Abstract
Vaginally administered antiviral agents may reduce the risk of HIV and HSV acquisition. Delivery of these drugs using intravaginal rings (IVRs) holds the potential benefits of improving adherence and decreasing systemic exposure, while maintaining steady-state drug levels in the vaginal tract. Elucidating how IVRs interact with the vaginal microbiome constitutes a critical step in evaluating the safety of these devices, as shifts the vaginal microbiome have been linked with several disease states. To date, clinical IVR trials have relied on culture-dependent methods that omit the high diversity of unculturable microbial population. Longitudinal, culture-independent characterization of the microbiota in vaginal samples from 6 women with recurrent genital HSV who used an acyclovir IVR was carried out and compared to the communities developing in biofilms on the IVR surface. The analysis utilized Illumina MiSeq sequence datasets generated from bar-coded amplicons of 16S rRNA gene fragments. Specific taxa in the vaginal communities of the study participants were found to be associated with the duration of recurrent genital HSV status and the number of HSV outbreaks. Taxonomic comparison of the vaginal and IVR biofilm communities did not reveal any significant differences, suggesting that the IVRs were not systematically enriched with members of the vaginal microbiome. Device usage did not alter the participants' vaginal microbial communities, within the confines of the current study design. Rigorous, molecular analysis of the effects of intravaginal devices on the corresponding microbial communities shows promise for integration with traditional approaches in the clinical evaluation of candidate products.
1. Introduction
In 2003, an estimated 536 million people worldwide aged 15-49 were living with herpes simplex virus type 2 (HSV-2) with an annual incidence of 23.6 million (Looker et al., 2008). Globally, HSV-2 is the most frequent cause of genital ulcer disease (Corey et al., 2004) and is associated with a three-fold increased risk for HIV-1 acquisition in women (Freeman et al., 2006). These epidemiological findings suggest that interventions against HSV-2 may have a key role in HIV prevention worldwide (Freeman et al., 2007). Daily oral valacyclovir (VACV), the 5′-L-valyl ester prodrug of the antiherpetic drug acyclovir (ACV), has been shown to prevent or delay genital recurrences by 85% (Patel et al., 1997) and to reduce the risk of transmission among HSV-2-discordant couples by 48% (Corey et al., 2004).
Topical application of ACV to the vagina is safe and has provided some clinical benefit for the treatment of primary or recurrent lesions by shortening their duration (Corey et al., 1982a; Corey et al., 1982b). We hypothesize that sustained delivery of ACV to the vaginal tract can provide an alternative approach to oral suppressive therapy and may protect against sexual HSV acquisition. Delivering ACV from intravaginal rings (IVRs) holds potential benefits of improved adherence and low systemic exposure while maintaining steady-state drug levels in the vaginal tract. We previously developed a pod-IVR technology (Baum et al., 2012) that can deliver multiple compounds independently in a controlled, sustained fashion with pseudo-zero order kinetics (Moss et al., 2012a; Moss et al., 2012b; Moss et al., 2012c). The safety and pharmacokinetics of pod-IVRs delivering ACV in combination with the nucleoside reverse transcriptase inhibitor (NRTI) tenofovir (TFV) were evaluated successfully in the rabbit and sheep models (Moss et al., 2012c). Acyclovir tissue penetration in both models was not impaired by vaginal mucous. We designed human silicone IVRs to release ACV and evaluated safety, pharmacokinetic, and surrogate efficacy in women with recurrent genital HSV, referenced herein as “genital herpes positive” (GHP), who switched their daily oral VACV suppression to the ACV pod-IVR for 7 and 14 days (Keller et al., 2012). This first-in-human study demonstrated that an IVR could safely deliver ACV and achieve comparable local mucosal levels to oral therapy without systemic absorption. The IVRs were well tolerated and there were no adverse events related to the ring, no abnormal colposcopic findings, and no reports of ring expulsions. There was no increase in pro-inflammatory and anti-inflammatory cytokine levels in cervicovaginal lavage (CVL) samples following IVR insertion. No HSV viral DNA was detected in vaginal tract swab samples during the course of the study. The median ex vivo anti-HSV activity of CVL fluids increased from 32% in samples collected just prior to IVR insertion to 58% in samples collected 7 days after IVR use, suggesting that sufficient ACV was present to augment the endogenous anti-HSV activity of cervicovaginal secretions, even though CVL represents an approximately 50-fold dilution of vaginal fluids.
The normal vaginal microbiota is believed to play an important protective role in maintaining the woman's health (Hillier, 2005). The Human Microbiome Project (HMP) (Peterson et al., 2009; Turnbaugh et al., 2007) is improving our understanding of the dynamics of vaginal bacteria in healthy individuals (Gajer et al., 2012; Ravel et al., 2011), and the pathogenic capabilities of key species that mediate poor health outcomes. In studies on IVRs delivering TFV in pig-tailed macaques, we used confocal laser scanning microscopy, fluorescence in situ hybridization, and scanning electron microscopy to investigate IVR colonization by polymicrobial biofilms (Gunawardana et al., 2011). Large areas of the ring surfaces were covered with monolayers of epithelial cells that supported two biofilm phenotypes, both with a broad diversity of associated bacterial cells. Similar results were obtained in our clinical evaluation of IVRs delivering ACV in GHP women (Keller et al., 2012). By Day 7, epithelial cell clusters had developed on the IVR surface, with little or no visible associated microbial growth. At Day 14, large areas of the ring surface were covered with a mat of epithelial cells that harbored the development of polymicrobial biofilms with similar morphological features to the biofilm phenotypes in our macaque studies.
