Abstract
Influenza virus is rich in variation and mutations. It would be very convenient for virus detection and isolation to histochemically detect viral infection regardless of variation and mutations. Here, we established a histochemical imaging assay for influenza virus sialidase activity in living cells by using a new fluorescent sialidase substrate, 2-(benzothiazol-2-yl)-4-bromophenyl 5-acetamido-3,5-dideoxy-α-D-glycero-D-galacto-2-nonulopyranosidonic acid (BTP3-Neu5Ac). The BTP3-Neu5Ac assay histochemically visualized influenza virus-infected cells regardless of viral hosts and subtypes. Influenza virus neuraminidase-expressed cells, viral focus formation, and virus-infected locations in mice lung tissues were easily, rapidly, and sensitively detected by the BTP3-Neu5Ac assay. Histochemical visualization with the BTP3-Neu5Ac assay is extremely useful for detection of influenza viruses without the need for fixation or a specific antibody. This novel assay should greatly improve the efficiency of detection, titration, and isolation of influenza viruses and might contribute to research on viral sialidase.
Influenza viruses are highly-infectious respiratory pathogens of humans, animals, and birds that cause a serious public health problems globally with social and economic impacts1. Influenza viruses cause diseases ranging from mild respiratory illness to fatal pneumonia and contribute significantly to morbidity and mortality worldwide, but etiological diagnosis based on clinical parameters is difficult2. Rapid detection and isolation of the virus are important for public health action, such as epidemic prediction and prevention3,4. For laboratory research and hygiene surveys, influenza infection detection has been based on the use of conventional methods such as observation of infected cell death5,6 viral antigen staining7,8, and viral gene detection. Since infection by some influenza viruses, mainly clinical isolates, is very difficult to detect due to weak ability to induce cell death, determination of infection only by observation of infected cell death is often inaccurate. Such a virus does not easily form plaques on an agarose overlay medium (generally called plaque assay), resulting in difficulty in virus titration and virus isolation. A method for immunochemical staining of the viral antigen is required for fixation pretreatment of cells and a specific antibody against each virus9. Because of the continuous variability of viral antigens, preparing a specific anti-virus antibody for every virus epidemic could be extremely hard work. A method for detection of the viral gene is the most sensitive and specific. However, the high variability of a viral gene might give rise to a mismatch of primer sequences. Also, a gene detection method requires costly equipment. An immunohistochemical method enables local detection of infected cells and regions, but a gene detection method does not. Local detectability of virus infection is a very important factor for obtaining virus infection information and for virus titration and virus isolation in laboratory research and hygiene surveys.
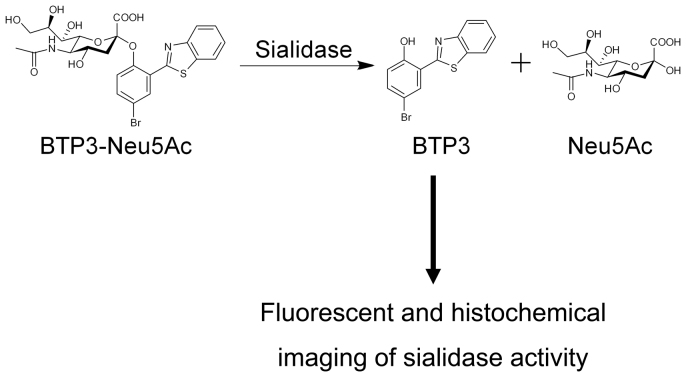
A benzothiazolylphenol derivative (named BTP3) is a crystalline, water-insoluble, acid-resistant, and fluorescently stable compound (Ex/Em = 372 nm/526 nm)10,11. The large Stokes shift of BTP3 is advantageous for preventing effects of excitation light, self-absorption, and noise fluorescence derived from cells and tissues. We have developed a new fluorescent sialidase substrate of BTP3 with N-acetylneuraminic acid (Neu5Ac), 2-(benzothiazol-2-yl)-4-bromophenyl 5-acetamido-3,5-dideoxy-α-D-glycero-D-galacto-2-nonulopyranosidonic acid (named BTP3-Neu5Ac), for performing fluorescent imaging of mammalian sialidase activity in tissues and cells, such as rat brain tissues and human cancer cells, that show elevated sialidase activity11. Water-soluble BTP3-Neu5Ac produces water-insoluble BTP3 by sialidase reaction. Although BTP3-Neu5Ac itself is not fluorescent, BTP3 should be locally deposited in a location of sialidase activity, enabling fluorescent and histochemical imaging of sialidase activity (Fig. 1). Influenza A and B viruses have a viral surface neuraminidase glycoprotein (NA), which shows high sialidase activity12 and is expressed on the surface membranes of infected cells13,14,15,16. In the late step of virus infection, the viral NA accumulates on the surface membranes of infected cells. This accumulation significantly increases sialidase activity on the surface membranes of infected cells, suggesting that BTP3-Neu5Ac would enable rapid and sensitive detection of infected cells. Although 4-methylumbelliferyl-Neu5Ac (4MU-Neu5Ac)17,18,19,20 and 1,2-dioxetane derivative of Neu5Ac (NA-star)21 are conventionally used as highly sensitive sialidase substrates, these substrates produce a water-soluble fluorescent and chemiluminescent compound, respectively, and cannot be used for local detection of sialidase activity in cells or tissues. Here, we show the usefulness of BTP3-Neu5Ac for histochemical imaging of sialidase activity on the infected cells with no need for cell fixation or a specific anti-virus antibody.
