Abstract
The intestinal immune system defends against pathogens and entry of excessive intestinal microbes; simultaneously, a state of immune tolerance to resident intestinal microbes must be maintained. Perturbation of this balance is associated with intestinal inflammation in various mouse models and is thought to predispose humans to inflammatory bowel disease (IBD). The innate immune system senses microbes; dendritic cells, macrophages, and epithelial cells produce an initial, rapid response. The immune system continuously monitors resident microbiota and utilizes constitutive antimicrobial mechanisms to maintain immune homeostasis. associations between IBD and genes that regulate microbial recognition and innate immune pathways, such as nucleotide oligomerization domain 2 (Nod2), genes that control autophagy (eg, ATG16L1, IRGM), and genes in the interleukin-23–T helper cell 17 pathway indicate the important roles of host-microbe interactions in regulating intestinal immune homeostasis. There is increasing evidence that intestinal microbes influence host immune development, immune responses, and susceptibility to human diseases such as IBD, diabetes mellitus, and obesity. Conversely, host factors can affect microbes, which in turn modulate disease susceptibility. We review the cell populations and mechanisms that mediate interactions between host defense and tolerance and how the dysregulation of host-microbe interactions leads to intestinal inflammation and IBD.
Keywords: Inflammatory Bowel Disease, Toll-Like Receptor, Pattern Recognition Receptor
The high density of microbes in the intestinal lumen leads to continuous communication between host cells and microbes. These interactions can be mutually beneficial or can have adverse effects and contribute to intestinal inflammation. The intestinal immune system must, on the one hand, defend against pathogens and entry of excessive intestinal microbes and, on the other hand, allow tolerance to resident intestinal microbes. Perturbation of this balance is thought to predispose to inflammatory bowel disease (IBD).
A major challenge for understanding the interactions between resident microbiota and mammalian hosts is the heterogeneity of microbial communities that can colonize the intestine and other sites. Different components of microbiota can have very different effects on the host; the composition of microbial communities can be influenced by a variety of factors, including diet, antibiotic therapy, environmental exposure to microorganisms, and sequential microbial colonization in the neonatal period. Certain species of bacteria, for example, can have large effects on the intestinal immune system, in part by altering the balance between populations of regulatory and effector T cells. However, the extent to which individual bacterial species or specific combinations of species affect the host is not clear. In addition, the concept that resident intestinal microbiota are largely tolerated by the immune system, whereas pathogenic microbes are targeted by host defense mechanisms, is not entirely correct. The immune system continuously monitors the resident microbiota, and certain antimicrobial mechanisms are constitutively engaged to prevent overgrowth of the colonizing microbes: this maintains what is loosely called immune homeostasis. Constitutive production of antimicrobial peptides by Paneth cells, mucus production by goblet cells, and the control of microbial attachment and invasion by secretory immunoglobulin (Ig) A are examples of immune defense mechanisms directed against microbes. It is not clear whether all bacterial and viral species are equally affected by these and other mechanisms or whether there is a greater degree of specificity in the host response whereby the defense mechanisms control the composition of microbiota and maintain immune homeostasis. The classical distinction between commensal and pathogenic microorganisms, although practically convenient, does not necessarily characterize the full spectrum of behaviors by these respective microorganisms towards the host. The host immune system could use unrecognized strategies to react to different classes of microbial communities. We review the cell populations and mechanisms that mediate host defense and tolerance and how dysregulation of host-microbe interactions can lead to intestinal inflammation and IBD.
Intestinal Immune Defenses Against Microbiota
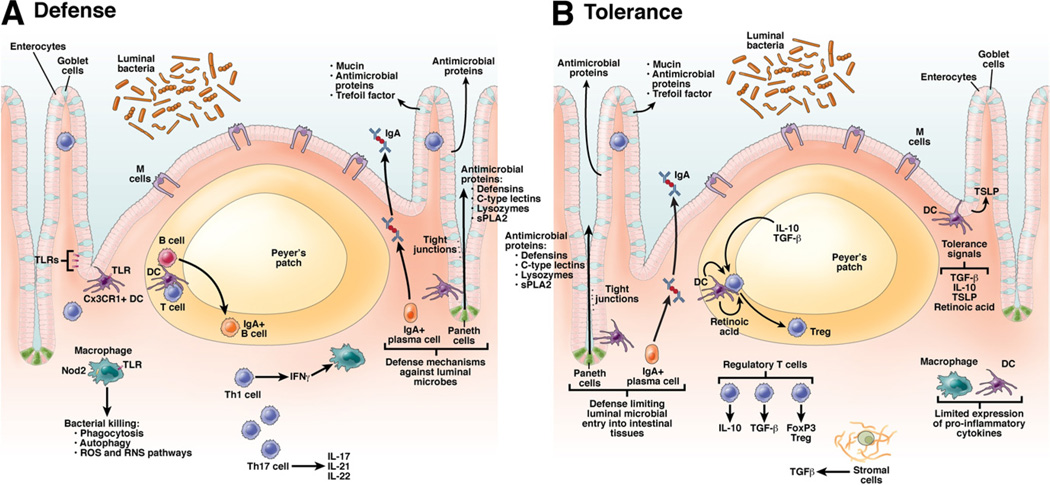
Intestinal epithelial, innate, and adaptive immune cells use defense mechanisms that involve pattern recognition receptors (PRRs). Initial recognition of microbes through PRR, in turn, activates cytokine and chemokine signaling pathways, antimicrobial killing (eg, antimicrobial proteins, phagocytosis, autophagy, reactive oxygen and nitrogen species), unfolded protein response, and initiation of adaptive T- and B-cell responses (Figure 1).