Our limited understanding of how the vaginal microbiome responds to topical delivery of antiviral candidates is a critical gap in developing these strategies for clinical evaluation. Here we describe the first culture-independent assessment on the bacterial colonization of IVRs in women and the concomitant effect on their vaginal microbiomes.
2. Materials and Methods
2.1. Participants and study design
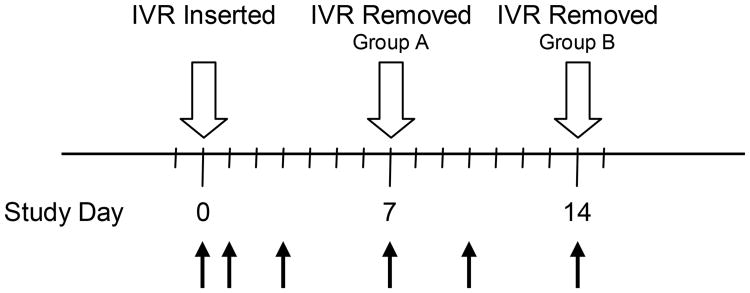
The participant characteristics and study design (Fig. 1) have been described in detail elsewhere (Keller et al., 2012). Briefly, 6 HIV-negative, GHP women who were willing to change their suppressive oral VACV to an ACV IVR were enrolled into a pharmacokinetic and safety study. In order to prevent ACV washout from the vaginal tract, IVR insertion occurred within 24 h of oral VACV dosing, which was discontinued during the study. The first three participants used an ACV IVR for 7 days and had cervicovaginal lavage (CVL) collected prior to IVR placement; 1 and 3 days post-insertion; and at Day 7 when the IVR was removed. The final three participants used an ACV IVR for 14 days, and the study visits were extended to include sampling at 10 days after IVR insertion and at Day 14 when the IVR was removed. The study design resulted in the collection of 30 CVL samples: 6 at IVR placement; 6 on Day 1; 6 on Day 3; 3 on Day 7; 3 on Day 10; 6 at IVR removal. None of the women displayed symptoms suggestive of active vaginal or sexually transmitted infection (STI) during the study. The women abstained from vaginal, anal and oral sex and the use of any vaginal products other than the ACV IVR for the duration of the study.
Fig. 1.
Study timelines and CVL sample collection points (black arrows). Participants used an ACV IVR from Day 0 (24 h after last dose of oral VACV) to Day 7 (Group A, n = 3) and Day 14 (Group B, n = 3). CVL was collected for microbial DNA isolation on: Group A, Days 0 (pre-dose), 1, 3, and 7 (upon IVR removal); Group B, Days 0 (pre-dose), 1, 3, 7, 10, and 14 (upon IVR removal).
2.2. Sample processing and microbial DNA isolation
ACV IVRs were removed aseptically on Day 7 or 14. The rings were cut into sections and portions of the segments without pods (i.e., unmedicated), were placed in 70% ethanol in water and stored at 4°C. CVL samples were placed on ice and clarified by centrifugation at 700 × g for 10 min at 4°C. The cell pellets together with a fraction of their supernatants (ca. 1 mL) were stored at -80°C.
Microbial DNA was isolated from a total of 36 samples (6 IVR and 30 CVL, see Fig. 1) according to the following methods. Unmedicated IVR segments were cut into small pieces using a pre-sterilized scalpel and DNA was extracted from these samples using the PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer's instructions. Frozen CVL samples were thawed on ice to afford a viscous fluid that was transferred into a microfuge tube and processed using the InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA, USA) kit with the following modifications to the manufacturer's instructions: sample to matrix volume ratio was 4:1 and the 56°C incubation time was extended to 40 minutes. Genomic DNA was isolated using two different methods, as the CVL samples contained inhibitors of PCR amplification that required a modified DNA extraction procedure.
Genomic microbial DNA isolated from the above samples was quantified using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies Corporation, Carlsbad, CA, USA) according to the manufacturer's instructions. The isolation procedures yielded between 9.7 and 22.5 μg (CVL, mean 15.1 μg) and between 3.3 and 35.5 μg (IVR, mean 14.5 μg) of high quality, PCR inhibitor-free, whole genomic DNA per vaginal sample.
2.3. DNA amplification and sequencing of 16S rRNA genes
Amplification and sequencing of the V4 hypervariable region of the 16S rRNA gene was performed using the validated, region-specific bacterial/archaeal primers 515F and 806R according to previously described methods (Caporaso et al., 2012) optimized for the Illumina MiSeq platform. 5′-Barcoded amplicons were generated in duplicate using Premix Ex Taq (TaKaRa, Otsu, Shiga, Japan) and a MyCycler thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA). The PCR conditions consisted of an initial denaturing step of 94°C for 2 min, followed by 7 cycles of 94°C for 30 s, 48°C for 30 s, and 72°C for 1 min, 28 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, and a final elongation step of 72°C for 10 min. Replicate reactions were pooled and the amplicons were separated by electrophoresis on 1.0% agarose gels. The amplicons were purified using QIAquick Gel Extraction kit (Qiagen, Valencia, CA, USA) according to manufacturer's instructions. The A260:A280 absorbance ratio was acquired with a SpectraMax® Plus Absorbance Microplate Reader (Molecular Devices, Sunnyvale, CA, USA) and used as an indicator of DNA purity. Amplicon DNA was quantified using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies Corporation, Carlsbad, CA, USA). Amplicon aliquots (100 ng) from all 36 samples were pooled and re-purified with the UltraClean® PCR Clean-Up Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA). The purified, pooled sample contained 8.8 μg of DNA with an A260:A280 ratio of 1.82 and was submitted for sequencing using the MiSeq platform (Illumina, Inc., San Diego, CA, USA) at the Advanced Genomics Facility, University of Colorado, Boulder, USA. The sequence data will be submitted to the European Molecular Biology Laboratory European Bioinformatics Institute (EBI) at the time of publication.
