might relate to several factors. First, because of the presence of multiple common alleles with small effects, almost all individuals carry some risk alleles. Second, the majority of individuals have multiple risk alleles close to the mean number of risk alleles in the overall population with the minority having extreme numbers of risk alleles as also reflected by the distribution of the cGRS in the current study (Figure 2B). Third, effect sizes between risk variants vary, which means that individuals having the same risk score may differ with regard to genetic risk unless GRSs were weighted as was the case in the current study. Finally, risk estimates on some of the variants may be imprecise as the number of studies is still relatively small. This might be interpreted as a failure of personalized medicine using genomics. On the contrary, the low level of genetic variance explained by common risk alleles identified to date suggests that much of the genetic predisposition is still undiscovered.42,43 Future studies will need to determine whether iterations of GRS incorporating additional common variants as well as low-frequency variants derived from whole-exome or whole-genome sequencing lead to clinically useful improvements in risk prediction.
The magnitude of effect of the cGRS in the combined case–control sample was higher than in the prospective population-based cohorts. This might be because of optimization of the cGRS in the derivation sample. Alternatively, the observed differences in OR might reflect different genetic architectures in clinic-based and incident cases from population-based cohorts. Compared with incident stroke cases from prospective population-based cohorts, clinic-based cases usually are younger7,44,45 and have more vascular risk factors including AF45 and hypertension.46 Another reason might be the relatively low incidence of stroke in the population-based cohorts resulting in reduced power. Of note, however, the cGRS was significantly associated with both prevalent and incident IS in the current study.
This study has several methodological strengths including replication of the cGRS in an independent case–control sample and validation in community-based samples that had been followed for extended time periods and provided a large number of incident events. Together, these samples represent one of the largest collections of IS cases with genome-wide data available to date.15 However, this study also has limitations. First, our GRS was based on SNPs that had been selected on the basis of predefined criteria. Thus, many variants with an effect on IS may have been excluded. We did not include variants published after January 2012 because of logistic challenges in obtaining calculated scores from all the replication and validation samples. This also includes variants from recently published loci associated with large artery stroke7,15 and coronary artery disease.10 Thus, our estimate on the predictive value and the explained variance of the cGRS likely is an underestimate of multilocus GRSs in IS. However, we consider it unlikely that the results would have been materially different with inclusion of those additional loci. Second, some quantitative traits could not be weighted for effect sizes because effects in the original studies had been reported on different scales. This likely resulted in an underestimation of effect sizes. Third, we did not analyze GRS for stroke subtypes because this information was not available in all validation cohorts. Finally, our sample consisted of white subjects of European descent. Thus our results may not be generalizable to other ethnicities.
We think our findings have clinical relevance because genetic testing is increasingly available and marketed to the public. We find no clinical use in constructing a multilocus panel of SNPs for stroke risk that extended to include variants acting on intermediate phenotypes such as hypertension or AF. It has been suggested previously that testing AF variants in well-defined populations such as patients with a cryptogenic stroke might aid in selecting subjects for further diagnostic procedures.16,47 The current study was not designed to address this. However, we found a wGRS for AF to have relatively small effect sizes in otherwise unselected clinic-based samples. Regardless of these results, any strategies aimed at testing common genetic variants to inform clinical decision making would need to be rigorously tested before moving to clinical practice.
In conclusion, we found a multilocus cGRS derived from GWAS for established risk factors for IS to be significantly associated with IS risk. Odds ratios in the highest and lowest quintile of the cGRS differed substantially. However, the power of the cGRS in predicting IS risk and hence its clinical use was found to be limited. Future alternative strategies of constructing GRS for IS and combining GRS with risk factor profiles and clinical information might eventually result in better risk prediction.