Figure 1.
Mechanisms of host defense and tolerance towards intestinal microbes. The intestinal environment modulates cellular differentiation in the immune system to control defense against pathogens and tolerance. (A) Defense mechanisms: Intestinal epithelial cells provide a physical barrier between the luminal microbes and the underlying intestinal tissues to control defense and tolerance. Specialized epithelial cells produce a mucus layer and secrete antimicrobial proteins that limit bacterial exposure to the epithelial cells. Production of large amounts of IgA provides additional protection from luminal microbiota. Innate microbial sensing by epithelial cells, DCs, and macrophages is mediated through PRRs such as TLRs and NLRs. Activation of PRRs on innate cells induces various pathways that mediate microbial killing and activate adaptive cells. DCs present antigens to naïve CD4+ T cells in secondary lymphoid organs (Peyer’s patches, mesenteric lymph nodes) where factors such as the phenotype of the antigen presenting cells and cytokine milieu modulate differentiation of CD4+ T-cell subsets (Th1, Th2, Th17, Treg) with characteristic cytokine and intestinal homing profiles. (B) Tolerance mechanisms: Defense mechanisms that limit microbial entry into intestinal tissues also serve as a mechanism of tolerance. Activation of PRRs on the unique populations of macrophages and DCs in the intestinal lamina propria does not result in secretion of proinflammatory cytokines, in contrast to similar activation of systemic innate cells. DC present antigen to T cells in the Peyer’s patches and mesenteric lymph nodes, which can lead to differentiation of Treg populations, regulated by IL-10, TGF-β, and retinoic acid. Thymic stromal lymphopoietin (TSLP) and other factors secreted by epithelial cells in the intestinal environment can contribute to tolerance of intestinal immune cells.
PRRs Recognize Microorganisms and Initiate Defense Mechanisms
PRRs initiate responses to and regulate microbial infections. PRRs recognize conserved structures of microorganisms, called pathogen-associated molecular patterns (PAMPs). Host PRRs include the family of Toll-like receptors (TLRs), C-type lectins, nucleotide-binding domain and leucine-rich repeat-containing receptors (NLRs), and retinoic acid-inducible gene I-like receptors.1 Differences between distinct PRRs include the repertoire of microbial and host ligands (eg, generated by tissue injury) recognized and the cellular locations surveyed (ie, cell surface, lysosomal, cytoplasmic). Distinct PRRs can, in turn, activate different signaling pathways.1 However, there is also overlap among PRR-initiated signaling pathways, such as in activation of the transcription factor nuclear factor-κB (NF-κB); defining unique roles for different PRRs is an important area of research. Requirements for PRRs can vary between intestinally introduced and systemically introduced pathogens. For example, intestinal, but not systemic, requirements have been shown for TLR2 in defense against Yersinia pseudotuberculosis2 and nucleotide oligomerization domain 2 (Nod 2) in defense against Listeria monocytogenes.3
The intestinal immune system uses many different mechanisms to regulate the high concentrations of resident microbes and protect the mucosal surface from pathogens. When one pathway is deficient, other immune pathways can compensate. For example, mice deficient in TLR signaling have increased penetration of intestinal microbiota but produce high titers of functional antibodies against resident intestinal microbiota that help compensate for the deficiency in innate immunity and allow mice to better survive infection.4 In the intestine, B cell-mediated antibody responses are unique in that they secrete high levels of IgA. The functions of intestinal IgA can overlap with those of innate immune responses in their ability to neutralize pathogenic molecules and microbes and regulate commensalism.5 Secreted IgA assists in defenses against various intestinal pathogens, such as Salmonella typhimurium and Giardia muris.5
It is not understood how the intestinal immune system can discriminate between resident intestinal microbes, with which it coexists, and pathogens, to which it must respond. Resident and pathogenic bacteria can make different modifications to PAMPs such as lipopolysaccharide (LPS).6 However, immune cells can have equal responses to comparable PAMPs expressed by resident intestinal microbes or pathogens, so there are likely to be other signals that contribute to the inflammatory response against intestinal pathogens; the nature of these signals is unresolved. One hypothesis is that pathogens cause tissue damage that activates an inflammatory response. During tissue injury and necrosis, extracellular matrix products and cellular products such as nucleic acids, uric acid, and mitochondrial components are released and can be sensed through various PRRs.7,8 Intestinal inflammation can be attenuated by blockade or deficiency of receptors that respond to these cellular products.8
Intestinal Epithelial Cell-Mediated Defense Mechanisms
The intestinal epithelium lies at the interface between the intestinal microbiome and the gastrointestinal-associated lymphoid tissues and provides a physical barrier against excessive entry of luminal microbiota. In addition to their barrier function, intestinal epithelial cells (IECs) actively defend against intestinal pathogens and limit penetration of resident intestinal microbes into tissues.