2.4. Microbial community analysis
The 16S rRNA gene sequences obtained from the MiSeq platform were processed through the open source software pipeline Quantitative Insights Into Microbial Ecology (QIIME) version 1.7.0-dev (Caporaso et al., 2010). Sequences were filtered for quality using established guidelines (Bokulich et al., 2013). Quality-filtered reads were demultiplexed, yielding 3,989,623 sequences total with an average length of 151 bases per read, and an average coverage of 110,822 sequences per sample. Sequences were binned into Operational Taxonomic Units (OTUs) based on 97% identity using UCLUST (Edgar, 2010) against the Greengenes reference database (McDonald et al., 2012) May 2013 release. The representative sequences for each OTU were compared against the Greengenes database for taxonomy assignment. Each sample's sequences was rarefied to a depth of 28,000 sequences per sample to reduce the effect of sequencing depth, and used for downstream analysis. This level of rarefaction was chosen to minimize the number of samples dropped from downstream analysis while maximizing the number of sequences allowed per sample. Following rarefaction, 1 sample (Subject60.CVL14.671842) was omitted from further analysis due to insufficient sequence coverage, yielding 35 samples and 581 OTUs in 980,000 sequences.
A Poisson embedding algorithm was employed to determine a 95% confidence interval for finding a new OTU (Lladser et al., 2011) and, consequently, to calculate the proportion of the overall community diversity captured by sequencing. The overall highest probability for discovering new OTUs in our samples was 1.295%, meaning that our sequencing had revealed 98.705% of the overall community diversity (Table S2). The β diversity of samples was measured using the weighted UniFrac metric (Lozupone and Knight, 2008), and the dimensionality reduction technique of Principal Coordinates Analysis (PCoA) was used to visualize the community differences. All correlations involving bacterial relative abundance were performed using the nonparametric Kendall's tau. The relative abundance of each taxon was calculated by dividing the sequences pertaining to a specific taxon by the total number of bacterial sequences for that sample. The statistical cutoff of P = 0.05 after False Discovery Rate (FDR) correction for multiple comparisons was used to define statistical significance when testing if taxa were significantly different between groups.
3. Results
The overarching study objective was to determine if IVRs delivering ACV alter the vaginal microbiota of GHP women and to elucidate how the device's surface becomes populated with vaginal bacteria at Day 7 and 14. The highly variable composition (Ravel et al., 2011) and dynamics (Gajer et al., 2012) of vaginal microbiota, together with the small number of participants in this preliminary study, led to a time-series design, where each woman served as her own control.
3.1. Sample size considerations
The statistical robustness of observed clinical effects based on interventions or treatments usually is determined through a power calculation. For microbiome studies, power calculations are very much an emerging area. Traditional power calculations based on parametric distributions (e.g., assuming that the frequency data can be analyzed by t-tests or ANOVA) are often more misleading than useful in this context. Although recent work using Dirichlet models has been informative (Holmes et al., 2012; La Rosa et al., 2012), these models also make certain parametric assumptions that are not always well-founded in microbial data. We therefore performed a rarefaction analysis (Fig. S3) on a per-sample basis and the HMP dataset (Aagaard et al., 2013), showing the accumulation of new diversity in both qualitative (unweighted UniFrac) and quantitative (weighted UniFrac) measures of β diversity. The analysis demonstrated that using the weighted UniFrac distance metric with 20 samples at 1000 sequences per sample covered more than 99.5% of total β diversity seen in the vaginal communities characterized by the HMP. The sample size used in this study therefore is sufficient to draw general conclusions about how IVRs affect the vaginal communities of GHP women.
Given the novelty of this report, there are few relevant precedents that can be used for group size comparison. Ravel et al. used 16S rRNA gene pyrosequencing to characterize the vaginal microbiota in samples from 35 healthy women in a two-week study of twice-daily application of 1 of 3 vaginal gel formulations: placebo (10 subjects), 6% cellulose sulfate (CS, 13 subjects), and 4% Nonoxynol-9 (N-9, 12 subjects) (Ravel et al., 2012). Despite the small cohort sizes, an inter-group comparison was possible and found that treatment with active microbicides shifted the vaginal microbiota toward a community type dominated by strict anaerobes and lacking significant numbers of Lactobacillus spp. These results support the cohort size used here for intra-group comparison, requiring less statistical power.
Demonstration of safety is a central component in the development of topical microbicides for the prevention of sexual HIV transmission and typically has involved evaluating general toxicology and irritation/inflammation in one or more animal species (Reichelderfer, 2002) along with a rudimentary (culture-dependent) assessment of the vaginal microflora in macaques (Van Rompay, 2010; Veazey, 2008). Clinical trial failures with N-9, Carraguard, C31G (Savvy), and CS underscore the urgent need for comprehensive, early microbicide efficacy and safety assessment prior to large, Phase II/III clinical trials (Keller and Herold, 2009). The application of a culture-independent analysis to determining the effect of the microbicide IVR on the vaginal microbiota therefore was performed during early safety evaluation of this novel device, using 6 subjects in an exploratory clinical trial. Future clinical evaluation of the product will be carried out in larger cohorts of women to gain a deeper understanding of potential, subtle effects on the vaginal microbiota.