Figure 1. Reaction scheme of BTP3-Neu5Ac for fluorescent and histochemical imaging of influenza virus sialidase activity.
Water-soluble BTP3-Neu5Ac is separated into Neu5Ac and BTP3 by sialidase activity. The water-insoluble fluorescent compound BTP3 is deposited at the location of sialidase activity and illuminates green fluorescence by UV irradiation (Ex/Em = 372 nm/526 nm).
Results
Fluorescent visualization of influenza A virus sialidase activity by staining with BTP3-Neu5Ac
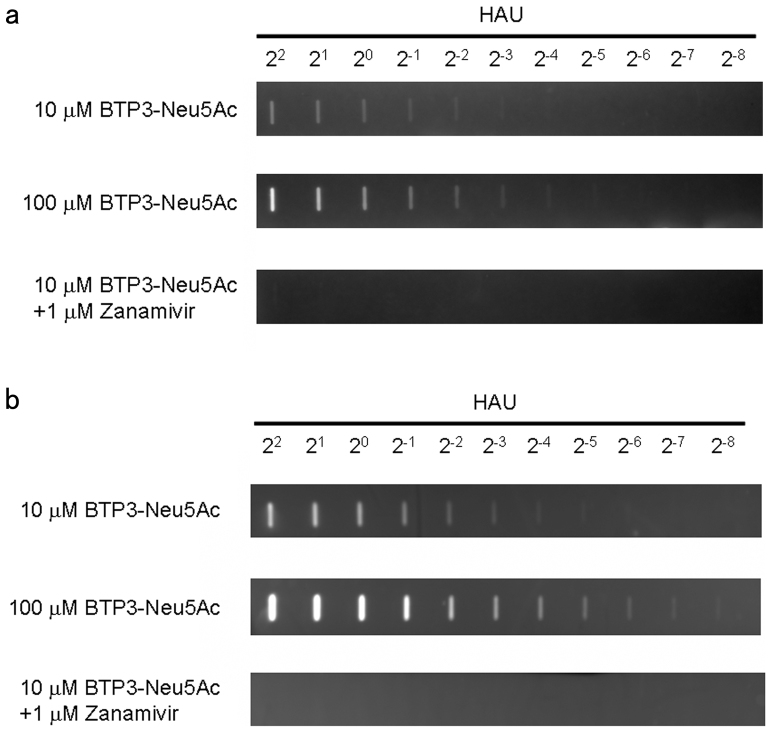
To confirm the substrate reactivity of BTP3-Neu5Ac for sialidase activity of influenza A virus NA, we performed fluorescent visualization of influenza A virus blotted on polyvinylidene difluoride (PVDF) membranes with BTP3-Neu5Ac. Virus-blotted membranes were incubated with 10 or 100 μM BTP3-Neu5Ac at 37°C for 10 min or 1 hr. The use of 10 μM and 100 μM BTP3-Neu5Ac for 10 min enabled fluorescent detection up to 2−3 and 2−4 in viral hemagglutination unit (HAU) titer, respectively (Fig. 2a). Also, the use of 10 μM and 100 μM BTP3-Neu5Ac for 1 hr enabled fluorescent detection up to 2−5 and more than 2−8 in viral HAU titer, respectively (Fig. 2b). The fluorescent detection was incubation time- and viral dose-dependent and was completely inhibited in the presence of zanamivir, a specific sialidase inhibitor of influenza A and B viruses (Fig. 2a and 2b). These results indicated that BTP3-Neu5Ac was a suitable substrate for very rapid (within 10 min) and sensitive detection of influenza A virus sialidase activity.
Figure 2. Fluorescent visualization of influenza A virus blotted on a PVDF membrane by using the BTP3-Neu5Ac assay.
A PVDF membrane was dot-blotted with 2-fold dilutions (22 to 2−8 HAU) of avian influenza A virus strain A/duck/Hong Kong/313/4/1978 (H5N3). The membrane was incubated with 10 and 100 μM BTP3-Neu5Ac at 37°C for 10 min (a) or 1 hr (b) in the absence or presence of 1 μM zanamivir.