IECs express PRRs and can recognize and respond to intestinal microbes through secretion of cytokines and antimicrobial proteins and up-regulation of surface molecules that mediate intercellular interactions. Studies of mice that have IEC-specific knockout of signals downstream of PRRs (eg, IKKγ, IKKβ) demonstrated that IECs have a fundamental role in intestinal immune homeostasis and responses to pathogens.9,10 Multiple intestinal epithelial lineages contribute to antimicrobial defenses. Enterocytes can produce enzymes, such as intestinal alkaline phosphatase and acyloxyacyl hydrolase, that modify LPS to reduce its inflammatory effects.6,11 Paneth cells produce antimicrobial proteins such as defensins and lectins,2,12 and goblet cells produce mucins, trefoil peptides, and resistin-like molecule β (RELMβ).13–16 Microbe sensing is required for optimal regulation of antimicrobial proteins, which, in turn, is mediated through PRR pathways, as demonstrated in studies of MyD88- and Nod2-deficient mice.3,12 In many cases, absence of these epithelial cell-derived antimicrobial pathways increases susceptibility to intestinal inflammation. Consistent with these observations, patients with IBD express lower levels of α-defensin than healthy individuals17,18; additional studies are necessary to define the cause of this decrease and its contribution to disease. In other cases, factors that mediate responses to pathogens can also increase susceptibility to intestinal inflammation; this is the case for RELMβ.16
Cellular Mechanisms of Microbial Killing
A number of pathways mediate the killing of microbes that occasionally breach the intestinal epithelial barrier. Macrophages in the intestinal lamina propria are highly effective in phagocytosis and killing of microbes.19,20 They achieve this, in part, through the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS), which is particularly important in regulating enteric infections. Mice that are defective in both ROS and RNS signaling pathways spontaneously develop large abscesses that contain predominantly enteric organisms, resulting in increased mortality.21 Studies of individuals with defects in these pathways have demonstrated their importance in regulating intestinal immune homeostasis. Patients with chronic granulomatous disease, which involves defects in the ROS pathway, have a predisposition to colitis.22
Autophagy is an important mechanism for clearance of intracellular components, including organelles, apoptotic bodies, and invading microbes (see this issue of Gastroenterology).23 Variants in 2 genes involved in autophagy (autophagy related protein 16-like 1 [ATG16L1] and immunity-related GTPase family M [IRGM]) have been associated with Crohn’s disease (CD)24; these variants cause loss of function in autophagy pathways.23,25 Autophagy contributes to microbial defenses through various mechanisms, including regulation of microbial killing, Paneth cell function, interleukin (IL) 1β production, and T-cell selection.23 Consistent with findings in mice with defects in ATG16L1 function, patients with CD who carry ATG16L1 risk alleles have Paneth cells with abnormal morphology, compared with CD patients who do not carry the ATG16L1 risk allele.26 Autophagy is activated through PRR, including Nod2,27 thereby highlighting pathways associated with susceptibility to CD that overlap.
IL-23 and Th17 Cells
The IL-23–T helper (Th) 17 cell pathway defends against microbial infection by intestinal and other pathogens.28 However, IL-23 and the cytokines produced upon activation of Th17 cells contribute to tissue inflammation in general, and to IBD specifically, demonstrated in several studies of patients and mice.28,29 These cytokines must therefore be carefully regulated during mucosal responses. IBD has been associated with variants in IL23R and genomic regions that include other genes in the IL-23–Th17 pathway (eg, IL12p40, STAT3, JAK2, CCR6, TNFSF15, Tyk2), indicating that it is an important regulatory component of intestinal immune homeostasis.24 Of note is that a number of these other genes are not unique to the IL-23–Th17 pathway, such that their contributions to altered susceptibility to IBD may be through various mechanisms. Further studies are needed to define the full spectrum of functional consequences of these IBD IL-23–Th17 pathway-associated polymorphisms.
Intestinal Immune Tolerance
Although the intestinal immune system must defend against pathogens, it must coexist with resident intestinal microbiota. This tolerance is mediated by multiple factors, including the intestinal microbiota themselves, intestinal epithelial and stromal cells, and innate and adaptive cells within the intestinal tissues. The mechanisms that control intestinal tolerance include those that minimize exposure to and immune recognition of intestinal microbiota and those that down-regulate immune responses through intracellular and intercellular mechanisms.
Limiting Intestinal Microbial Exposure
Exposure to intestinal microbiota is minimized by the defense mechanisms described above (Figure 1). Intestinal mucus, produced by goblet cells, limits penetration of microbes into intestinal and systemic tissues. Mice with deletion of mucin 2 (MUC2), a major mucin component, develop intestinal inflammation,13 and a genomic region that includes MUC19 has been associated with CD.24 Adaptive immune responses, in particular the secretion of intestinal IgA, also limit penetration of intestinal microbes into host tissues.5
The physical barrier formed by intestinal epithelial cells that prevents excessive entry of luminal microbiota is maintained through intercellular junctions, of which tight junctions are a critical component. Tight junctions are disrupted during inflammation, which contributes to paracellular permeability and a cycle of microbial penetration into intestinal tissues.30 Patients with IBD have an increase in intestinal paracellular permeability and defects in tight junction regulation30; it is not clear whether these features arise secondary to inflammation or are primary events in disease pathogenesis. Interestingly, selective perturbation of intestinal tight junction function in mice increases intestinal permeability but does not result in spontaneous colitis. However, the intestinal lamina propria of these mice express increased levels of proinflammatory cytokines, and the mice have increased susceptibility to T cell-mediated colitis.31 Therefore, a combination of breaches in the intestinal barrier and dysregulation of either intestinal immune defense or tolerance might result in predisposition to IBD.
