Abstract
Probiotic bacteria are microorganisms that benefit the host by preventing or ameliorating disease. However, little information is known regarding the scientific rationale for using probiotics as alternative medicine. The purpose of this paper is to investigate the mechanisms of probiotic beneficial effects on intestinal cell homeostasis. We now report that one such probiotic, Lactobacillus rhamnosus GG (LGG), prevents cytokine-induced apoptosis in two different intestinal epithelial cell models. Culture of LGG with either mouse or human colon cells activates the anti-apoptotic Akt/protein kinase B. This model probiotic also inhibits activation of the pro-apoptotic p38/mitogen-activated protein kinase by tumor necrosis factor, interleukin-1α, or γ-interferon. Furthermore, products recovered from LGG culture broth supernatant show concentration-dependent activation of Akt and inhibition of cytokine-induced apoptosis. These observations suggest a novel mechanism of communication between probiotic microorganisms and epithelia that increases survival of intestinal cells normally found in an environment of pro-apoptotic cytokines.
The human gastrointestinal microflora establishes at birth and acquires through a series of colonizations more than 400 bacterial species in the adult (1). Recent evidence clearly demonstrates that commensal bacteria regulate intestinal epithelial development and function (2, 3), and interruption of these interactions results in pathological conditions. Probiotics, as described by Lilly and Stillwell (4), are living microorganisms with low or no pathogenicity that exert beneficial effects on the health of the host. In human clinical trials the non-pathogenic Gram-positive commensal Lactobacilli found in the human and mouse gastrointestinal tracts have been considered as probiotics with beneficial health effects, including enhanced lymphocyte proliferation (5), innate and acquired immunity (6), and anti-inflammatory cytokine production (7). Lactobacillus rhamnosus GG (LGG),1 a bacterium used in the production of yogurt, is one of the best-studied probiotic bacteria in clinical trials. It is effective in preventing and treating diarrhea (8), recurrent Clostridium difficile (9), primary rotavirus infections (10), and atopic dermatitis (11). Therefore, manipulation of enteric microflora by selective living microorganisms termed probiotics are promoted by practitioners of alternative health measures.
There is increasing evidence that probiotic Lactobacillus species play a role in the treatment of inflammatory bowel disease (IBD). In this disease, abnormal immune responses cause inflammation that appears to be stimulated by elements of the enteric microenviroment, including normal and pathogenic organisms (12). Reduction of Lactobacillus and Bifidobacteria has been found in colonic biopsies of patients with IBD (13, 14). Although conventional therapeutic strategies focus on suppression or modulation of host immunity, modification of enteric microflora by probiotics may have a role in treating IBD. Several Lactobacillus strains prevent or attenuate intestinal inflammation in the interleukin (IL)-10−/− mouse model, which spontaneously develops enterocolitis (15, 16). LGG increases anti-inflammatory IL-10 levels in children (7). Furthermore, LGG has been suggested as adjunctive therapy for Crohn’s disease (17).
Putative mechanisms of the action of probiotics include regulation of cytokine production (18), such as stimulation of mononuclear cells to generate both pro- and anti-inflammatory cytokines (19, 20), enhancement of secretory IgA secretion (21), production of antibacterial substances (22, 23), an increase in the integrity of the intestinal barrier (24), and competition with pathogenic microorganisms for enterocyte binding (25). Yet almost nothing is known regarding the basic molecular mechanisms of probiotic regulation of intestinal epithelial health.
Two common features of IBD involve increased cytokine production (26, 27) and increased apoptosis of intestinal cells (28). We have reported that a central cytokine in the pathogenesis of IBD, tumor necrosis factor (TNF) (27), regulates both anti- and pro-apoptotic signaling pathways, and the balance between these two kinds of signals determines the cell fate (29). Therefore, the purpose of the present study is to investigate the effects of LGG on cytokine-regulated signaling pathways determining intestinal cell fate.
We now report that LGG prevents cytokine-induced apoptosis in mouse or human intestinal epithelial cells. Culture of LGG with colon cells activates the anti-apoptotic Akt/protein kinase B. Further, this model probiotic inhibits activation of the pro-apoptotic p38/mitogen-activated protein (MAP) kinase by TNF, IL-1α, or γ-interferon (IFN). Therefore, LGG promotes intestinal epithelial cell survival through regulation of both anti- and pro-apoptotic signal transduction pathways. These novel observations provide insight into the rationale for LGG as a potential treatment for IBD.
EXPERIMENTAL PROCEDURES
Bacterial Culture and Supernatant Preparation
LGG (American Type Culture Collection 53103) was incubated in Lactobacillus MRS broth at 37 °C for 24 h, then diluted in MRS broth, and incubated at 37 °C to reach log phase with the density determined as 0.5 at A600 (20). LGG was precipitated from MRS broth (1000 × g, 15 min) and washed twice with phosphate-buffered saline. Heat-killed (hk) LGG was prepared by boiling LGG in phosphate-buffered saline for 1 h and then washing it with phosphate-buffered saline. Lactobacillus acidophilus (American Type Culture Collection 393) and Lactobacillus casei (American Type Culture Collection 4356) were cultured per ATCC guidelines. 107 colony-forming units/ml LGG, L. casei, or L. acidophilus in RPMI cell culture medium were used to treat cells. Salmonella pullorum (provided by Andrew Neish, Emory University, Atlanta, GA) was prepared and incubated with cells as described (30).