Histochemical visualization of virus-infected cells by using the BTP3-Neu5Ac assay
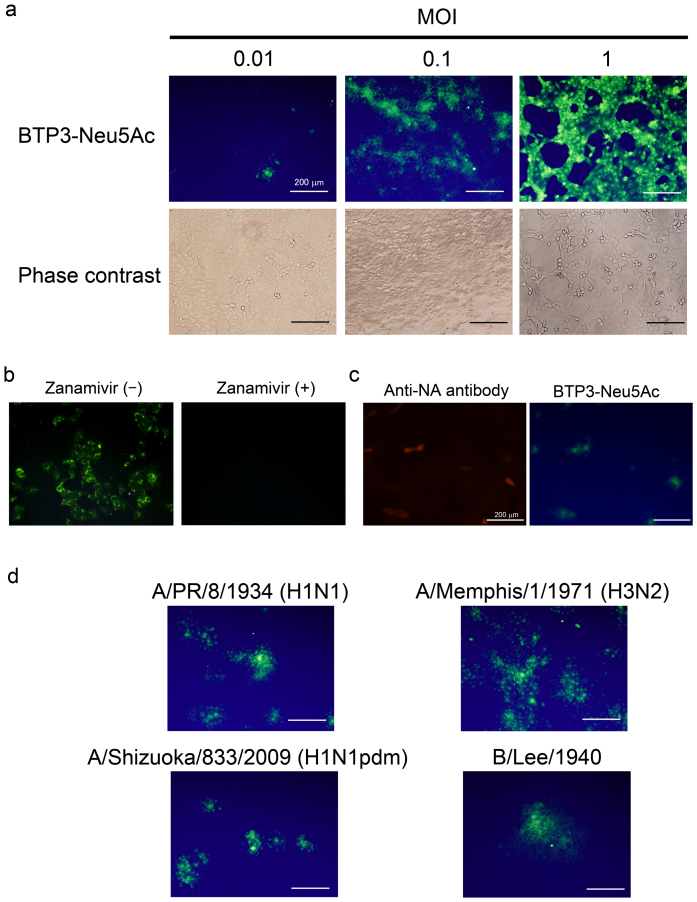
We examined the usefulness of BTP3-Neu5Ac for histochemical visualization of influenza virus-infected cells. Madin-Darby canine kidney (MDCK) cells were infected with avian influenza A virus strain A/duck/Hong Kong/313/4/1978 (H5N3) for 12 hr and incubated with BTP3-Neu5Ac at 37°C for 10 min. The infected cells were fluorescently and histochemically visualized in a virus dose-dependent manner (Fig. 3a). The fluorescence was completely inhibited in the presence of zanamivir (Fig. 3b), indicating that the fluorescence with BTP3-Neu5Ac was dependent on the viral sialidase activity but not on endogenous mammalian sialidase activity. Moreover, a fluorescence image with BTP3-Neu5Ac coincided with an immunohistochemical image with anti-influenza A virus NA monoclonal antibody (Fig. 3c). Sialidase activity of the fixed cells expressing NA remained approximately 66% of that of the non-fixed cells expressing NA (see Supplementary Fig. S1 online). Cells infected with old human influenza A virus strains [A/PR/8/1934 (H1N1) and A/Memphis/1/1971 (H3N2)], a recent clinical strain [A/Shizuoka/833/2009 (H1N1pdm)], and influenza B virus strain (B/Lee/1940) were clearly detected by the BTP3-Neu5Ac assay (Fig. 3d). The results indicate that the BTP3-Neu5Ac assay is applicable to sialidase activities of all influenza viruses, regardless of the host, surface antigens (subtypes), and internal antigens (A and B types). We also obtained high magnification images of intracellular staining of fixed infected cells with BTP3-Neu5Ac by using confocal laser microscopy. In infected cells at 8 hr postinfection, distribution of viral NA proteins coincided with fluorescence of BTP3 (Fig. 4).
Figure 3. Histochemical visualization of virus-infected cells by using the BTP3-Neu5Ac assay.
(a) MDCK cells were infected with avian influenza A virus strain A/duck/Hong Kong/313/4/1978 (H5N3) at a multiplicity of infection (MOI) of 0.01 to 1. After culture for 12 hr, the infected cells were incubated with 10 μM BTP3-Neu5Ac at 37°C for 10 min. (b) MDCK cells were infected with A/duck/Hong Kong/313/4/1978 (H5N3) and cultured for 12 hr. The infected cells were incubated with 10 μM BTP3-Neu5Ac in the absence or presence of zanamivir at 37°C for 10 min. (c) MDCK cells were infected with A/duck/Hong Kong/313/4/1978 (H5N3) and cultured for 10 hr. The infected cells were fixed with 4% paraformaldehyde-PBS and immunostained with mouse anti-NA monoclonal antibody (red). Then the immunostained cells were incubated with 10 μM BTP3-Neu5Ac at 37°C for 3 min (green). (d) MDCK cells were infected with influenza A virus strains [A/PR/8/1934 (H1N1), A/Memphis/1/1971 (H3N2), and A/Shizuoka/833/2009 (H1N1pdm)], and influenza B virus strain (B/Lee/1940). After culture for 12 hr, the infected cells were incubated with 10 μM BTP3-Neu5Ac at 37°C for 10 min. Scale bars indicate 200 μm.
Figure 4. Intracellular staining of infected cells with BTP3-Neu5Ac.
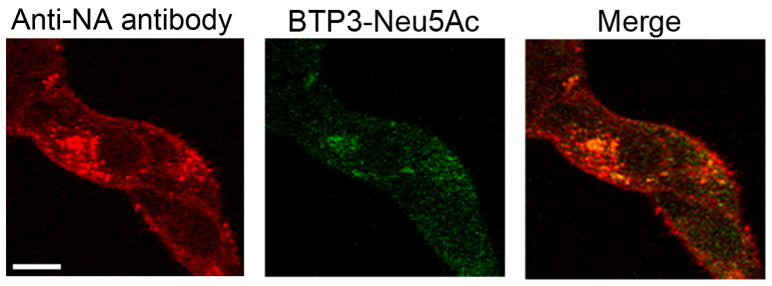
The cells were infected with A/duck/Hong Kong/313/4/1978 (H5N3) and incubated at 37°C for 8 hr. The infected cells were fixed with paraformaldehyde and permeabilized. The cells were double-stained by anti-NA antibody and BTP3-Neu5Ac. Fluorescent images were obtained by confocal laser microscopy. A bar indicates 10 μm.