Following injury, the abilities to repair and regenerate the epithelium constitute important mechanisms for controlling and ultimately resolving the inflammatory response. Maintenance of epithelial cell function and restitution during inflammation depend on a number factors, including growth factors (eg, trefoil factor),14,15 innate signals,32 cytokines (eg, IL-18, IL-22),33,34 and regulation of endoplasmic reticulum stress.35
Active Down-regulation of the Immune Response
Host-microbial interactions in the intestinal environment can down-regulate inflammatory responses (Figure 1). This occurs through regulation of PRR expression and responsiveness, secretion of inhibitory mediators, and modulation of transcription and expression of factors in intracellular signaling pathways within distinct intestinal immune cells.
Regulating the expression levels, distribution, and distribution-dependent responses through PRR is one mechanism of actively down-regulating immune responses. Intestinal immune tolerance, as well as intestinal immune responses in general, are developmental processes that depend, in part, on microbial signals. For example, fetal IECs respond to LPS stimulation by activating NF-κB and secreting chemokines, whereas these responses are lost from neonatal and adult IECs.36 These postnatal changes in IEC activity are associated with post-transcriptional down-regulation of IL-1 receptor-associated kinase 1 and depend on microbial colonization; disruption of this process increases susceptibility of premature infants to necrotizing enterocolitis.36 In the post-natal period, apical epithelial cell responses are differentially regulated from basolateral responses, given the proximity of the apical surface to the intestinal lumen. For example, TLR5 expression and flagellin-mediated NF-κB activation are restricted to the basolateral epithelial cell membrane.37 In addition, whereas basolateral IEC signaling by TLR9 activates NF-κB, apical stimulation of TLR9 does not.38
Characteristics of the intestinal immune environment include high levels of the anti-inflammatory proteins IL-10, transforming growth factor (TGF)- β, and retinoic acid. Multiple cell populations contribute to (eg, epithelial, stromal, innate, and adaptive cells) and respond to these anti-inflammatory mediators. Mice deficient in IL-10 and TGF-β develop spontaneous colitis.39 In addition to its key role in intestinal homing, retinoic acid is an important regulator of T-regulatory cell (Treg) differentiation in the intestine.40,41 Studies in human tissues or in patients with IBD have also demonstrated the importance of these secreted proteins in disease pathogenesis. For example, studies of human colon explants demonstrated that mucosal IL-10 and TGF-β have important roles in preventing LPS-mediated, interferon-γ-induced, epithelial damage.42 Genetic studies have associated IL-10 with IBD,43 indicating the importance of this pathway in mediating human intestinal homeostasis.
Resident intestinal microbes participate directly in intestinal immune tolerance through a combination of mechanisms that includes regulation of NF-κB and ubiquitin-proteasome pathways44,45 and induction of anti-inflammatory cytokine secretion and specific immune cell subsets (eg, Tregs).46 Bacterial interactions with dietary substances can produce products such as short-chain fatty acids that down-regulate intestinal inflammatory responses.47 Consistent with these findings, mice deficient in responses to short-chain fatty acids are more susceptible to dextran sodium sulfate-induced colitis,47 and bacterial protective factors (eg, polysaccharide A) can attenuate T cell-mediated and trinitrobenzene sulfonic acid-induced colitis.46 These studies indicate that intestinal resident microbes directly induce host immune tolerance.
Innate Signaling Regulates Intestinal Tolerance by Adaptive T Cells
Adaptive immune cells are important for intestinal tolerance and are regulated, indirectly and directly, through innate pathways. Exposure to antigens through the intestine induces T-cell tolerance. Lamina propria dendritic cells (DCs), especially those that are CD103-positive, and macrophages from mice produce IL-10, TGF-β, and retinoic acid, which lead to differentiation and maintenance of forkhead box P3 (Foxp3)+ Tregs.40,41,48 Dysregulation of tolerogenic intestinal DCs decreases intestinal lamina propria Tregs and increased frequencies of Th1 and Th17 cells.49 Some aspects of innate regulation of adaptive tolerance may only be apparent during inflammation. For example, innate cell production of IL-10, specifically during intestinal inflammation, is required to maintain optimal Treg activity.50 Innate receptors can also have direct roles in adaptive immune responses. For example, activation of TLR4 on Tregs increases their suppressor functions.51 Consistent with this, some studies have shown that MyD88−/− regulatory T cells do not protect against colitis as well as wild-type regulatory T cells.52 TLR4 expressed on effector T cells can also down-regulate effector T-cell responses, and its absence from these cells results in earlier and more severe colitis in mice.53
Balancing Defense and Tolerance
Unique Populations of Intestinal Innate Immune Cells
We have increased our understanding of the diversity and unique nature of intestinal innate immune cells. The distinct characteristics of these cells are shaped by the intestinal environment, which includes cell-cell and host-microbe interactions54 and anti-inflammatory cytokines. Diverse subsets of lamina propria DCs have been identified based on cell surface markers (eg, CD103), chemokine receptors (eg, CX3CR1, CCR7), cytokines produced, and distinct functions.55,56 These distinct functions include luminal antigen sampling, T-cell stimulatory and differentiation capacities (eg, Th1, Th17, and Treg and intestinal homing), microbial uptake, pathogen defenses, and dead cell clearance.48,55–59 In addition to resident DC and macrophage subsets, during infection and injury, peripheral inflammatory monocytes are recruited to the intestine where they regulate intestinal pathogens; recruitment of the monocytes requires alternative trafficking molecules such as CCR2.60 A balance of these innate cell subsets is required to maintain intestinal immune homeostasis.