3.2. Classification of vaginal microbiota
Ravel et al. have shown that the vaginal bacterial communities of 396 asymptomatic women could be broadly classified into 5 major community state types (CSTs) (Ravel et al., 2011). Two of the women in the current study had vaginal microbiota dominated by Lactobacillus iners, one dominated by L. helveticus, and the remaining 3 woman had more diverse vaginal communities, consisting mainly of Lactobacillus, Proteobacteria, the Bacteroidetes genus Prevotella, and the Actinobacteria Gardnerella vaginalis in varying proportions (Fig. S1 and Fig. 2). The vaginal bacterial communities of 5 participants are consistent with 2 of the 5 proposed major CSTs (Fig. S1). One of the study participants, however, had a vaginal community that was dominated (91% relative abundance) by Lactobacillus helveticus, a community structure that does not match with any of the CSTs. Furthermore, our analysis suggests that L. crispatus was not present in any of the samples although this species makes up a dominant member of one of the CSTs (Ravel et al., 2011). Larger cohorts of GHP women will be needed in future analyses to determine if these taxon differences are a consequence of GHP status, VACV suppressive therapy, or merely reflect the small sample size. The vaginal bacterial communities of the 6 study participants cluster primarily by individual (Fig. 2), an expected result given the large interpersonal variation observed across body sites (Ursell et al., 2012).
Fig. 2.
Weighted UniFrac distances plotted in PCoA space comparing the vaginal microbial communities from the 6 GHP study participants (represented by blue, red, yellow, orange, green, and dark purple) reveals clustering by individual that is strongly dictated by specific taxa. The labeled light purple spheres represent taxa, and the proximity of colored participant samples to the taxa spheres is indicative of increased membership of that taxon in a given community. The larger the taxon sphere, the greater that taxon's overall abundance. Three women had communities dominated by Lactobacillus iners, one was dominated by L. helveticus, and two other women had communities with a shared proportion of Atopobium vaginae and Gardnerella vaginalis in addition to Lactobacilli. The vaginal communities represented by the green and dark purple spheres are so closely related that they cannot be distinguished from this vantage point in PCoA space.
3.3. Specific taxa are associated with duration of GHP and with number of HSV outbreaks
Potential correlations between the relative abundance of specific taxa and GHP status were investigated. To reduce the number of spurious correlations, only taxa that were present in at least 5 CVL samples with at least 2 representative sequences were included in the analysis. The first correlation was between the relative abundance of bacterial taxa and the duration of GHP status (median: 4.0 years, 2-8 year range) in the 6 study participants (Table S3A). Three of the most significant taxa that could be identified at the species level, for which the changes were statistically significant at the most stringent level, included Ureaplasma parvum (tau = 0.619, P = 0.0001), Gardnerella vaginalis (tau = 0.552, P = 0.00059), and Atopobium vaginae (tau = 0.534, P = 0.00071) (Fig. 3), and all exhibited a positive correlation between duration of GHP status and relative abundance. Many other taxa, identifiable to the genus level, also showed a positive correlation between duration of GHP and relative abundance including Pseudomonas and Sphingomonas, while some Lactobacillus taxa were negatively associated with GHP duration and relative abundance (Table S3A).
Fig. 3.
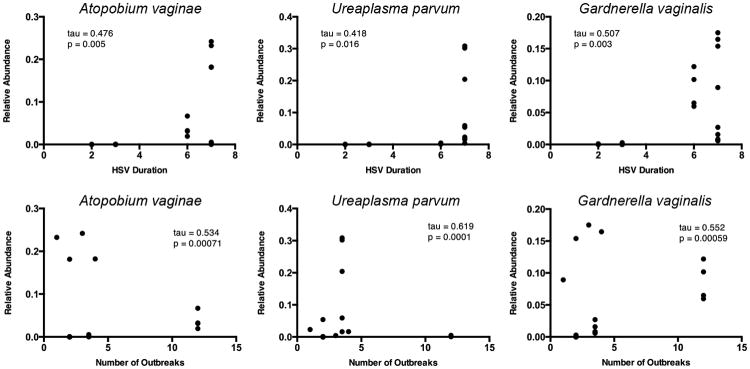
The relative abundance of taxa was significantly correlated with GHP duration and the number of related outbreaks. An increased abundance of the species-level taxa including Atopobium vaginae, Ureaplasma parvum, and Gardnerella vaginalis, was associated with a longer duration of GHP as well as an increase in the number of HSV outbreaks across the 6 subjects. Other taxa, specific only to the genus level, are reported in Table S3A and S3B.
There also was a positive correlation between the number of HSV outbreaks and the relative abundance of the above taxa: U. parvum (tau = 0.418, P = 0.016), G. vaginalis (tau = 0.507, P = 0.003), and A. vaginae (tau = 0.476, P = 0.005) (Fig. 3). Additional correlations with genus level taxa also were observed (Table S3B). While the small sample size precludes any definitive, generalizable conclusions concerning GHP status and the nature of associated organisms, the results are consistent with previous reports that identified a correlation between GHP women and an altered vaginal microflora (Cherpes et al., 2008; Kaul et al., 2007).