Supernatant recovered from LGG culture broth (LGG-s) was generated by centrifuging (1000 × g, 15 min) and filtering (0.2 μm) LGG culture in MRS broth; then the filtrate was concentrated using Centricon Plus-20 (5–100 kDa, Millipore, Bedford, MA) by centrifugation (4000 × g, 1 h) following guidelines from the manufacturer. Protein concentrations were determined by DC protein assay (Bio-Rad) using MRS broth as the control. LGG-conditioned cell culture medium (LGG-cm) was generated by incubating washed LGG (107 colony-forming units/ml) in RPMI cell culture medium at 37 °C for 2 h; the medium was centrifuged twice (1000 × g, 15 min) and then filtered (0.2 μm).
Cell Culture
Young adult mouse colon (YAMC) cells were grown in RPMI with 5% fetal bovine serum on collagen-coated culture dishes as described previously (31). Clonal cell lines stably expressing pFLAG-cDNA3-kinase-inactive (ki) kinase suppressor of Ras (KSR) or pFLAG-cDNA3-wild-type KSR (provided by Richard Kolesnick, Memorial Sloan-Kettering Cancer Center, New York, NY) were generated as described (32). The kiKSR plasmid expresses a kinase-inactive dominant-negative KSR as previously described (32, 33). Clonal cells were cultured in the presence of G418 (100 μg/ml) until 24 h prior to the experiments. The human colonic epithelial carcinoma cell line, HT 29 cells, was grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. All cells were serum-starved (0.5%) at 37 °C for 24 h before experiments.
Preparation of Cellular Lysates
Lysates were prepared from cells treated with murine TNF, IL-1α, or γ-IFN (Pepro Tech, Inc., Rocky Hill, NJ) for the indicated times in the presence or absence of LGG, LGG-s, LGG-cm, hk-LGG, phosphoinositide (PI) 3-kinase inhibitors, wortmannin (Sigma) or LY294002 or p38 inhibitors (Biomol, Plymouth Meeting, PA) SB203580 or SB202190. Cell monolayers were washed twice with ice-cold phosphate-buffered saline and then scraped into cell lysis buffer (20 mm HEPES (pH 7.5), 1 mm orthovandate, 50 mm β-glycerophosphate, 10 mm sodium pyrophosphate with leupeptin (10 μg/ml), aprotinin (10 μg/ml), phenylmethylsulfonyl fluoride (18 μg/ml), and 1% Triton X-100). The scraped suspensions were centrifuged (14,000 × g, 10 min) at 4 °C, and protein content was determined using DC protein assay (Bio-Rad). Equal amounts of cellular lysate protein were mixed with Laemmli sample buffer and separated by SDS-PAGE for Western blot analysis with anti-phospho-Akt, anti-Akt, anti-phospho-p38, anti-p38, anti-IκBα, anti-phospho stress-activated protein kinase/c-Jun amino-terminal kinase (SAPK/JNK), anti-SAPK/JNK (Cell Signaling Technology, Beverly, MA), or anti-phospho extracellular signal-regulated kinase (ERK)1/ERK2/MAP kinase (Promega, Madison, WI) antibodies.
Apoptosis Assay
YAMC and clonal cell lines were cultured on collagen-coated or HT29 cells on non-coated chamber glass slides and were prepared as described above. Following treatment, apoptotic cells were labeled by ApopTag in situ apoptosis detection kits (Intergen, Purchase, NY) using terminal deoxynucleotidyltransferase for detection of positive cells following guidelines from the manufacturer. Apoptotic cells were labeled by fluorescein isothiocyanate-conjugated anti-digoxigenin or anti-digoxigenin peroxidase conjugate and 3,3′-diaminobenzidine as substrate and were then dehydrated and mounted using Vectashield mounting medium. Slides were counterstained with 4,6-diamidino-2-phenylindole (DAPI) by using 1 μg/ml DAPI in mounting medium. The cells were observed by fluorescence microscopy or differential interference contrast. Apoptotic terminal deoxynucleotidyltransferase-mediated dUTP nick-end-labeling (TUNEL) positive cells were determined by counting at least 200 cells in randomly chosen fields and expressing them as a percentage of the total number of cells counted.
Caspase activity was detected in living cells using the sulforhodamine multi-caspase activity kit (Biomol) following guidelines from the manufacturer. Active caspase enzyme in living cells was labeled with cell-permeable sulforhodamine-conjugated valylalanylaspartic acid fluoromethyl ketone, an inhibitor of caspase activity that binds to active caspase enzyme. Cells with increased caspase activity were detected by fluorescence microscopy.