Transcriptional profile analyses of mouse intestinal DCs49 and macrophages48 and human intestinal macrophages61 identified large differences from peripheral counterparts, revealing unique intestinal innate cell phenotypes and tolerogenic pathways. For example, in contrast to peripheral monocytes, certain intestinal macrophages do not secrete cytokines upon stimulation through PRRs.19,61 Other macrophage subsets might selectively secrete anti-inflammatory cytokines such as IL-10.48 The decreased secretion of cytokines is likely regulated at multiple levels. For example, select PRRs and components of PRR recognition of ligands and signaling are down-regulated in human intestinal macrophages, compared with peripheral cells.19,20,61 Intestinal macrophage signaling through the PRR is also down-regulated via increased expression of inhibitory proteins such as suppressor of cytokine signaling 1 and sterile and Armadillo motif-containing protein.61 Additional cellular proteins and pathways that are important in down-regulation of intestinal innate cell responses include A20, the membrane protein β-catenin, and the phosphotidylinositol-3-kinase pathway.49,62,63 Dysregulation of these pathways in mice results in either spontaneous colitis62 or increased susceptibility to experimental colitis.49 In patients with IBD, lamina propria DCs and macrophages produce increased amounts of inflammatory cytokines,20,59 compared with cells of healthy individuals, consistent with dysregulated tolerance. In addition to down-regulation of inflammatory pathways, analyses of transcriptional profiles of intestinal innate cells revealed increased transcription of pathways required for antimicrobial defenses. This observation was consistent with the need to limit inflammatory responses and tissue injury while simultaneously mediating bacterial killing.
Nod2 Mediates Host Recognition of Microbes, Functioning in Defense and Tolerance
Of genetic variants associated with CD, polymorphisms in Nod2 confer the greatest risk. Cells of individuals who carry the major CD-associated Nod2 polymorphisms cannot signal through its gene product.64 Loss-of-function Nod2 polymorphisms increase the risk for CD, but CD still does not develop in most individuals with the polymorphism, so additional risk factors, such as genetic and environmental factors, must contribute to development of CD.
Nod2 regulates intestinal immune defense and tolerance. Nod2 is an NLR that is activated by peptidoglycan, a component of gram-positive and gram-negative bacteria.64 A modified muramyl dipeptide component of peptidoglycan expressed in mycobacteria has been shown to have particularly strong stimulatory capacity.65 More recently, Nod2 was found to be activated by specific viruses,66 which is significant given the increasing recognition of the role that viruses (eg, norovirus) have in intestinal inflammation in mice.67 Nod2 is expressed on various cell populations, including myeloid-derived cells, epithelial cells, and endothelial cells. Nod2 stimulation in DCs and macrophages results in activation of the NF-κB pathway. Nod2 signaling increases intestinal defenses via secretion of proinflammatory cytokines, induction of antimicrobial proteins, generation of ROS, enhanced killing of microbes, and maturation of antigen presenting cells that have the ability to activate T cells.64 There is evidence that Nod2 is expressed on T cells and has a direct role in T-cell functions such as regulation of cytokine production and clearance of pathogens.68 Mice deficient in Nod2 have an increased load of resident intestinal microbiota in the terminal ileum and increased susceptibility to intestinal pathogens such as Listeria monocytogenes, Salmonella typhimurium, and Helicobacter hepaticus.3,64,69
In the intestinal environment, Nod2 can also contribute to down-regulation of inflammatory responses, supporting its role in tolerance.70 This likely occurs, in part, via the ability of Nod2 to down-regulate its own signaling during intestinal inflammatory responses and that of other PRRs, after a period of prolonged microbial stimulation.70–72 Moreover, Nod2 promotes survival of human Tregs; individuals with CD-associated Nod2 polymorphisms have reduced numbers of intestinal lamina propria Tregs.73 Innate immune cells from carriers of the Nod2 risk allele produce lower levels of cytokines following acute stimulation with Nod2 ligands and have lost the ability to down-regulate cytokine secretion under conditions of chronic Nod2 stimulation.64,70–72 Therefore, Nod2 loss-of-function polymorphisms might increase the risk of CD through the dual functions of Nod2 in intestinal immune defense and tolerance.