3.4. Microbial biofilms developing on the IVRs consist of communities reflective of those in the vaginal tract
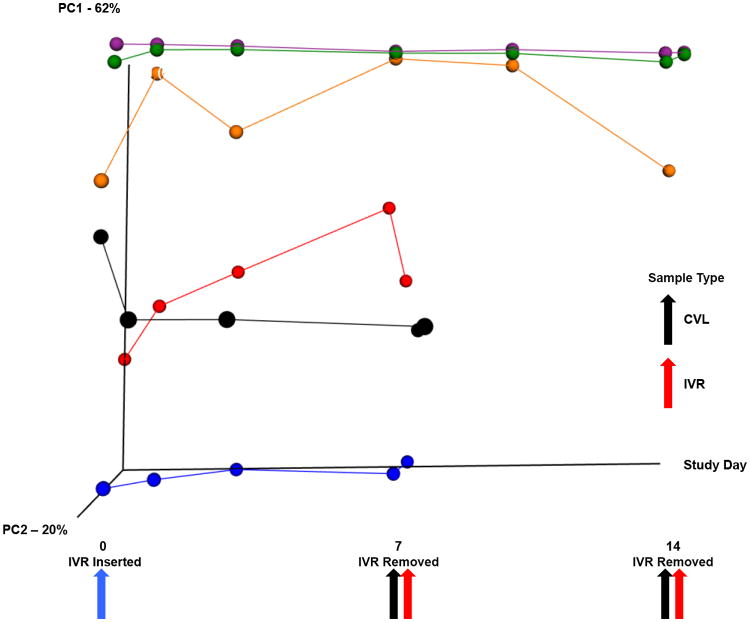
At Days 7 and 14, the IVR surface was covered with microbial biofilms at different stages of development (Keller et al., 2012). The structure of these sessile communities was found to be representative of the corresponding communities established on the hosts' vaginal epithelium. A weighted UniFrac β diversity plot showed an overlap of vaginal (CVL) and IVR samples, and taxonomic comparison revealed that no taxa were significantly different between within any one participant across all time points, but differences between participants were observed (Fig. 4). Comparing the microbial communities developing on the IVRs (Day 7 or Day 14) with the corresponding vaginal microbiota in samples on the day of IVR removal did not identify any taxa that were different across all participants, suggesting that the IVR biofilms were not systematically enriched with any members of the vaginal microbiome. These results also suggest that no significant, selective biases were introduced as a result of the different DNA extraction methods used for CVL and IVR samples (see 2.2.). There were no significant taxonomic differences in the microbial communities between the IVR biofilms that were removed on Day 7 compared with those removed on Day 14, nor was the α diversity of the IVR samples different from the vaginal (CVL) samples (Fig. S2).
Fig. 4.
Temporal dynamics of the vaginal microbial communities. The β diversity for vaginal (CVL) and IVR samples from the 6 subjects is plotted against time. Each colored trace, and correspondingly colored circles, represents one subject who had the IVR removed either at Day 7 or Day 14 (see Fig. 1). The blue arrow points to CVL samples taken at the time of IVR insertion. The black arrows represent the CVL samples taken at the time of IVR removal (red arrows). The subject represented by the orange line and spheres has an omitted CVL sample at Day 14 as it did not meet minimum 16S processing standards.
3.5. IVRs do not significantly alter the hosts' vaginal communities over time
Comparing the vaginal (CVL) and IVR samples on Days 7 and 14 assessed the effect of IVR administration length on community composition. Vaginal samples from participants who received the IVR for 14 days contained significantly fewer members from the phyla Proteobacteria and Firmicutes and from the genera Bacillus and Sphingomonas compared to participants who received the IVR for 7 days (relative abundance 3.58×10-05 versus 0, P = 9.91×10-07; relative abundance 3.58×10-05 versus 0, P = 4.96×10-07). The observed differences in vaginal microbiota were reflective of the large interpersonal differences, and not apparently driven by IVR usage.
Studies on the dynamics of the human vaginal microbiota have shown that some communities can shift over short time periods while others remain relatively stable (Brotman, 2011; Gajer et al., 2012). In order to determine the effect of the IVR's presence on the temporal dynamics of the vaginal communities, the β diversity in each participant's IVR and vaginal samples was plotted over time (Fig. 4). The CVL and IVR samples were connected by a solid line through PCoA space, with the first point representing the starting CVL microbial community prior to IVR administration, and the final point representing the IVR microbial community. Some of these microbial communities remained stable over the study period, denoted by the steady, level lines through PCoA space, while other exhibited more variability. If the introduction of the IVR caused a systematic disturbance in the vaginal microbiota, a large shift in the tracing lines in the same direction in PCoA space for all 6 patients would be expected. However, no large shift was apparent in the PCoA plots over time, likely because no taxa were significantly different in the vaginal samples collected prior to IVR administration compared with the corresponding samples collected when the IVR was removed. This finding further suggests that the presence of the IVR is not selecting for any specific taxa across all subjects, nor is the IVR pushing the vaginal communities of these GHP women to a new CST. It is possible that IVRs are causing a more subtle change in the vaginal communities over time that cannot be elucidated with our current sample size, and future confirmation for this finding likely will require larger cohorts.
4. Discussion
Culture-based evaluation of IVR effects on the vaginal microbiota has played an important role in early clinical assessment of candidate products for 40 years. In 1973, Henzl et al. evaluated early reservoir-IVRs delivering the hormonal contraceptive chlormadinone acetate in 12 women over 3 consecutive menstrual cycles (Henzl et al., 1973). The microbiologic examinations of the vaginal secretions consisted of direct smears stained with Gram's stain and a special stain for Trichomonas vaginalis. In addition, the secretions were evaluated by cultivation on broad-spectrum media for aerobic and anaerobic microbes and a selective media for mycoses, Trichomonas vaginalis, and Neiserria spp. In the early 1980s, Population Council reservoir-IVRs (Jackanicz, 1981) delivering the contraceptive levonorgestrel (LNG) in combination with estradiol (E2) were studied for contraceptive effectiveness and acceptability in multicentered trials involving 1,103 ring users (Sivin et al., 1981a). Increased vaginal discharge, potentially due to alteration of the vaginal microbiome, was identified as the most frequently voiced complaint by users (Sivin et al., 1981a, b). This possibility was disproven by Schwan and colleagues in a group of 17 subjects who used the Population Council LNG-E2 IVR for 6 months (Schwan et al., 1983). Culture-dependent methods were used to characterize the aerobic and anaerobic bacteria, yeast, and mycoplasma/ureaplasma in vaginal secretions collected from the cervix and posterior fornix before and after treatment with the IVR. No significant difference was observed between both groups and it was concluded that alterations in the vaginal bacterial ecology through IVR usage was not the cause of the increased discharge.