RESULTS
LGG Prevents Cytokine-induced Apoptosis in Intestinal Epithelial Cells
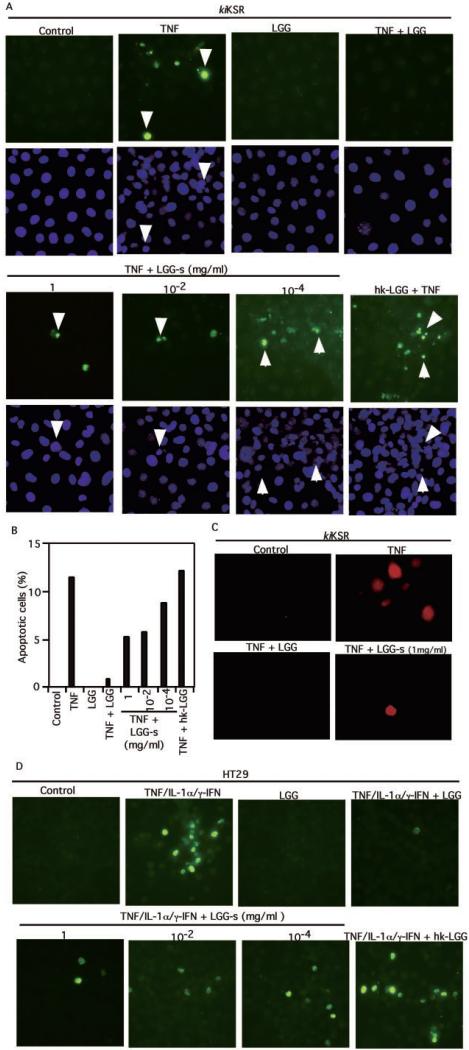
Pathological concentrations of TNF inhibit intestinal cell proliferation without inducing apoptosis in YAMC cells (29, 31). Because dominant-negative kiKSR expression induces apoptosis in TNF-treated intestinal cells (29), we used kiKSR-expressing colon cells to test our hypothesis that probiotics prevent TNF-induced apoptosis through regulation of signal transduction pathways. We found that TNF-stimulated apoptosis detected by TUNEL staining in kiKSR-expressing YAMC cells was inhibited by co-culture with viable LGG but was not hkLGG (Fig. 1, A and B). Furthermore, in a model of human intestinal cell apoptosis the effect of a “cytokine mixture” combination of TNF, IL-1α, and γ-IFN is reversed by LGG (Fig. 1D). DAPI staining shows nuclear condensation of the TUNEL-positive intestinal cells undergoing apoptosis (Fig. 1A). To further demonstrate that the observed LGG effect is caused by the inhibition of apoptosis, we used a multi-caspase activity assay in living cells. The increased caspase activity of cells undergoing apoptosis is blocked by viable LGG (Fig. 1C). Interestingly, cell survival is also enhanced by LGG-s, and this inhibitory effect on apoptosis is concentration-dependent (Fig. 1, A–D).
Fig. 1. LGG inhibits cytokine-induced apoptosis in intestinal epithelial cells.
kiKSR-expressing YAMC cells (A–C) or human colonic epithelial carcinoma cell line (HT29) cells (D) were treated with TNF (100 ng/ml) or the “cytokine mixture” combination of TNF (100 ng/ml), IL-1α (10 ng/ml), and γ-IFN (100 ng/ml), respectively, for 6 h in the presence or absence of a 1-h pretreatment with viable LGG, hk-LGG, or concentrated supernatant recovered from LGG culture MRS broth (LGG-s) at the concentrations indicated. Then, cells were fixed for terminal deoxynucleotidyltransferase TUNEL with apoptotic nuclei labeled with fluorescein isothiocyanate and DAPI staining (A and D). Fluorescein isothiocyanate- and DAPI-labeled images were taken from the same field. The percentage of cells undergoing apoptosis from a representative experiment is shown in B. Caspase activity in living cells was detected using a multi-caspase activity assay kit (C). Arrows indicate representative apoptotic nuclei. All experiments in this and subsequent figures were performed on at least three separate occasions.