Cross Talk Between Host and Microbes
Bidirectional, host-microbe interactions in the intestine can benefit each organism or have adverse effects that contribute to intestinal inflammation.
Intestinal Microbes Influence Host Immune Development and Responses
The intestinal microbiota has an important role in the development of the intestinal immune system: it primes systemic innate immune responses and regulates development of autoimmune and inflammatory diseases. As such, germ-free mice and mice that are deficient in microbe recognition pathways have defects in intestinal mucosal immune development and function, including in development of isolated lymphoid follicles, function of the intestinal epithelium, production of IgA, differentiation of T-cell subsets, and induction of pathways that down-regulate intestinal inflammation.5,32,38,74,75 Moreover, intestinal colonization is required for the optimal expression of PRR and signaling pathways required for intestinal immune function (eg, Nod2).69 Distinct intestinal microbes and/or their products can differentially regulate T-cell polarization. For example, induction of Tregs and IL-10 can be mediated through the polysaccharide A component of intestinal resident microbe, Bacterioides fragilis.46 In contrast, the enriched IL-17–producing T-cell population observed in the intestine depends on intestinal microbiota and microbe-derived factors such as adenosine 5′-triphosphate.76 There is increasing evidence that specific intestinal microbiota, such as segmented filamentous bacteria, induce intestinal Th17 cytokines.77 Failure of this induction increases susceptibility to infection with intestinal pathogens such as Citrobacter rodentium in mice.77
Alterations in intestinal colonization or in host-microbe interactions can modulate disease in mouse models of colitis39 and in systemic diseases, such as metabolic syndrome and diabetes.78,79 Mice raised in a germ-free environment or deficient in PRR pathways are frequently protected from colitis,39,80 indicating that intestinal inflammation requires intestinal microbes. However, this concept incompletely captures the complexity of intestinal host-microbe interactions. For example, mice with perturbations in certain host-microbe recognition pathways develop spontaneous colitis (eg, TLR5−/− mice).75 In other cases, the inflammation that develops in experimental models of colitis is more severe under germ-free or PRR pathway-deficient conditions.32 The requirement for interaction between host and resident luminal microbes to maintain intestinal epithelial homeostasis might account for these differences,32 which indicate the diverse effects of intestinal host-microbe interactions on intestinal immune regulation.
Importantly, changes in diet, use of antibiotics, and intestinal colonization (eg, eradication of intestinal helminthes),81,82 have likely modified intestinal microbial communities and contributed to the increased prevalence of IBD during the past century. Alterations in intestinal microbiota have been identified in patients with IBD and are associated with prognostic outcomes.83 Patients with CD have increased colonization of ileal mucosa by adherent, invasive Escherichia coli.84 Patients with either CD or ulcerative colitis have less diversity in colonization among members of the phyla Firmicutes and Bacteroidetes than control individuals.83 Changes in intestinal micro-biota observed in IBD patients might be primary pathogenic factors, secondary responses to an inflammatory environment, or alterations induced by genetic differences.
Host Factors Can Affect Microbial Composition to Determine Disease Susceptibility
Studies in animal models have shown that differences in host conditions (eg, obesity, disease) and factors that regulate immunity (eg, MyD88-, Nod2-, Tbet-, defensin-, CD1d- and IgA-deficient mice)69,74,79,85–87,88 affect the microbial composition of the intestine and thereby contribute to disease susceptibility. To study the effects of modifying the composition of intestinal microbes, researchers have investigated the effects of transferring microbiota between different mice models. Transfer of microbiota from obese mice, compared with lean mice, into germ-free mice resulted in increased body fat.87 Bacteria transferred from MyD88−/− nonobese diabetic mice, raised in a specific pathogen-free facility, attenuated diabetes in germ-free recipients.79 Transfer of bacteria from mice that lack the transcription factor Tbet, which controls lineage commitment of Th1 cells, into wild-type mice leads to intestinal inflammation.85 These experimental systems have provided specific examples of how differences in host genetics modify intestinal microbiota and determine disease susceptibility. These concepts require further study in patients with IBD to better understand pathogenesis and develop new therapies.
Acknowledgments
The authors thank Judy H. Cho and Eric Elton for critical reading of the manuscript.
Funding
Supported by grants from the National Institute of Health: R01DK077905, DK-P30-34989, and U19-AI082713 (C.A.) and the Howard Hughes Medical Institute (R.M.).
Abbreviations used in this paper:
- CD
Crohn’s disease
- DC
dendritic cells
- IBD
inflammatory bowel disease
- IECs
intestinal epithelial cells
- Ig
immunoglobulin
- IL
interleukin
- LPS
lipopolysaccharide
- NF-κB
nuclear factor-κB
- NLR
nucleotide-binding domain and leucine-rich repeat-containing receptors
- NOD2
nucleotide oligomerization domain
- PAMPs
pathogen-associated molecular patterns
- PRR
pattern recognition receptor
- RELMβ
resistin-like molecule β
- ROS
reactive oxygen species
- TGF
transforming growth factor
- TLR
Toll-like receptor
- Treg
T regulatory cells.