Histochemical visualization of genetically NA-expressed cells by using the BTP3-Neu5Ac assay
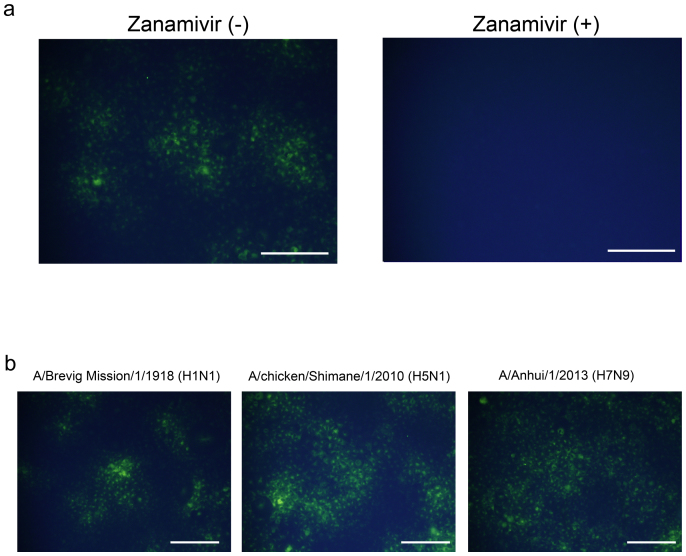
We checked the usefulness of the BTP3-Neu5Ac assay for histochemical visualization of genetically NA-expressed cells. We transfected the viral NA gene into African green monkey kidney (COS7) cells and stained the NA-expressed cells with 10 μM BTP3-Neu5Ac. The NA-expressed cells of A/duck/Hong Kong/313/4/1978 (H5N3) were histochemically visualized at 37°C for 5 min. The fluorescence was completely inhibited in the presence of zanamivir, indicating that the fluorescence with BTP3-Neu5Ac was dependent on sialidase activity of genetically expressed NA (Fig. 5a). All NA-expressed cells, including cells expressing 1918 pandemic Spanish flu NA [A/Brevig Mission/1/1918 (H1N1)], highly pathogenic avian influenza A virus NA [A/chicken/Shimane/1/2010 (H5N1)], and H7N9 avian influenza A virus NA isolated from humans in 2013 [A/Anhui/1/2013 (H7N9)], were clearly detected by the BTP3-Neu5Ac assay (Fig. 5b). These results strongly suggest that the BTP3-Neu5Ac assay is applicable to sialidase activities of new subtype (pandemic) viruses among humans and to avian viruses including highly pathogenic avian viruses.
Figure 5. Histochemical visualization of genetically NA-expressed cells by using the BTP3-Neu5Ac assay.
(a) COS7 cells were transfected with the NA gene of A/duck/Hong Kong/313/4/1978 (H5N3). After culture for 24 hr, the transfected cells were incubated with 10 μM BTP3-Neu5Ac in the absence or presence of 1 μM zanamivir at 37°C for 5 min. (b) COS7 cells were transfected with each NA gene of A/Brevig Mission/1/1918 (H1N1), A/chicken/Shimane/1/2010 (H5N1), and A/Anhui/1/2013 (H7N9). After culture for 24 hr, the transfected cells were incubated with 10 μM BTP3-Neu5Ac at 37°C for 5 min. Scale bars indicate 200 μm.
Histochemical visualization of viral focus formation by using the BTP3-Neu5Ac assay
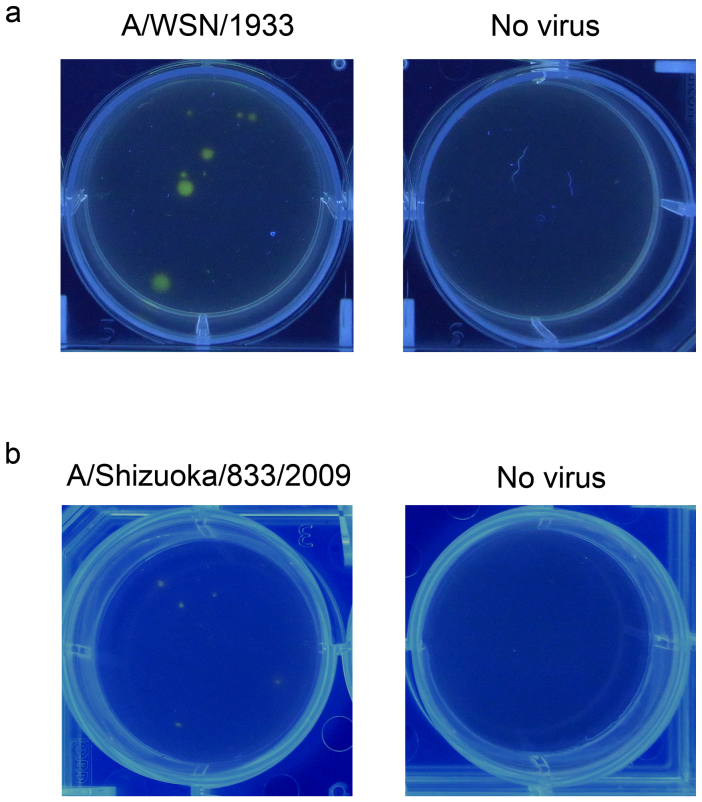
The plaque assay is a traditional method for virus titration and isolation. Influenza A and B viruses form plaques on an agarose-overlaid cell monolayer through cell destruction by virus-induced apoptosis. However, some of the viruses do not show obvious sizes of plaques. Therefore, we examined the usefulness of BTP3-Neu5Ac for fluorescence visualization of viral focus formation. MDCK cells were infected with influenza A virus strain A/WSN/1933 (H1N1) and cultured for 2 days after overlaying on an agarose-containing medium. By dropping BTP3-Neu5Ac solution onto the agarose-containing medium, localized clusters of the virus-infected cells (foci) at the bottom of a plate were fluorescently and distinctly visualized under UV irradiation (Fig. 6a). Furthermore, the viruses isolated from the BTP3-stained focus retained viral growth ability (data not shown), confirming the useful availability of BTP3-Neu5Ac for virus isolation. Most clinical influenza virus isolates do not form clear plaques. We also confirmed that the BTP3-Neu5Ac assay was applicable to fluorescence visualization of viral foci of the recent clinical influenza A virus strain A/Shizuoka/833/2009 (H1N1pdm), which did not show clear plaque formation (Fig. 6b). The BTP3-Neu5Ac assay would be a powerful tool for detection of viruses showing weak or no clear plaque-forming ability in the traditional plaque assay, contributing to the effectiveness of virus titration and isolation of such viruses.