LGG Regulates Signaling Pathways That Determine Cell Fate
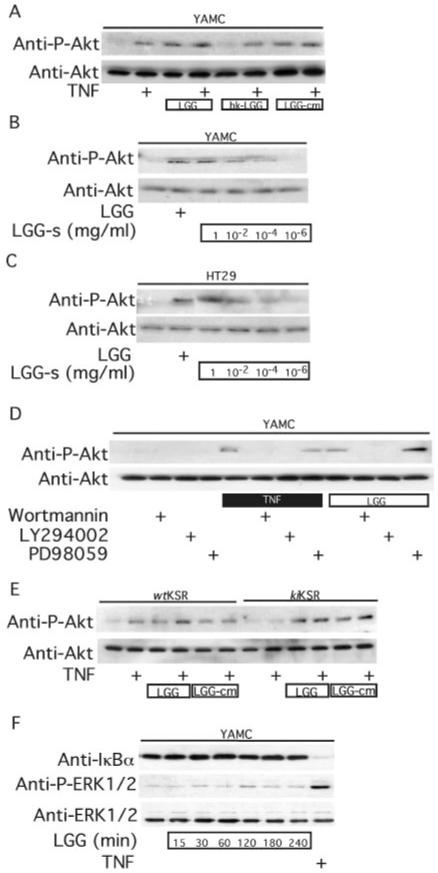
The balance between anti-apoptotic and pro-apoptotic signal transduction pathways regulates the fate of cells exposed to various stimuli including TNF. Therefore, we studied the effects of LGG on signal transduction pathways that regulate cell fate. LGG or LGG-cm, but not heat-killed LGG, stimulates anti-apoptotic Akt activation determined by antibody against Ser(P)-473-Akt (Fig. 2A). Akt activation is also enhanced by LGG-s in a concentration-dependent manner (Fig. 2, B and C). This activation by either LGG or TNF is blocked by the PI 3-kinase inhibitors, LY294002 or wortmannin, but not by the MEK1 inhibitor, PD98059 (Fig. 2D). Whereas kiKSR expression inhibits TNF activation of Akt, both LGG and LGG-cm stimulate Akt phosphorylation in these cells (Fig. 2E), indicating that TNF and LGG utilize different mechanisms to activate PI 3-kinase upstream of Akt. Nuclear factor (NF) κB and ERK/MAP kinase activation are important TNF-activated anti-apoptotic signal transduction pathways in intestinal cells (29). TNF stimulates both degradation of the inhibitor of κB and activation of ERK1/ERK2. However, LGG does not effect activation of either of these anti-apoptotic molecules (Fig. 2F), which indicates that LGG inhibition of TNF-induced apoptosis is not caused by the disruption of TNF receptor binding activities.
Fig. 2. LGG or factors recovered in LGG-cm or LGG-s stimulate Akt activation.
YAMC (A, B, D, F), HT29 (C), and kiKSR-expressing cells (E) were treated with viable LGG, hk-LGG, LGG-cm, or LGG-s as indicated for 1 h followed by TNF (100 ng/ml, 15 min) treatment. PI 3-kinase inhibitors, wortmannin (100 nm) or LY294002 (10 μm), or MEK1 inhibitor PD98059 (10 μm) was used to pre-treat cells for 1 h before TNF or LGG treatment (D). Akt activation was detected by Western blot analysis of cellular lysates with anti-Ser(P)-473-Akt and anti-Akt antibodies. YAMC cells were treated with LGG for the indicated times, and IκBα degradation and ERK1/2 activation were detected by Western blot analysis of cellular lysates with anti-IκBα or anti-phospho (P) ERK1/2 antibodies (F).
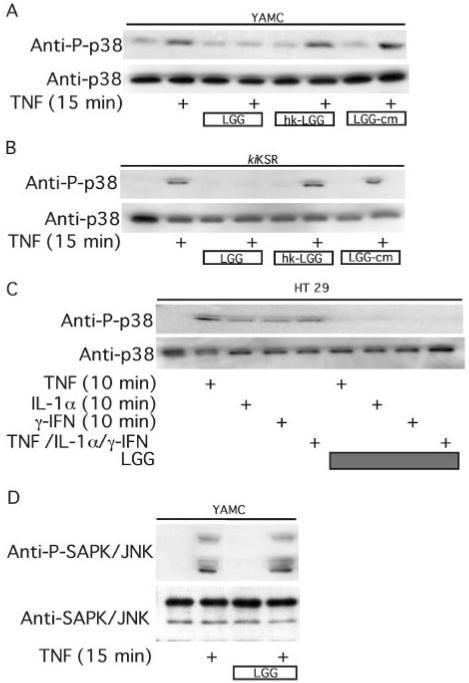
By using an antibody to phospho-p38 we determined that viable LGG inhibits TNF-induced activation of this pro-apoptotic MAP kinase (Fig. 3, A–C). Unlike Akt activation, inhibition of p38 is not reproduced by factors present in LGG-cm (Fig. 3, A and B). The cytokine-activated pro-apoptotic SAPK/JNK activities are not affected by LGG (Fig. 3D). Collectively these data indicate that LGG promotes survival of intestinal epithelial cells exposed to inflammatory cytokines through activation of specific anti-apoptotic and inhibition of pro-apoptotic signals.
Fig. 3. LGG blocks cytokine-induced p38 phosphorylation.
YAMC (A, D), kiKSR (B), or HT29 (C) cells were treated with TNF (100 ng/ml), IL-1α (10 ng/ml), or γ-IFN (100 ng/ml) as indicated in the presence or absence of a 1-h pretreatment with viable LGG, hk-LGG, or LGG-cm. Cellular lysates were prepared for Western blot analysis with anti-Thr(P)-p38, anti-p38, anti-phospho-SAPK/JNK, and anti-SAPK/JNK antibodies.