Biographies

Clara Abraham

Ruslan Medzhitov
Footnotes
Conflicts of interest
The authors disclose no conflicts.
References
- 1.Rakoff-Nahoum S, Medzhitov R. Innate immune recognition of the indigenous microbial flora. Mucosal Immunol. 2008;1(Suppl 1):S10–S14. doi: 10.1038/mi.2008.49. [DOI] [PubMed] [Google Scholar]
- 2.Dessein R, Gironella M, Vignal C, et al. Toll-like receptor 2 is critical for induction of Reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis. Gut. 2009;58:771–776. doi: 10.1136/gut.2008.168443. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 4.Slack E, Hapfelmeier S, Stecher B, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macpherson AJ, McCoy KD, Johansen FE, et al. The immune geography of IgA induction and function. Mucosal Immunol. 2008;1:11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 6.Munford RS, Varley AW. Shield as signal: lipopolysaccharides and the evolution of immunity to gram-negative bacteria. PLoS Pathog. 2006;2:e67. doi: 10.1371/journal.ppat.0020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, Evans JE, Rock KL. Molecular identification of a danger signal that alerts the immune system to dying cells. Nature. 2003;425:516–521. doi: 10.1038/nature01991. [DOI] [PubMed] [Google Scholar]
- 8.Cavassani KA, Ishii M, Wen H, et al. TLR3 is an endogenous sensor of tissue necrosis during acute inflammatory events. J Exp Med. 2008;205:2609–2621. doi: 10.1084/jem.20081370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nenci A, Becker C, Wullaert A, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 10.Zaph C, Troy AE, Taylor BC, et al. Epithelial-cell-intrinsic IKK-β expression regulates intestinal immune homeostasis. Nature. 2007;446:552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 11.Bates JM, Akerlund J, Mittge E, et al. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaishnava S, Behrendt CL, Ismail AS, et al. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van der Sluis M, De Koning BA, De Bruijn AC, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 14.Taupin D, Podolsky DK. Trefoil factors: initiators of mucosal healing. Nat Rev Mol Cell Biol. 2003;4:721–732. doi: 10.1038/nrm1203. [DOI] [PubMed] [Google Scholar]
- 15.Furuta GT, Turner JR, Taylor CT, et al. Hypoxia-inducible factor 1-dependent induction of intestinal trefoil factor protects barrier function during hypoxia. J Exp Med. 2001;193:1027–1034. doi: 10.1084/jem.193.9.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McVay LD, Keilbaugh SA, Wong TM, et al. Absence of bacterially induced RELMβ reduces injury in the dextran sodium sulfate model of colitis. J Clin Invest. 2006;116:2914–2923. doi: 10.1172/JCI28121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wehkamp J, Salzman NH, Porter E, et al. Reduced Paneth cell α-defensins in ileal Crohn’s disease. Proc Natl Acad Sci U S A. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simms LA, Doecke JD, Walsh MD, et al. Reduced α-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn’s disease. Gut. 2008;57:903–910. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 19.Smythies LE, Sellers M, Clements RH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber B, Saurer L, Mueller C. Intestinal macrophages: differentiation and involvement in intestinal immunopathologies. Semin Immunopathol. 2009;31:171–184. doi: 10.1007/s00281-009-0156-5. [DOI] [PubMed] [Google Scholar]
- 21.Shiloh MU, MacMicking JD, Nicholson S, et al. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity. 1999;10:29–38. doi: 10.1016/s1074-7613(00)80004-7. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal S, Mayer L. Gastrointestinal manifestations in primary immune disorders. Inflamm Bowel Dis. 2010;16:703–711. doi: 10.1002/ibd.21040. [DOI] [PubMed] [Google Scholar]
- 23.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 28.Abraham C, Cho J. Interleukin-23/Th17 pathways and inflammatory bowel disease. Inflamm Bowel Dis. 2009 doi: 10.1002/ibd.20894. [DOI] [PubMed] [Google Scholar]
- 29.Hue S, Ahern P, Buonocore S, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner JR. Molecular basis of epithelial barrier regulation: from basic mechanisms to clinical application. Am J Pathol. 2006;169:1901–1909. doi: 10.2353/ajpath.2006.060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su L, Shen L, Clayburgh DR, et al. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136:551–563. doi: 10.1053/j.gastro.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Zaki MH, Boyd KL, Vogel P, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugimoto K, Ogawa A, Mizoguchi E, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaser A, Lee AH, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lotz M, Gutle D, Walther S, et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med. 2006;203:973–984. doi: 10.1084/jem.20050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gewirtz AT, Navas TA, Lyons S, et al. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 38.Lee J, Mo JH, Katakura K, et al. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat Cell Biol. 2006;8:1327–1336. doi: 10.1038/ncb1500. [DOI] [PubMed] [Google Scholar]