Figure 6. Fluorescent visualization of viral focus formation using BTP3-Neu5Ac.
MDCK cells were infected with influenza A virus strain A/WSN/1933 (H1N1) (a) or A/Shizuoka/833/2009 (H1N1pdm) (b). The infected cells were overlaid with a serum-free medium (SFM) containing 0.8% agarose and 2 μg/ml acetylated trypsin. After culture for 2 days (a) or 3 days (b), 100 μl of 200 μM BTP3-Neu5Ac was dropped onto the overlaid agarose-containing medium. After incubation at 37°C for 15 min, the plate was observed under UV irradiation at 354 nm.
Histochemical visualization of virus-infected tissue in the mouse lung by using the BTP3-Neu5Ac assay
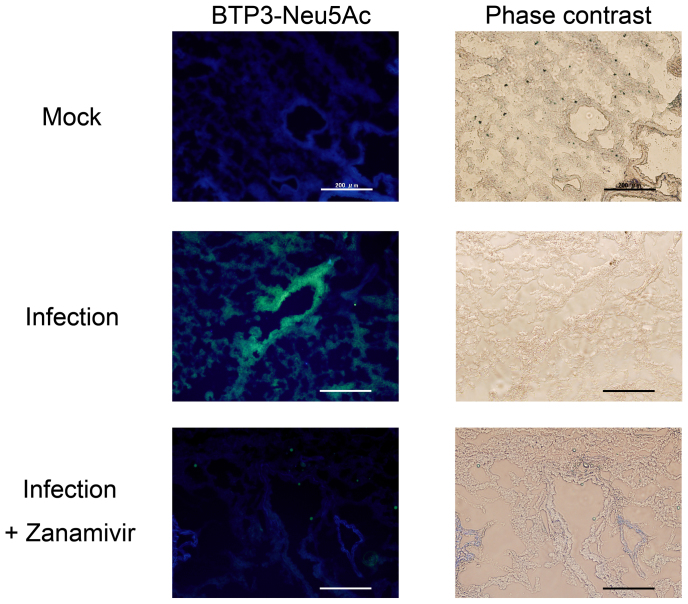
We examined the usefulness of BTP3-Neu5Ac for histochemical visualization of virus-infected tissues in vivo. Mice were intranasally infected with influenza A virus strain A/PR/8/1934 (H1N1). After fixation with paraformaldehyde, lung tissues at 1 day postinfection were sliced into 10-μm-thick sections and incubated with BTP3-Neu5Ac at 37°C for 20 min. Distinct green fluorescence was observed at bronchiole mucosa in virus-infected lung sections but not in mock-infected lung sections (Fig. 7). When the virus-infected lung sections were incubated with 20 μM BTP3-Neu5Ac in the presence of 1 μM zanamivir, the fluorescence with BTP3-Neu5Ac was completely inhibited (Fig. 7), confirming specificity of the BTP3-Neu5Ac assay for influenza A virus sialidase. These results indicated that the imaging assay of sialidase activity using BTP3-Neu5Ac is also applicable to virus-infected tissues in vivo.
Figure 7. Histochemical visualization of virus-infected tissue in the mouse lung by using the BTP3-Neu5Ac assay.
Mice were intranasally infected with influenza A virus strain A/PR/8/1934 (H1N1). After fixation with 4% paraformaldehyde-PBS, lung sections at 1 day postinfection were incubated with 20 μM BTP3-Neu5Ac at 37°C for 20 min. Mock lung sections were from mice intranasally inoculated with PBS only. Scale bars indicate 200 μm. The lung sections were incubated with 20 μM BTP3-Neu5Ac in the presence of 1 μM zanamivir to confirm specificity of the BTP3-Neu5Ac assay for influenza A virus sialidase. Scale bars indicate 200 μm.
Discussion
Histochemical detection of influenza virus infection provides very important information for both laboratory research and hygiene survey. Such detection has conventionally been performed by an immunochemical method, which requires a specific anti-virus antibody in addition to cell fixation. In the present study, we established an easy, rapid, and sensitive histochemical detection method using a novel fluorescent sialidase substrate, BTP3-Neu5Ac, with no need for a specific anti-virus antibody or cell fixation. Generally, an immunohistochemical method takes at least 1 hr (30 min each for primary and secondary antibody reactions) until complete detection of antigens, whereas the BTP3-Neu5Ac assay is almost completed within 5 to 20 min. The protocol is also much easier because only replacement of the culture supernatant with a BTP3-Neu5Ac-containing medium or buffer is needed for the BTP3-Neu5Ac assay, unlike the conventional immunohistochemical method. RT-PCR is usually the most sensitive and specific method for virus gene detection and virus subtype elucidation. However, this method is unsuitable for histochemical detection of virus infection. Since the RT-PCR method needs an extremely specific primer to detect the target gene, detection for new subtype virus occurrence is predicted to be somewhat inconvenient. The BTP3-Neu5Ac assay should be applicable to all influenza viruses possessing sialidase activity, regardless of subtype, host, and gene variability. The RT-PCR method needs a more complicated protocol compared to the BTP3-Neu5Ac assay. Thus, the new assay has many advantages compared to the conventional methods for detection of virus infection. Fluorescence staining also has general advantages: it is more sensitive than observation with visible light and can be used for multi-color staining, though fluorescent observation of BTP3 requires preparation of appropriate excitation and emission filters, and UV excitation provokes large autofluorescence, which can be distinguished from fluorescence of BTP3 by its large Stokes shift.