While culture-based methods have continued to be used in clinical evaluation of IVRs (Davies et al., 1992), the recent introduction of high-throughput, culture-independent molecular methods (Caporaso et al., 2012) has made it possible to collect hundreds of thousands of sequences spanning hundreds of samples. The so-called democratization of sequencing (Tringe and Hugenholtz, 2008) has continued to be fueled by ever decreasing sequencing costs, notably on several Illumina platforms (Caporaso et al., 2012). The Illumina sequencing platform returns on the order of 100 million sequencing reads per flowcell and therefore supports unprecedented sequencing depth (Caporaso et al., 2011) enabling the detection of very rare phylotypes (Binladen et al., 2007). Deep sequencing methods allow important parameters for describing microbial community composition to be determined, including the species richness within each sample (α diversity), and the diversity shared between multiple environments (β diversity) (Kuczynski et al., 2010; Lozupone et al., 2007). We have used bar-coded Illumina sequence datasets generated from 16S rRNA gene fragments to study the response of the vaginal microbiomes of GHP women to pod-IVRs medicated with ACV. This analysis allowed the vaginal communities developing in microbial biofilms on the IVR surface to be compared for the first time to the corresponding vaginal microbiomes. Our results suggest that rigorous, molecular analysis of the effects of intravaginal devices on the corresponding microbial communities shows promise for integration with traditional approaches in the clinical evaluation of candidate products.
5. Conclusion
Despite the small sample size (n = 6), this study supports the preliminary safety of the ACV pod-IVR as there were no detectable and systematic changes in the vaginal microbiota in response to the devices. Although microbial biofilms were readily detected on the IVR surface, the composition of these sessile communities was similar to that of the corresponding vaginal microbiome. These observations need to be tempered by the exploratory nature of the study. Future research will need to investigate the impact of the IVR on the vaginal microbiota over a complete menstrual cycle and how the device affects vaginal microbiomes representative of all major CSTs (Ravel et al., 2011). These important questions will be addressed in larger, follow-on clinical studies.
Supplementary Material
Highlight.
Intravaginal rings (IVRs) delivering the antiviral acyclovir were evaluated clinically.
The participants were women with recurrent genital HSV in a longitudinal study.
Culture-independent characterization of the microbiota was carried out.
Specific taxa were associated with the duration of recurrent genital HSV status.
This highly sensitive experimental approach promises to provide improved safety data for IVRs.
Acknowledgments
Funding support from Rose Hills Foundation for research fellowships (SC, MM) is gratefully acknowledged. Support for LKU was provided by the Signaling and Cellular Recognition Training Grant (T32 GM08759). The authors thank Auritec Pharmaceuticals, Inc. for making the clinical samples available to this study.
Footnotes
Appendix A. Supplementary data: Appendix S1.PDF. Supplementary data, including 2 tables and 3 figures.
Table S3.XLSX. Kendall's tau correlations between (A) duration of GHP status and OTU abundance and (C) number of HSV outbreaks and OTU abundance.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aagaard K, Petrosino P, Keitel W, Watson M, Katancik J, Garcia N, Patel S, Cutting M, Madden T, Hamilton H, Harris E, Gevers D, Simone G, McInnes P, Versalovic J. The Human Microbiome Project Strategy for Comprehensive Sampling of the Human Microbiome and Why It Matters. FASEB J. 2013;23:1012–1022. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum MM, Butkyavichene I, Gilman J, Kennedy S, Kopin E, Malone AM, Nguyen C, Smith TJ, Friend DR, Clark MR, Moss JA. An Intravaginal Ring for the Simultaneous Delivery of Multiple Drugs. J Pharm Sci. 2012;101:2833–2843. doi: 10.1002/jps.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binladen J, Gilbert MTP, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. The Use of Coded PCR Primers Enables High-throughput Sequencing of Multiple Homolog Amplification Products by 454 Parallel Sequencing. PLoS One. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering Vastly Improves Diversity Estimates from Illumina Amplicon Sequencing. Nat Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotman RM. Vaginal Microbiome and Sexually Transmitted Infections: an Epidemiologic Perspective. J Clin Invest. 2011;121:4610–4617. doi: 10.1172/JCI57172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME Allows Analysis of High-throughput Community Sequencing Data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc Natl Acad Sci U S A. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherpes TL, Hillier SL, Meyn LA, Busch JL, Krohn MA. A Delicate Balance: Risk Factors for Acquisition of Bacterial Vaginosis Include Sexual Activity, Absence of Hydrogen Peroxide-producing Lactobacilli, Black Race, and Positive Herpes Simplex Virus Type 2 Serology. Sex Transm Dis. 2008;35:78–83. doi: 10.1097/OLQ.0b013e318156a5d0. [DOI] [PubMed] [Google Scholar]