Probiotic Bacterium Regulation of Cell Survival Is Strain-specific
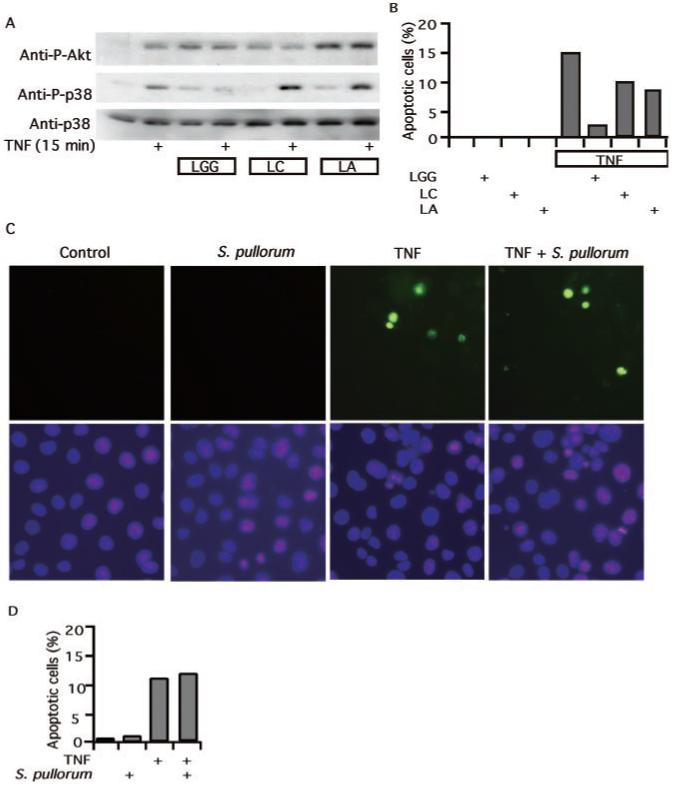
These findings raise an important question. Is this anti-apoptotic response unique to LGG, or is it common to other probiotic commensal or non-pathogenic intestinal microflora? To address this question we studied the effects of two other Gram-positive strains of Lactobacillus, L. acidophilus and L. casei, known to enhance lymphocyte proliferation and immunity (5, 6) or macrophage activation (34), respectively. Whereas these two bacteria stimulate Akt activation, they show no inhibition of TNF-stimulated p38 (Fig. 4A). Accordingly, their inhibitory effect on TNF-induced apoptosis is significantly reduced compared with LGG (Fig. 4B). Furthermore, we studied two Gram-negative bacteria derived from colon flora isolates, Salmonella typhimurium PhoPc and S. pullorum, that have been shown to regulate TNF-stimulated signal transduction pathways in intestinal cells (30). Neither of these attenuate TNF-induced apoptosis in colon cells (Fig. 4, C and D). Therefore, reversal of cytokine-induced apoptosis by LGG does not extend to all commensal or non-pathogenic intestinal microflora. This evidence suggests that a need for caution in selecting “probiotics” for clinical study is warranted and may explain variability in results of clinical trials investigating different probiotics.
Fig. 4. Regulation of signal transduction and apoptosis in intestinal cells by commensal or non-pathogenic bacteria.
YAMC cells were treated with LGG, L. casei (LC), or L. acidophilus (LA) for 1 h followed by 15 min of TNF (100 ng/ml) treatment as indicated in A. Akt and p38 activation was detected as before. LC, LA (B), or S. pullorum (C and D), which were prepared as indicated under “Experimental Procedures,” were incubated with kiKSR-expressing cells for 1 h prior to the addition of TNF (100 ng/ml) and then treated for 6 h. The cells were prepared for TUNEL staining as before (C). The percentage of cells undergoing apoptosis in a representative experiment is shown (B and D).
Akt and p38 MAP Kinase Determine the Fate of Intestinal Epithelial Cells Exposed to TNF
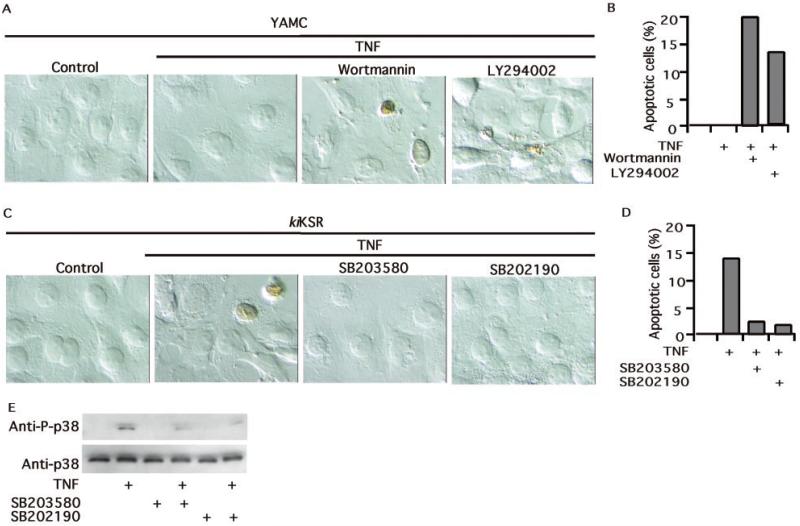
To determine the relative importance of Akt activation and inhibition of p38 activity in regulating the fate of intestinal cells exposed to TNF, we used pharmacological inhibitors. A blockade of Akt activation by PI 3-kinase inhibitors (as shown in Fig. 2D) increases apoptosis in YAMC cells treated with TNF (Fig. 5, A and B), whereas inhibitors of p38 block apoptosis in TNF-treated kiKSR-expressing cells (Fig. 5, C and D). As expected, both p38 inhibitors reduce p38 activation at concentrations that were studied (Fig. 5E). These results are consistent with the hypothesis that activation of Akt and inhibition of p38 by LGG are central to its anti-apoptotic effect.