Many hygiene survey facilities often use RT-PCR for primary virus detection. However, for the purpose of virus isolation, some facilities are still using optical microscopic observation of virus-induced cell death in cell culture to which each clinical sample has been added for primary virus detection. In such facilities, the BTP3-Neu5Ac assay would be an extremely convenient tool for primary virus screening because of its easiness, rapidness, and sensitiveness. In virus titration and isolation, the BTP3-Neu5Ac assay would also be useful for clear visualization of the viral foci, even though viral plaques are often invisible due to weak virus-inducing apoptosis ability, which is often observed on clinical isolates. Since BTP3 and BTP3-Neu5Ac show no cytotoxicity11, virus isolation and proliferation can be started immediately and directly from a fluorescently visualized foci. In the case of screening of influenza A and B viruses by using the BTP3-Neu5Ac assay, the virus can be confirmed with inhibition of fluorescence by zanamivir, which is a specific sialidase inhibitor against influenza A and B viruses. A zanamivir-resistant virus or a sialidase-deficient virus has not yet become epidemic in humans, birds, pigs, and horses. For that reason, in these species, there is no natural influenza A or B virus that can escape from detection with the BTP3-Neu5Ac assay.
To fluorescently visualize virus infection in living cells, influenza A viruses carrying a reporter gene such as green fluorescent protein (GFP) have been created by reverse genetics22,23,24. For example, once cells are infected with a virus carrying GFP as a reporter gene, the infected cells are confirmed by green fluorescence from GFP expression, with no need for fixation or an anti-virus antibody. However, this method is only applicable to genetically artificial viruses. On the other hand, it is expected that the use of BTP3-Neu5Ac will enable detection of all influenza A and B virus infections showing sialidase activity in living cells, regardless of whether the strains are natural or artificial, with no need for fixation or an anti-virus antibody.
We previously demonstrated fluorescent histochemical visualization of the sialidase activity of influenza A virus using 5-bromo-4-chloroindol-3-yl-Neu5Ac (X-Neu5Ac) and Fast Red Violet LB25. However, this method has two major disadvantages: (i) its sensitivity to sialidase activity is very low due to the requirement of two reactions of sialidase and oxidation before staining, and nonspecific signals are therefore easily detected due to the long reaction time, and (ii) oxidation reaction to generate a fluorescent compound is less efficient at pH of more than 5. This condition is unsuitable for sialidase of most seasonal human and swine viruses and highly pathogenic H5N1 avian viruses, which lose sialidase activity at pH below 5 because of their low-pH-unstable NA26,27,28,29,30,31. Since BTP3-Neu5Ac is a rapid and sensitive sialidase substrate of influenza virus, independent of pH, there is no problem regarding these issues for X-Neu5Ac10.
In the present study, we fixed cells with paraformaldehyde for immunohistochemical staining (Fig. 3c) and for infected mouse tissue staining (Fig. 7) before incubation with BTP3-Neu5Ac, and we detected sialidase activity of the fixed cells with BTP3-Neu5Ac. All other assays were performed in non-fixed live cells. We also previously reported that viral sialidase activity was detected after fixation with paraformaldehyde25. Moreover, sialidase activity of the fixed cells expressing NA remained approximately 66% of that of the non-fixed cells expressing NA (see Supplementary Fig. S1 online). Paraformaldehyde is thought to sustain protein structure to some extent, enabling determination of protein function such as enzyme activity.
Sialidase activity of influenza virus NA is known to facilitate release of progeny viruses from the surface membranes of infected cells and to prevent self-aggregation among progeny viruses by cleavage of sialic acid from sialo-glycoconjugates on the surface membranes of infected cells and on viral surface glycoproteins. However, some studies have suggested that NA plays an important role for not only these steps but also for the cell entry step of a virus under the condition of receptor attachment and endocytosis25,29,31,32,33,34. To investigate novel roles of viral sialidase activity, it is necessary to develop a breakthrough method to trace viral sialidase activity within the infected cells, especially during the process of viral cell entry. We are trying to establish such a method using BTP3-Neu5Ac derivatives. For both virological research and hygiene surveys, if BTP3-Neu5Ac becomes commercially available, it would be extremely useful tool for easy, rapid, and sensitive histochemical detection of influenza A and B virus infections in vitro and in vivo. We are now studying the usefulness of BTP3-Neu5Ac for other viruses that have sialidase, such as Newcastle disease virus, Sendai virus, mumps virus, and human parainfluenza virus. Even if these viruses are detected by the BTP3-Neu5Ac assay, influenza A and B viruses can be distinguished from other virus infections by fluorescent inhibition using a specific sialidase inhibitor such as zanamivir. Further studies using BTP3-Neu5Ac should greatly improve the efficiency of detection, titration, and isolation of influenza A and B viruses and may contribute to progress in research on their NAs.