- 39.Elson CO, Cong Y, McCracken VJ, et al. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–276. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 40.Sun CM, Hall JA, Blank RB, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mucida D, Park Y, Kim G, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 42.Jarry A, Bossard C, Bou-Hanna C, et al. Mucosal IL-10 and TGF-beta play crucial roles in preventing LPS-driven, IFN-γ-mediated epithelial damage in human colon explants. J Clin Invest. 2008;118:1132–1142. doi: 10.1172/JCI32140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franke A, McGovern DP, Barrett JC, et al. Genome-wide metaanalysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neish AS, Gewirtz AT, Zeng H, et al. Prokaryotic regulation of epithelial responses by inhibition of IκB-α ubiquitination. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 45.Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 46.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 47.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denning TL, Wang YC, Patel SR, et al. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T-cell responses. Nat Immunol. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 49.Manicassamy S, Reizis B, Ravindran R, et al. Activation of β-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science. 329:849–853. doi: 10.1126/science.1188510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murai M, Turovskaya O, Kim G, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caramalho I, Lopes-Carvalho T, Ostler D, et al. Regulatory T cells selectively express toll-like receptors and are activated by lipopolysaccharide. J Exp Med. 2003;197:403–411. doi: 10.1084/jem.20021633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomita T, Kanai T, Fujii T, et al. MyD88-dependent pathway in T cells directly modulates the expansion of colitogenic CD4+ T cells in chronic colitis. J Immunol. 2008;180:5291–5299. doi: 10.4049/jimmunol.180.8.5291. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Navajas JM, Fine S, Law J, et al. TLR4 signaling in effector CD4+ T cells regulates TCR activation and experimental colitis in mice. J Clin Invest. 2010;120:570–581. doi: 10.1172/JCI40055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 55.Varol C, Vallon-Eberhard A, Elinav E, et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 56.Bogunovic M, Ginhoux F, Helft J, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rescigno M, Urbano M, Valzasina B, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 58.Niess JH, Brand S, Gu X, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 59.Kamada N, Hisamatsu T, Okamoto S, et al. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-γ axis. J Clin Invest. 2008;118:2269–2280. doi: 10.1172/JCI34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunay IR, Damatta RA, Fux B, et al. Gr1(+) inflammatory monocytes are required for mucosal resistance to the pathogen Toxoplasma gondii. Immunity. 2008;29:306–317. doi: 10.1016/j.immuni.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smythies LE, Shen R, Bimczok D, et al. Inflammation anergy in human intestinal macrophages is due to Smad-induced IκBα expression and NF-κB inactivation. J Biol Chem. 2010;285:19593–19604. doi: 10.1074/jbc.M109.069955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee EG, Boone DL, Chai S, et al. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uno JK, Rao KN, Matsuoka K, et al. Altered macrophage function contributes to colitis in mice defective in the phosphoinositide-3 kinase subunit p110δ. Gastroenterology. 2010;139:1642–1653. doi: 10.1053/j.gastro.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abraham C, Cho JH. Functional consequences of NOD2 (CARD15) mutations. Inflamm Bowel Dis. 2006;12:641–650. doi: 10.1097/01.MIB.0000225332.83861.5f. [DOI] [PubMed] [Google Scholar]
- 65.Coulombe F, Divangahi M, Veyrier F, et al. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J Exp Med. 2009;206:1709–1716. doi: 10.1084/jem.20081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sabbah A, Chang TH, Harnack R, et al. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cadwell K, Patel KK, Maloney NS, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shaw MH, Reimer T, Sanchez-Valdepenas C, et al. T cell-intrinsic role of Nod2 in promoting type 1 immunity to Toxoplasma gondii. Nat Immunol. 2009;10:1267–1274. doi: 10.1038/ni.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petnicki-Ocwieja T, Hrncir T, Liu YJ, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watanabe T, Asano N, Murray PJ, et al. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J Clin Invest. 2008;118:545–559. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hedl M, Li J, Cho JH, et al. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc Natl Acad Sci U S A. 2007;104:19440–19445. doi: 10.1073/pnas.0706097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hedl M, Abraham C. Secretory mediators as a critical mechanism for Nod2-mediated tolerance in human macrophages. Gastroenterology. 2011;140:231–241. doi: 10.1053/j.gastro.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rahman MK, Midtling EH, Svingen PA, et al. The pathogen recognition receptor NOD2 regulates human FOXP3+ T cell survival. J Immunol. 2010;184:7247–7256. doi: 10.4049/jimmunol.0901479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bouskra D, Brezillon C, Berard M, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 75.Vijay-Kumar M, Sanders CJ, Taylor RT, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Atarashi K, Nishimura J, Shima T, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 77.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rakoff-Nahoum S, Hao L, Medzhitov R. Role of Toll-like receptors in spontaneous commensal-dependent colitis. Immunity. 2006;25:319–329. doi: 10.1016/j.immuni.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 81.Weinstock JV. Helminths and mucosal immune modulation. Ann N Y Acad Sci. 2006;1072:356–364. doi: 10.1196/annals.1326.033. [DOI] [PubMed] [Google Scholar]
- 82.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Frank DN, St, Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barnich N, Carvalho FA, Glasser AL, et al. CEACAM6 acts as a receptor for adherent-invasive E coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garrett WS, Lord GM, Punit S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salzman NH, Hung K, Haribhai D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 88.Nieuwenhuis EE, Matsumoto T, Lindenbergh D, et al. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J Clin Invest. 2009;119:1241–1250. doi: 10.1172/JCI36509. [DOI] [PMC free article] [PubMed] [Google Scholar]