Fig. 5. Akt functions as an inhibitor of apoptosis, whereas p38 promotes apoptosis in cells exposed to TNF.
YAMC cells (A and B) or kiKSR-expressing YAMC cells (C–E) were treated with TNF for 6 h in the absence or presence of 1-h pretreatment with wortmannin (100 nm), LY294002 (10 μm), SB203580 (10 μm), or SB202190 (10 μm) as indicated. TUNEL staining was used to identify apoptotic nuclei by peroxidase. The cells were visualized by differential interference contrast microscopy (A and C). The percentage of cells undergoing apoptosis from representative experiments is shown (B and D). p38 phosphorylation was detected as described in Fig. 3.
DISCUSSION
The lumen of the gastrointestinal tract has evolved with a complex ecosystem that includes commensal and pathogenic bacteria with the capability of cross-species communication (3, 35). Lilly and Stillwell (4) described microbes that had potential benefit to their hosts as probiotics. We have studied the relationships between Lactobacillus GG, as a model probiotic, and the intestinal epithelial cell, the point of first contact with the host. Our novel findings demonstrate that LGG promotes the survival of intestinal epithelial cells through the activation of the anti-apoptotic Akt/protein kinase B and inhibition of the pro-apoptotic p38 MAP kinase. LGG produces a factor that activates Akt independent of bacterial cellular interaction.
It is possible that other commensal organisms with potential probiotic activity (6, 30, 36, 37) could prevent cytokine-induced apoptosis. Although some of those we tested reduced apoptosis, none were as effective as LGG, and LGG appeared unique in its ability to block p38 MAP kinase activation. These findings extend our understanding of the complexity of the host commensal relationship and suggest that intestinal cell survival in its cytokine-rich environment may be regulated by the presence of beneficial microbes. This effect appears to require live bacteria, because either heat treatment (Figs. 2 and 3) or glutaraldehyde fixation (data not shown) prevents LGG regulation of Akt or p38 MAP kinase.
The mechanisms by which intestinal bacteria modulate the inflammatory process are complex and incompletely understood. Probiotic bacteria interact with three components of the gastrointestinal tract including intestinal epithelial cells, luminal flora, and the mucosal immune cells. It is known that probiotic bacteria can regulate lymphocyte cytokine production (18-20). Nonpathogenic Salmonella species can regulate the immune response by blocking NFκB activation and IL-8 production (30). However, these Salmonella species do not alter apoptotic response to cytokine (Fig. 4, B and C), nor does LGG inhibit NFκB activation (data not shown). We did not detect NFκB activation by either inhibitor of κB degradation (Fig. 2F) or NFκB p65 subunit nuclear translocation up to 6 h after LGG treatment (data not shown). Although we did not specifically study IL-8 secretion in our cells, it is increased in CaCo2 cells treated with Lactobacillus (38). A commensal Bifidobacterium strain regulates intestinal cell gene expression (3) and promotes its own survival within the host (2). Clearly, the mechanisms of activating intestinal cell signal transduction pathways by commensal bacteria are relevant to the maintenance of this important physical and functional mucosal barrier.
Our current understanding of cell survival suggests that the balance between pro-apoptotic and anti-apoptotic signals generated by cytokines regulates apoptosis (39). Anti-apoptotic signaling pathways initiated by TNF, other cytokines, and growth factors include NFκB (40), extracellular ERK1/ERK2/MAP kinase (41), and PI 3-kinase/Akt (42). By contrast, other members of the MAP kinase family, including SAPK/JNK and p38 function as pro-apoptotic signals for a number of cell types (41). KSR is an essential kinase in TNF signal transduction, determining intestinal epithelial cell fate by regulating the anti-apoptotic NFκB, ERK/MAP kinase, and Akt pathways (29). However, neither TNF activation of p38 MAP kinase nor SAPK/JNK is inhibited by disruption of KSR signaling. Despite blocking all the anti-apoptotic signals mediated through KSR, LGG still promotes intestinal cell survival (Fig. 1). This effect appears to require blocking p38 MAP kinase activation (Fig. 5B), because Akt activation by the LGG supernatant is insufficient to fully reverse apoptosis (Fig. 1B).
Surprisingly, factors recovered from bacterial culture or conditioned cell culture media reduce apoptosis and activate Akt. Production of these factors does not require bacterial-intestinal cell contact. Previously studies have reported that some Lactobacillus strains produce proteinaceous (23) or protease-insensitive (22) anti-bacterial compounds. However, we are unaware of reports indicating that bacterially produced factors regulate either Akt signal transduction or cell survival pathways. Thus far we have identified two proteins in LGG culture broth with molecular sizes of ~80 and 42 kDa based on SDS-PAGE. These two factors are heat- and protease-sensitive, and their activities are recoverable by electroelution from polyacrylamide gels (data not shown). Clearly, recovery and further characterization of these factors is a high priority.