Methods
Cells and viruses
MDCK cells were grown in minimum essential medium (MEM) supplemented with 5% fetal bovine serum. COS7 cells were grown in Dulbecco's MEM supplemented with 10% fetal bovine serum. Influenza A virus strains, A/duck/Hong Kong/313/4/1978 (H5N3), A/WSN/1933 (H1N1), A/PR/8/1934 (H1N1), and A/Memphis/1/1971 (H3N2), were propagated in the allantoic sacs of 10-day-old embryonated eggs and purified by sucrose density gradient centrifugation as described previously35. Influenza A virus strain A/Shizuoka/833/2009 (H1N1pdm) and influenza B virus strain B/Lee/1940 were grown in MDCK cells. Hemagglutination units (HAU) of the viruses were determined as described previously36. HAU were expressed as the highest dilution of the virus suspension giving complete agglutination of guinea pig erythrocytes.
Histochemical visualization of influenza A virus blotted on a membrane by using the BTP3-Neu5Ac assay
PVDF membrane was activated with methanol and washed twice 10 min each time. Two-fold dilutions of virus suspensions were dot-blotted on PVDF membranes with a Hybri. Slot 24 slot blotting apparatus (Core Life Sciences, California, USA). The virus blotted on the PVDF membrane was washed three times and incubated with 10 or 100 μM BTP3-Neu5Ac at 37°C for 10 min or 1 hr. Images of the virus-blotted membrane were obtained during UV irradiation by using Lumivision Pro HR (AISIN SEIKI Co., Ltd., Aichi, Japan) with a DR655 green enhancer filter. To confirm specificity of the BTP3-Neu5Ac assay for detection of influenza virus sialidase activity, the virus-blotted membrane was incubated with 10 μM BTP3-Neu5Ac in the presence of 1 μM zanamivir, a specific sialidase inhibitor of influenza A and B viruses.
Focus-forming assay
Virus titers were determined by an endpoint dilution and immunohistochemical focus-forming assay as described previously29. Briefly, viruses were serially diluted in a serum-free medium, Hybridoma-SFM (SFM; Invitrogen Corp., California, USA). MDCK cells on a 6-well plate were infected with 1 ml of the virus dilutions at 37°C for 30 min. After washing with phosphate buffered saline (PBS), the cells were cultured in SFM containing 1.2% Avicel (FMC BioPolymer, Pennsylvania, USA) and 2 μg/ml acetylated trypsin for an additional 24 hr37. After fixing with methanol, cells were incubated with anti-influenza A virus nucleoprotein (NP) monoclonal antibody (4E6)38, followed by incubation with horseradish peroxidase-conjugated goat anti-mouse IgG + M. The infected cells were stained as described previously14,29,35. Virus titers [focus-forming units (ffu)] were measured by counting viral foci.
Histochemical visualization of infected cells by using the BTP3-Neu5Ac assay
MDCK cells on a 24-well plate (1 × 105 cells/well) were cultured for 24 hr. The cells were infected with influenza A virus strain A/duck/Hong Kong/313/4/1978 (H5N3) at a multiplicity of infection (MOI) of 0.01 to 1 in 250 μl/well of SFM at 37°C for 30 min. After washing with PBS again, the cells were cultured in 500 μl/well of SFM at 37°C for an additional 12 hr. After wash with PBS, the infected cells were incubated with 10 μM BTP3-Neu5Ac in SFM at 250 μl/well at 37°C for 10 min. For an additional experiment using zanamivir, MDCK cells were infected with A/duck/Hong Kong/313/4/1978 (H5N3) at an MOI of 0.1 and cultured in SFM at 37°C for an additional 12 hr. The infected cells were incubated with 10 μM BTP3-Neu5Ac in the absence or presence of 1 μM zanamivir at 37°C for 10 min. For immunohistochemical detection of infected cells, MDCK cells were infected with A/duck/Hong Kong/313/4/1978 (H5N3) at an MOI of 0.001 and cultured at 37°C for 12 hr. The infected cells were fixed with 4% paraformaldehyde-PBS at room temperature for 10 min and incubated with mouse anti-A/duck/Hong Kong/313/4/1978 (H5N3) NA monoclonal antibody (4D12D5; no NA neutralizing activity), generated as described previously39, at room temperature for 30 min. The infected cells were incubated with tetramethyl rhodamine (TRITC)-conjugated goat anti-mouse IgG secondary antibody (Sigma-Aldrich Corp., Missouri, USA). After immunohistochemical reaction, the infected cells were incubated with 10 μM BTP3-Neu5Ac at 37°C for 3 min. For additional 4 influenza A and B virus strains [A/PR/8/1934 (H1N1), A/Memphis/1/1971 (H3N2), A/Shizuoka/833/2009 (H1N1pdm), and B/Lee/1940], MDCK cells were infected with each virus at an MOI of 0.01 to 0.1 and cultured at 37°C for 12 hr. The infected cells were incubated with 10 μM BTP3-Neu5Ac at 37°C for 10 min. Fluorescent images were obtained during UV irradiation by using an IX71 fluorescence microscope (Olympus Co., Ltd., Tokyo, Japan) equipped with a fluorescent filter (U-MWU2, DM400, BP336-385, and BA420 for BTP3 fluorescence or U-MWIG3, DM570, BP530–550, and BA575IF for TRITC fluorescence).