Supplementary Material
Acknowledgments
Sources of Funding
Australian Stroke Genetics Collaboration Australian population control data were derived from the Hunter Community Study. We also thank the University of Newcastle for funding and the men and women of the Hunter region who participated in this study. This research was funded by grants from the Australian National and Medical Health Research Council (NHMRC project grant ID: 569257), the Australian National Heart Foundation (NHF project grant ID: G 04S 1623), the University of Newcastle, the Gladys M. Brawn Fellowship scheme, and the Vincent Fairfax Family Foundation in Australia. E.G. Holliday is supported by the Australian NHMRC Fellowship scheme. Bio-Repository of DNA in Stroke is partly funded by a Senior Fellowship from the Department of Health (United Kingdom) to P. Sharma, the Henry Smith Charity, and the UK-India Education Research Initiative from the British Council. Genetics of Early Onset Stroke Study, Baltimore, MD, was supported by the National Institutes of Health (NIH) Genes, Environment and Health Initiative (GEI) Grant U01 HG004436, as part of the Gene-Environment Association Studies (GENEVA) consortium under GEI, with additional support provided by the Mid-Atlantic Nutrition and Obesity Research Center (P30 DK072488), and the Office of Research and Development, Medical Research Service, and the Baltimore Geriatrics Research, Education, and Clinical Center of the Department of Veterans Affairs. Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research, which is fully funded through a federal contract from the NIH to the Johns Hopkins University (contract number HHSN268200782096C). Assistance with data cleaning was provided by the GENEVA Coordinating Center (U01 HG 004446; PI Bruce S Weir). Study recruitment and assembly of datasets were supported by a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control and Prevention, and by grants from the National Institute of Neurological Disorders and Stroke (NINDS) and the NIH Office of Research on Women’s Health(R01 NS45012, U01 NS069208-01). Ischemic Stroke Genetics Study (ISGS)/Siblings With Ischemic Stroke Study (SWISS) was supported in part by the Intramural Research Program of the National Institute on Aging (NIA), NIH project Z01 AG-000954-06. ISGS/SWISS used samples and clinical data from the NIH-NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds), human subjects protocol numbers 2003–081 and 2004–147. ISGS/SWISS used stroke-free participants from the Baltimore Longitudinal Study of Aging (BLSA) as controls. The inclusion of BLSA samples was supported in part by the Intramural Research Program of the NIA, NIH project Z01 AG-000015-50, human subjects protocol number 2003–078. The ISGS study was funded by NIH-NINDS grant R01 NS-42733 (J.F. Meschia). The SWISS study was funded by NIH-NINDS grant R01 NS-39987 (J.F. Meschia). This study used the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (http://biowulf.nih.gov). MGH Genes Affecting Stroke Risk and Outcome Study (MGH-GASROS) was supported by the NINDS (U01 NS069208), the American Heart Association (AHA)/Bugher Foundation Centers for Stroke Prevention Research 0775010 N, the NIH and the National Heart, Lung, and Blood Institute’s (NHLBI) SNP Typing for Association with Multiple Phenotypes from Existing Epidemiological Data genomics research program (R01 HL087676), and a grant from the National Center for Research Resources. The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research resources. Funding for the Wellcome Trust Case Control Consortium 2 was provided by the Wellcome Trust (grants 085475/B/08/Z and085475/Z/08/Z). The Stroke Association provided additional support for collection of some of the St George’s, London cases. The Oxford cases were collected as part of the Oxford Vascular Study, which is funded by the MRC, Stroke Association, Dunhill Medical Trust, National Institute of Health Research (NIHR), and the NIHR Biomedical Research Centre, Oxford. The Edinburgh Stroke Study was supported by the Wellcome Trust (clinician scientist award to C. Sudlow), and the Binks Trust. Sample processing occurred in the Genetics Core Laboratory of the Wellcome Trust Clinical Research Facility, Western General Hospital, Edinburgh. Much of the neuroimaging occurred in the Scottish Funding Council Brain Imaging Research Centre (www.sbirc.ed.ac.uk), Division of Clinical Neurosciences, University of Edinburgh, a core area of the Wellcome Trust Clinical Research Facility and part of the Scottish Imaging Network—A Platform for Scientific Excellence collaboration (www.sinapse.ac.uk), funded by the Scottish Funding Council and the Chief Scientist Office. Collection of the Munich cases and data analysis was supported by the Vascular Dementia Research Foundation. The Rotterdam study was supported by The Netherlands Organization of Scientific Research (175.010.2005.011), the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research Netherlands Consortium for Healthy Ageing(050-060-810), Nederlandse Hartstichting (2009B102), the Erasmus Medical Center and Erasmus University, Rotterdam, the Netherlands Organization for Health Research and Development, the Research Institute for Diseases in the Elderly, the Ministry of Education, Culture, and Science, the Ministry for Health, Welfare, and Sports, the European Commission, and the Municipality of Rotterdam to the Rotterdam Study. The Atherosclerosis Risk in Communities Study (ARIC) is performed as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C,HHSN268201100008C, HHSN268201100009C, HHSN268201100010C,HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute contractU01HG004402; NIH contract HHSN268200625226C, and NHLBI contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020,N01-HC-55021, N01-HC-55022, and grants R01-HL087641, U01 HL096917(Mosley). Infrastructure was partly supported by grant number UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. ARIC analyses performed as part of this project were supported by grant HL-093029 to M. Fornage. This Cardiovascular Health Study (CHS) research was supported by theNHLBI contracts N01-HC-85079, N01-HC-85080, N01-HC-85081, N01-HC-85082, N01-HC-85083, N01-HC-85084, N01-HC-85085, N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133,N01-HC-85239, and by HHSN268201200036C and NHLBI grants HL080295,HL087652, and HL105756 with additional contribution from the NINDS. Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the NIA. See also http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping at Cedars-Sinai Medical Center was supported in part by theNational Center for Research Resources, grant UL1RR033176 and is now at the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, in addition to the National Institute of Diabetes and Digestive and Kidney Disease grant DK063491 to the Southern California Diabetes Endocrinology Research Center. Framingham Heart Study (FHS) research was supported by the NHLBI’s Framingham Heart Study (contract no. N01-HC-25195) and its contract with Affymetrix, Inc, for genotyping services (contract no. N02-HL-6-4278) and grants (U01 HL096917 and R01 HL093029). A portion of this research used the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. Analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource project. This study was also supported by grants from the NINDS (NS17950) and the National Institute of Aging (AG08122,AG033193). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS, the NHLBI, the NIA, the NIH, or the AHA.