- Corey L, Benedetti JK, Critchlow CW, Remington MR, Winter CA, Fahnlander AL, Smith K, Salter DL, Keeney RE, Davis LG, Hintz MA, Connor JD, Holmes KK. Double-blind Controlled Trial of Topical Acyclovir in Genital Herpes Simplex Virus Infections. Am J Med. 1982a;73:326–334. doi: 10.1016/0002-9343(82)90117-6. [DOI] [PubMed] [Google Scholar]
- Corey L, Nahmias AJ, Guinan ME, Benedetti JK, Critchlow CW, Holmes KK. A Trial of Topical Acyclovir in Genital Herpes Simplex Virus Infections. N Engl J Med. 1982b;306:1313–1319. doi: 10.1056/NEJM198206033062201. [DOI] [PubMed] [Google Scholar]
- Corey L, Wald A, Patel R, Sacks SL, Tyring SK, Warren T, Douglas JM, Paavonen J, Morrow RA, Beutner KR, Stratchounsky LS, Mertz G, Keene ON, Watson HA, Tait D, Vargas-Cortes M. Once-daily Valacyclovir to Reduce the Risk of Transmission of Genital Herpes. N Engl J Med. 2004;350:11–20. doi: 10.1056/NEJMoa035144. [DOI] [PubMed] [Google Scholar]
- Davies GC, Feng LX, Newton JR, Dieben TOM, Coelinghbennink HJT. The Effects of A Combined Contraceptive Vaginal Ring Releasing Ethinylestradiol and 3-Ketodesogestrel on Vaginal Flora. Contraception. 1992;45:511–518. doi: 10.1016/0010-7824(92)90163-n. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Freeman EE, Orroth KK, White RG, Glynn JR, Bakker R, Boily MC, Habbema D, Buve A, Hayes RJ. Proportion of New HIV Infections Attributable to Herpes Simplex 2 Increases over Time: Simulations of the Changing Role of Sexually Transmitted Infections in sub-Saharan African HIV Epidemics. Sex Transm Infect. 2007;83:I17–I24. doi: 10.1136/sti.2006.023549. [DOI] [PubMed] [Google Scholar]
- Freeman EE, Weiss HA, Glynn JR, Cross PL, Whitworth JA, Hayes RJ. Herpes Simplex Virus 2 Infection Increases HIV Acquisition in Men and Women: Systematic Review and Meta-analysis of Longitudinal Studies. AIDS. 2006;20:73–83. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- Gajer P, Brotman RM, Bai GY, Sakamoto J, Schuette UME, Zhong X, Koenig SSK, Fu L, Ma ZSS, Zhou X, Abdo Z, Forney LJ, Ravel J. Temporal Dynamics of the Human Vaginal Microbiota. Sci Transl Med. 2012;4:132ra152. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardana M, Moss JA, Smith TJ, Kennedy S, Kopin E, Nguyen C, Malone AM, Rabe L, Schaudinn C, Webster P, Srinivasan P, Sweeney ED, Smith JM, Baum MM. Microbial Biofilms on the Surface of Intravaginal Rings Worn in Non-human Primates. J Med Microbiol. 2011;60:828–837. doi: 10.1099/jmm.0.028225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzl MR, Mishell DR, Velazque Jg, Leitch WE. Basic Studies for Prolonged Progestogen Administration by Vaginal Devices. Am J Obstet Gynecol. 1973;117:101–106. [Google Scholar]
- Hillier SL. The Complexity of Microbial Diversity in Bacterial Vaginosis. N Engl J Med. 2005;353:1886–1887. doi: 10.1056/NEJMp058191. [DOI] [PubMed] [Google Scholar]
- Holmes I, Harris K, Quince C. Dirichlet Multinomial Mixtures: Generative Models for Microbial Metagenomics. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackanicz TM. Levonorgestrel and Estradiol Release from an Improved Contraceptive Vaginal Ring. Contraception. 1981;24:323–339. doi: 10.1016/0010-7824(81)90002-0. [DOI] [PubMed] [Google Scholar]
- Kaul R, Nagelkerke NJ, Kimani J, Ngugi E, Bwayo JJ, MacDonald KS, Rebbaprgada A, Fonck K, Temmerman M, Ronald AR, Moses S. Prevalent Herpes Simplex Virus Type 2 Infection is Associated with Altered Vaginal Flora and an Increased Susceptibility to Multiple Sexually Transmitted Infections. J Infect Dis. 2007;196:1692–1697. doi: 10.1086/522006. [DOI] [PubMed] [Google Scholar]
- Keller MJ, Herold BC. Understanding Basic Mechanisms and Optimizing Assays to Evaluate the Efficacy of Vaginal Microbicides. Sex Transm Dis. 2009;36:S92–S95. doi: 10.1097/OLQ.0b013e318199417d. [DOI] [PubMed] [Google Scholar]
- Keller MJ, Malone AM, Carpenter CA, Lo Y, Huang M, Corey L, Willis R, Nguyen C, Kennedy S, Gunawardana M, Guerrero D, Moss JA, Baum MM, Smith TJ, Herold BC. Safety and Pharmacokinetics of Acyclovir in Women Following Release From a Silicone Elastomer Vaginal Ring. J Antimicrob Chemother. 2012;67:2005–2012. doi: 10.1093/jac/dks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J, Costello EK, Nemergut DR, Zaneveld J, Lauber CL, Knights D, Koren O, Fierer N, Kelley ST, Ley RE, Gordon JI, Knight R. Direct Sequencing of the Human Microbiome Readily Reveals Community Differences. Genome Biol. 2010;11:210. doi: 10.1186/gb-2010-11-5-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa PS, Brooks JP, Deych E, Boone EL, Edwards DJ, Wang Q, Sodergren E, Weinstock G, Shannon WD. Hypothesis Testing and Power Calculations for Taxonomic-Based Human Microbiome Data. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lladser ME, Gouet R, Reeder J. Extrapolation of Urn Models via Poissonization: Accurate Measurements of the Microbial Unknown. PLoS One. 2011;6:e21105. doi: 10.1371/journal.pone.0021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looker KJ, Gamett GP, Schmid GP. An Estimate of the Global Prevalence and Incidence of Herpes Simplex Virus Type 2 Infection. Bull World Health Organ. 