Our evaluation of LGG as a model probiotic organism reveals an important and novel relationship between intestinal epithelial cell survival and selective microflora. Clearly, this single epithelial layer lining the intestine exists in a relatively hostile environment of pathogenic organisms, toxic waste by-products, and cytokines. This report adds to our understanding of the signal transduction pathways in the intestinal epithelial cell that are regulated by probiotics and commensal organisms to include intestinal cell survival in addition to intestinal homeostasis (3, 30). Therefore, further studies of the molecular basis for probiotic regulation of intestinal epithelium are relevant to understanding normal development as well as the pathogenesis of inflammatory bowel disease.
Acknowledgments
We thank Andrew Neish (Emory University) for S. pullorum and S. typhimurium PhoPc; Valio Ltd. (Helsinki, Finland) for LGG; Sam Wells, Han Liang, Wei Tong, Guinn Wilson, and Mark Frey for excellent technical assistance; the Vanderbilt University Medical Center Imaging Core Research Laboratory (CA68485); and Graham Carpenter, Steve Hanks, and Raymond DuBois for helpful discussions and critical review of the manuscript.
Footnotes
This work was supported by a Children’s Digestive Health and Nutrition Foundation/Nestle Nutrition Grant (to F. Y.), National Institutes of Health Grants DK10105 (to F. Y.) and DK56008 (to D. B. P.), and the Vanderbilt University Digestive Disease Research Center (DK58404).
The abbreviations used are: LGG, Lactobacillus rhamnosus GG; ERK, extracellular signal-regulated kinase; IBD, inflammatory bowel disease; IFN, interferon; IL, interleukin; ki, kinase-inactive; KSR, kinase suppressor of Ras; LGG-cm, LGG conditioned medium; LGG-s, supernatant recovered from LGG culture broth; MAP, mitogen activated protein; NFκB, nuclear factor κB; PI, phosphoinositide; SAPK/JNK, stress-activated protein kinase/c-Jun amino-terminal kinase; TNF, tumor necrosis factor; YAMC, young adult mouse colon; hk, heat killed; DAPI, 4,6-diamidino-2-phenylindole; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling.
REFERENCES
- 1.Berg RD. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 2.Bry L, Falk PG, Midtvedt T, Gordon JI. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 3.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 4.Lilly DM, Stillwell RH. Science. 1965;147:747–748. doi: 10.1126/science.147.3659.747. [DOI] [PubMed] [Google Scholar]
- 5.Kirjavainen PV, El-Nezami HS, Salminen SJ, Ahokas JT, Wright PF. FEMS Immunol. Med. Microbiol. 1999;26:131–135. doi: 10.1111/j.1574-695X.1999.tb01380.x. [DOI] [PubMed] [Google Scholar]
- 6.Gill HS, Rutherfurd KJ, Prasad J, Gopal PK. Br. J. Nutr. 2000;83:167–176. doi: 10.1017/s0007114500000210. [DOI] [PubMed] [Google Scholar]
- 7.Pessi T, Sutas Y, Hurme M, Isolauri E. Clin. Exp. Allergy. 2000;30:1804–1808. doi: 10.1046/j.1365-2222.2000.00948.x. [DOI] [PubMed] [Google Scholar]
- 8.Marteau PR, de Vrese M, Cellier CJ, Schrezenmeir J. Am. J. Clin. Nutr. 2001;73:430S–436S. doi: 10.1093/ajcn/73.2.430s. [DOI] [PubMed] [Google Scholar]
- 9.Gorbach SL, Chang TW, Goldin B. Lancet. 1987;2:1519. doi: 10.1016/s0140-6736(87)92646-8. [DOI] [PubMed] [Google Scholar]
- 10.Szajewska H, Kotowska M, Mrukowicz JZ, Armanska M, Mikolajczyk W. J. Pediatr. 2001;138:361–365. doi: 10.1067/mpd.2001.111321. [DOI] [PubMed] [Google Scholar]
- 11.Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Lancet. 2001;357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 12.Shanahan F. Science. 2000;289:1311–1312. doi: 10.1126/science.289.5483.1311. [DOI] [PubMed] [Google Scholar]
- 13.Fabia R, Ar’Rajab A, Johansson ML, Andersson R, Willen R, Jeppsson B, Molin G, Bengmark S. Digestion. 1993;54:248–255. doi: 10.1159/000201045. [DOI] [PubMed] [Google Scholar]
- 14.Favier C, Neut C, Mizon C, Cortot A, Colombel JF, Mizon J. Dig. Dis. Sci. 1997;42:817–822. doi: 10.1023/a:1018876400528. [DOI] [PubMed] [Google Scholar]
- 15.Madsen KL, Doyle JS, Jewell LD, Tavernini MM, Fedorak RN. Gastroenterology. 1999;116:1107–1114. doi: 10.1016/s0016-5085(99)70013-2. [DOI] [PubMed] [Google Scholar]