References
- 1.Bonita R. Epidemiology of stroke. Lancet. 1992;339:342–344. doi: 10.1016/0140-6736(92)91658-u. [DOI] [PubMed] [Google Scholar]
- 2.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 3.Dichgans M. Genetics of ischaemic stroke. Lancet Neurol. 2007;6:149–161. doi: 10.1016/S1474-4422(07)70028-5. [DOI] [PubMed] [Google Scholar]
- 4.Bevan S, Traylor M, Adib-Samii P, Malik R, Paul NL, Jackson C, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43:3161–3167. doi: 10.1161/STROKEAHA.112.665760. [DOI] [PubMed] [Google Scholar]
- 5.Dichgans M, Malik R, König IR, Rosand J, Clarke R, Gretarsdottir S, et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. [Accessed December 21, 2013];Stroke. 2013 Nov 21; doi: 10.1161/STROKEAHA.113.002707. http://stroke.ahajournals.org/content/early/2013/11/21/STROKEAHA.113.002707.long. [DOI] [PMC free article] [PubMed]
- 6.Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, Burgess AI, Pirinen M, et al. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holliday EG, Maguire JM, Evans TJ, Koblar SA, Jannes J, Sturm JW, et al. Australian Stroke Genetics Collaborative; International Stroke Genetics Consortium; Wellcome Trust Case Control Consortium 2. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet. 2012;44:1147–1151. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Cardiogenics; CARDIoGRAM Consortium. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CARDIoGRAMplusC4D Consortium, Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gschwendtner A, Bevan S, Cole JW, Plourde A, Matarin M, Ross-Adams H, et al. International Stroke Genetics Consortium. Sequence variants on chromosome 9p21.3 confer risk for atherosclerotic stroke. Ann Neurol. 2009;65:531–539. doi: 10.1002/ana.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams FM, Carter AM, Hysi PG, Surdulescu G, Hodgkiss D, Soranzo N, et al. EuroCLOT Investigators; Wellcome Trust Case Control Consortium 2; MOnica Risk, Genetics, Archiving and Monograph; MetaStroke; International Stroke Genetics Consortium. Ischemic stroke is associated with the ABO locus: the EuroCLOT study. Ann Neurol. 2013;73:16–31. doi: 10.1002/ana.23838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, et al. Australian Stroke Genetics Collaborative, Wellcome Trust Case Control Consortium 2 (WTCCC2); International Stroke Genetics Consortium. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, Gschwendtner A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–409. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 17.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Agostino RB, Wolf PA, Belanger AJ, Kannel WB. Stroke risk profile: adjustment for antihypertensive medication. The Framingham Study. Stroke. 1994;25:40–43. doi: 10.1161/01.str.25.1.40. [DOI] [PubMed] [Google Scholar]
- 19.Goliasch G, Kleber ME, Richter B, Plischke M, Hoke M, Haschemi A, et al. Routinely available biomarkers improve prediction of long-term mortality in stable coronary artery disease: the Vienna and Ludwigshafen Coronary Artery Disease (VILCAD) risk score. Eur Heart J. 2012;33:2282–2289. doi: 10.1093/eurheartj/ehs164. [DOI] [PubMed] [Google Scholar]
- 20.Ky B, French B, Levy WC, Sweitzer NK, Fang JC, Wu AH, et al. Multiple biomarkers for risk prediction in chronic heart failure. Circ Heart Fail. 2012;5:183–190. doi: 10.1161/CIRCHEARTFAILURE.111.965020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roman MJ, Kizer JR, Best LG, Lee ET, Howard BV, Shara NM, et al. Vascular biomarkers in the prediction of clinical cardiovascular disease: the Strong Heart Study. Hypertension. 2012;59:29–35. doi: 10.1161/HYPERTENSIONAHA.111.181925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes MF, Saarela O, Stritzke J, Kee F, Silander K, Klopp N, et al. Genetic markers enhance coronary risk prediction in men: the MORGAM prospective cohorts. PLoS One. 2012;7:e40922. doi: 10.1371/journal.pone.0040922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, SharmaA, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 25.Davies RW, Dandona S, Stewart AF, Chen L, Ellis SG, Tang WH, et al. Improved prediction of cardiovascular disease based on a panel of single nucleotide polymorphisms identified through genome-wide association studies. Circ Cardiovasc Genet. 2010;3:468–474. doi: 10.1161/CIRCGENETICS.110.946269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. [Accessed August 2, 2013];Stroke Genetic Risk. 23 and Me. Https://www.23andme.Com/health/stroke/.