2008;86:805–812. doi: 10.2471/BLT.07.046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and Qualitative Beta Diversity Measures Lead to Different Insights into Factors that Structure Microbial Communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Species Divergence and the Measurement of Microbial Diversity. FEMS Microbiol Rev. 2008;32:557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich JA, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An Improved Greengenes Taxonomy with Explicit Ranks for Ecological and Evolutionary Analyses of Bacteria and Archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss JA, Baum MM, Malone AM, Kennedy S, Kopin E, Nguyen C, Gilman J, Butkyavichene I, Willis R, Vincent KL, Motamedi M, Smith TJ. Tenofovir and Tenofovir Disoproxil Pharmacokinetics from Intravaginal Rings. AIDS. 2012a;26:707–710. doi: 10.1097/QAD.0b013e3283509abb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss JA, Malone AM, Smith TJ, Butkyavichene I, Cortez C, Gilman J, Kennedy S, Kopin E, Nguyen C, Sinha P, Hendry RM, Guenthner P, Holder A, Martin A, McNicholl J, Mitchell J, Pau CP, Srinivasan P, Smith JM, Baum MM. Safety and Pharmacokinetics of Intravaginal Rings Delivering Tenofovir in Pig-tailed Macaques. Antimicrob Agents Chemother. 2012b;56:5952–5960. doi: 10.1128/AAC.01198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss JA, Malone AM, Smith TJ, Kennedy S, Kopin E, Nguyen C, Gilman J, Butkyavichene I, Vincent KL, Motamedi M, Friend DR, Clark MR, Baum MM. Simultaneous Delivery of Tenofovir and Acyclovir via an Intravaginal Ring. Antimicrob Agents Chemother. 2012c;56:875–882. doi: 10.1128/AAC.05662-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R, Bodsworth NJ, Woolley P, Peters B, Vejlsgaard G, Saari S, Gibb A, Robinson J. Valaciclovir for the Suppression of Recurrent Genital HSV Infection: A Placebo Controlled Study of Once Daily Therapy. Genitourin Med. 1997;73:105–109. doi: 10.1136/sti.73.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M Grp, N.H.W. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. Vaginal Microbiome of Reproductive-age Women. Proc Natl Acad Sci U S A. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J, Gajer P, Fu L, Mauck CK, Koenig SSK, Sakamoto J, Motsinger-Reif AA, Doncel GF, Zeichner SL. Twice-daily Application of HIV Microbicides Alters the Vaginal Microbiota. mBio. 2012;3:e00370–00312. doi: 10.1128/mBio.00370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelderfer P. Toxicology and Safety Testing, The Science of Microbicides: Accelerating Development. The Rockefeller Foundation; New York: 2002. pp. 37–39. [Google Scholar]
- Schwan A, Ahren T, Victor A. Effects of Contraceptive Vaginal Ring Treatment on Vaginal Bacteriology and Cytology. Contraception. 1983;28:341–347. doi: 10.1016/0010-7824(83)90036-7. [DOI] [PubMed] [Google Scholar]
- Sivin I, Mishell DR, Victor A, Diaz S, Alvarezsanchez F, Nielsen NC, Akinla O, Pyorala T, Coutinho E, Faundes A, Roy S, Brenner PF, Ahren T, Pavez M, Brache V, Giwaosagie OF, Fasan MO, Zausnerguelman B, Darze E, Dasilva JCG, Diaz J, Jackanicz TM, Stern J, Nash HA. A Multi-center Study of Levonorgestrel-estradiol Contraceptive Vaginal Rings .1. Use Effectiveness - An International Comparative Trial. Contraception. 1981a;24:341–358. doi: 10.1016/0010-7824(81)90003-2. [DOI] [PubMed] [Google Scholar]
- Sivin I, Mishell DR, Victor A, Diaz S, Alvarezsanchez F, Nielsen NC, Akinla O, Pyorala T, Coutinho E, Faundes A, Roy S, Brenner PF, Ahren T, Pavez M, Brache V, Giwaosagie OF, Fasan MO, Zausnerguelman B, Darze E, Dasilva JCG, Diaz J, Jackanicz TM, Stern J, Nash HA. A Multi-Center Study of Levonorgestrel-Estradiol Contraceptive Vaginal Rings .2. Subjective and Objective Measures of Effects - An International Comparative Trial. Contraception. 1981b;24:359–376. doi: 10.1016/0010-7824(81)90004-4. [DOI] [PubMed] [Google Scholar]
- Stoner KA, Reighard SD, Miguel RDV, Landsittel D, Cosentino LA, Kant JA, Cherpes TL. Recalcitrance of Bacterial Vaginosis among Herpes Simplex Virus Type 2 Seropositive Women. J Obstet Gynaecol Res. 2012;38:77–83. doi: 10.1111/j.1447-0756.2011.01697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, Hugenholtz P. A Renaissance for the Pioneering 16S rRNA Gene. Curr Opin Microbiol. 2008;11:442–446. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The Human Microbiome Project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell LK, Clemente JC, Rideout JR, Gevers D, Caporaso JG, Knight R. The Interpersonal and Intrapersonal Diversity of Human-associated Microbiota in Key Body Sites. J Allergy Clin Immunol. 2012;129:1204–1208. doi: 10.1016/j.jaci.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rompay KKA. Evaluation of Antiretrovirals in Animal Models of HIV Infection. Antiviral Res. 2010;85:159–175. doi: 10.1016/j.antiviral.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Veazey RS. Microbicide Safety/Efficacy Studies in Animals - Macaques and Small Animal Models. Curr Opin HIV AIDS. 2008;3:567–573. doi: 10.1097/COH.0b013e32830891bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.