- 16.Schultz M, Veltkamp C, Dieleman LA, Grenther WB, Wyrick PB, Tonkonogy SL, Sartor RB. Inflamm. Bowel Dis. 2002;8:71–80. doi: 10.1097/00054725-200203000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Malin M, Suomalainen H, Saxelin M, Isolauri E. Ann. Nutr. Metab. 1996;40:137–145. doi: 10.1159/000177907. [DOI] [PubMed] [Google Scholar]
- 18.Maassen CB, van Holten-Neelen C, Balk F, den Bak-Glashouwer MJ, Leer RJ, Laman JD, Boersma WJ, Claassen E. Vaccine. 2000;18:2613–2623. doi: 10.1016/s0264-410x(99)00378-3. [DOI] [PubMed] [Google Scholar]
- 19.Miettinen M, Matikainen S, Vuopio-Varkila J, Pirhonen J, Varkila K, Kurimoto M, Julkunen I. Infect. Immun. 1998;66:6058–6062. doi: 10.1128/iai.66.12.6058-6062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miettinen M, Vuopio-Varkila J, Varkila K. Infect. Immun. 1996;64:5403–5405. doi: 10.1128/iai.64.12.5403-5405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigues AC, Cara DC, Fretez SH, Cunha FQ, Vieira EC, Nicoli JR, Vieira LQ. J. Appl. Microbiol. 2000;89:404–414. doi: 10.1046/j.1365-2672.2000.01128.x. [DOI] [PubMed] [Google Scholar]
- 22.Zamfir M, Callewaert R, Cornea PC, Savu L, Vatafu I, De Vuyst L. J. Appl. Microbiol. 1999;87:923–931. doi: 10.1046/j.1365-2672.1999.00950.x. [DOI] [PubMed] [Google Scholar]
- 23.Bernet-Camard MF, Lievin V, Brassart D, Neeser JR, Servin AL, Hudault S. Appl. Environ. Microbiol. 1997;63:2747–2753. doi: 10.1128/aem.63.7.2747-2753.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gotteland M, Cruchet S, Verbeke S. Aliment Pharmacol. Ther. 2001;15:11–17. doi: 10.1046/j.1365-2036.2001.00898.x. [DOI] [PubMed] [Google Scholar]
- 25.Bernet MF, Brassart D, Neeser JR, Servin AL. Gut. 1994;35:483–489. doi: 10.1136/gut.35.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breese EJ, Michie CA, Nicholls SW, Murch SH, Williams CB, Domizio P, Walker-Smith JA, MacDonald TT. Gastroenterology. 1994;106:1455–1466. doi: 10.1016/0016-5085(94)90398-0. [DOI] [PubMed] [Google Scholar]
- 27.Sartor RB. Gastroenterol. Clin. N. Am. 1995;24:475–507. [PubMed] [Google Scholar]
- 28.Iwamoto M, Koji T, Makiyama K, Kobayashi N, Nakane PK. J. Pathol. 1996;180:152–159. doi: 10.1002/(SICI)1096-9896(199610)180:2<152::AID-PATH649>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 29.Yan F, John SK, Polk DB. Cancer Res. 2001;61:8668–8675. [PubMed] [Google Scholar]
- 30.Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, Karmali V, Rao AS, Madara JL. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 31.Kaiser GC, Polk DB. Gastroenterolog. 1997;112:1231–1240. doi: 10.1016/s0016-5085(97)70135-5. [DOI] [PubMed] [Google Scholar]
- 32.Yan F, Polk DB. Cancer Res. 2001;61:963–969. [PubMed] [Google Scholar]
- 33.Zhang Y, Yao B, Delikat S, Bayuomy S, Lin X, Basu S, McGinley M, Chan-Hui P, Lichenstein H, Kolesnick R. Cell. 1997;89:63–72. doi: 10.1016/s0092-8674(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 34.Perdigon G, de Macias ME, Alvarez S, Oliver G, de Ruiz Holgado AA. Infect. Immun. 1986;53:404–410. doi: 10.1128/iai.53.2.404-410.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tieng V, Le Bouguenec C, du Merle L, Bertheau P, Desreumaux P, Janin A, Charron D, Toubert A. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2977–2982. doi: 10.1073/pnas.032668099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirjavainen PV, ElNezami HS, Salminen SJ, Ahokas JT, Wright PFA. Clin. Diagn. Lab Immunol. 1999;6:799–802. doi: 10.1128/cdli.6.6.799-802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peranen J, Auvinen P, Vitra H, Wepf R, Simons K. J. Cell Biol. 1996;135:153–167. doi: 10.1083/jcb.135.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haller D, Bode C, Hammes WP, Pfeifer AM, Schiffrin EJ, Blum S. Gut. 2000;47:79–87. doi: 10.1136/gut.47.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Budd RC. J. Clin. Invest. 2002;109:437–441. doi: 10.1172/JCI15077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 41.Xia Z, Dickens M, Rainegeaud J, Davis RJ, Greenberg ME. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 42.Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]