- 27.Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. CHARGE Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein LB, Bushnell CD, Adams RJ, Appel LJ, Braun LT, Chaturvedi S, et al. American Heart Association Stroke Council; Council on Cardiovascular Nursing; Council on Epidemiology and Prevention; Council for High Blood Pressure Research, Council on Peripheral Vascular Disease, and Interdisciplinary Council on Quality of Care and Outcomes Research. Guidelines for the primary prevention of stroke: a guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2011;42:517–584. doi: 10.1161/STR.0b013e3181fcb238. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd-Jones DM. Cardiovascular risk prediction: basic concepts, current status, and future directions. Circulation. 2010;121:1768–1777. doi: 10.1161/CIRCULATIONAHA.109.849166. [DOI] [PubMed] [Google Scholar]
- 30.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei Z, Wang K, Qu HQ, Zhang H, Bradfield J, Kim C, et al. From disease association to risk assessment: an optimistic view from genome-wide association studies on type 1 diabetes. PLoS Genet. 2009;5:e1000678. doi: 10.1371/journal.pgen.1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park JH. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet. 2013;45:400–405. 405e1. doi: 10.1038/ng.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148:839–843. doi: 10.1148/radiology.148.3.6878708. [DOI] [PubMed] [Google Scholar]
- 35.Pencina MJ, D’Agostino RB, Sr, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lumley T, Kronmal RA, Cushman M, Manolio TA, Goldstein S. A stroke prediction score in the elderly: validation and Web-based application. J Clin Epidemiol. 2002;55:129–136. doi: 10.1016/s0895-4356(01)00434-6. [DOI] [PubMed] [Google Scholar]
- 37.Wacholder S, Hartge P, Prentice R, Garcia-Closas M, Feigelson HS, Diver WR, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 39.Meigs JB, Shrader P, Sullivan LM, McAteer JB, Fox CS, Dupuis J, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brautbar A, Pompeii LA, Dehghan A, Ngwa JS, Nambi V, Virani SS, et al. A genetic risk score based on direct associations with coronary heart disease improves coronary heart disease risk prediction in the Atherosclerosis Risk in Communities (ARIC), but not in the Rotterdam and Framingham Offspring, Studies. Atherosclerosis. 2012;223:421–426. doi: 10.1016/j.atherosclerosis.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Jager PL, Chibnik LB, Cui J, Reischl J, Lehr S, Simon KC, et al. Steering committee of the BENEFIT study; Steering committee of the BEYOND study; Steering committee of the LTF study; Steering committee of the CCR1 study. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1111–1119. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kraft P, Hunter DJ. Genetic risk prediction–are we there yet? N Engl J Med. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 44.Schulz UG, Flossmann E, Rothwell PM. Heritability of ischemic stroke in relation to age, vascular risk factors, and subtypes of incident stroke in population-based studies. Stroke. 2004;35:819–824. doi: 10.1161/01.STR.0000121646.23955.0f. [DOI] [PubMed] [Google Scholar]
- 45.Schulz UG, Rothwell PM. Differences in vascular risk factors between etiological subtypes of ischemic stroke: importance of population-based studies. Stroke. 2003;34:2050–2059. doi: 10.1161/01.STR.0000079818.08343.8C. [DOI] [PubMed] [Google Scholar]
- 46.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meschia JF. Decoding cryptogenic cardioembolism. Ann Neurol. 2008;64:364–366. doi: 10.1002/ana.21470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.