Intracellular double-staining of infected cells with anti-NA antibody and BTP3-Neu5Ac
MDCK cells in an 8-well Teflon printed glass slide plate (1 × 103 cells/well) were cultured at 37°C for 24 hr. The cells were infected with A/duck/Hong Kong/313/4/1978 (H5N3) at an MOI of 100 at 37°C for 30 min. After washing with PBS, the cells were cultured in SFM at 37°C for 8 hr. The infected cells were fixed with 4% paraformaldehyde-PBS at room temperature for 30 min and permeabilized with 0.05% TritonX-100-PBS at room temperature for 30 min. The cells were incubated with 10 μM BTP3-Neu5Ac at room temperature for 30 min. After washing with PBS, the cells were incubated with mouse anti-NA monoclonal antibody (4D12D5) against A/duck/Hong Kong/313/4/1978 (H5N3) at room temperature for 30 min. The cells were incubated with TRITC-conjugated goat anti-mouse IgG secondary antibody (Sigma-Aldrich Corp., Missouri, USA). After adding SlowFade Gold antifade reagent (Invitrogen Corp., California, USA), fluorescent images were obtained by using an LSM510meta confocal laser microscope (Carl Zeiss, Oberkochen, Germany).
Histochemical visualization of genetically NA-expressed cells by using the BTP3-Neu5Ac assay
We used an expression plasmid vector containing the NA gene from influenza A virus strains A/duck/Hong Kong/313/4/1978 (H5N3), A/Brevig Mission/1/1918 (H1N1), A/chicken/Shimane/1/2010 (H5N1), and A/Anhui/1/2013 (H7N9). The NA gene of A/duck/Hong Kong/313/4/1978 (H5N3) was amplified from viral RNA by RT-PCR method and inserted into EcoR I site of the expression pCAGGS/MCS vector27,28. The pCAGGS vector containing the NA gene of A/Brevig Mission/1/1918 (H1N1) has been described previously29. The NA gene of A/chicken/Shimane/1/2010 (H5N1) was amplified from viral RNA by RT-PCR method, which was kindly provided by Dr. Yuko Uchida and Dr. Takehiko Saito (National Institute of Animal Health in Japan), and inserted into the restriction enzyme sites between EcoR I site and Sph I site of the pCAGGS/MCS vector. The NA gene of A/Anhui/1/2013 (H7N9) was PCR-amplified from pHH21 vector containing the NA gene, which was kindly provided by Dr. Yoshihiro Kawaoka (University of Wisconsin and University of Tokyo), and inserted into the restriction enzyme sites between Kpn I site and Sph I site of the pCAGGS/MCS vector. Primer sequences are available upon request.
COS7 cells on a 48-well plate (5 × 104 cells/well) were cultured for 24 hr. The cells were transfected with the pCAGGS vector containing the NA gene (50 ng/well) using transfection reagent Trans-IT LT1 (Mirus, Wisconsin, USA), according to manufacturer's instructions. After 24 hr, the transfected cells were incubated with 150 μl/well of SFM containing 10 μM BTP3-Neu5Ac at room temperature for 5 min. Fluorescent images were obtained by using a fluorescent microscope.
Fluorescent focus-forming assay using BTP3-Neu5Ac
A monolayer of MDCK cells in a 6-well plate was infected with serial dilutions of influenza A virus strain A/WSN/1933 (H1N1) or A/Shizuoka/833/2009 (H1N1pdm) for 30 min and covered with 2 ml/well of an overlay SFM medium containing 0.8% agarose and 2 μg/ml acetylated trypsin. After 2 or 3 days at 37°C, 100 μl of 200 μM BTP3-Neu5Ac was dropped onto the overlay medium and incubated at 37°C for 20 min. The plate was observed under a UV illuminator at 354 nm.
Histochemical visualization of lungs from influenza A virus-infected mice by using the BTP3-Neu5Ac assay
Specific pathogen-free 6-to-8-week-old female mice (BALB/c) were intranasally infected with 25 μl (1 × 107 ffu)/mouse of influenza A virus strain A/PR/8/1934 (H1N1). Lung tissues from virus-infected mice at 1 day postinfection were fixed with 4% paraformaldehyde-PBS at room temperature for 10 min and embedded in Tissue-Tek O.C.T. compound (Sakura Finetechnical Co., Ltd., Tokyo, Japan). After being frozen, sections were cut at 10 μm in thickness at −25°C, affixed on glass slides, and fixed in 4% paraformaldehyde-PBS for 15 min. After wash with PBS, the lung sections were incubated with 20 μM BTP3-Neu5Ac in 10 mM acetate buffer (pH 6.0) containing 1 mM calcium chloride at 37°C for 20 min. To confirm viral sialidase specificity of BTP3-Neu5Ac for fluorescent imaging of virus-infected tissues, a viral sialidase inhibitory experiment using 1 μM zanamivir was performed simultaneously. All animal experiment procedures were carried out with institutional approval.
Author Contributions
Y. Kurebayashi designed and performed experiments, analyzed data, and wrote the main manuscript text as the co-first author. T.T. designed and supervised experiments, analyzed data, and wrote the main manuscript text as the co-first author. S.T. and M.T. designed and performed experiments. T.A., T. Sato and Y.M. performed experiments. A.M. and H.K. advised on animal experiments. T.O. and K.I. designed and synthesized BTP3-Neu5Ac. Y.U. and T. Saito provided H5N1 virus gene; Y. Kawaoka provided H7N9 virus gene; T.Y. and F.K. provided H1N1pdm virus. R.T. and M.V.I. provided zanamivir. T.S. designed and supervised experiments, and reviewed and submitted the manuscript as the corresponding author.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by a grant form Nakatani Foundation for Advancement of Measuring Technologies in Biomedical Engineering, the Kurata Memorial Hitachi Science and Technology Foundation, the Futaba Electronics Memorial Foundation, Tokai Foundation for Technology, Takahashi Industrial and Economic Research Foundation, MEXT/JSPS KAKENHI Grant (challenging Exploratory Research; 26670064), in part, by a MEXT/JSPS KAKENHI Grant (Scientific Research C; 23590549).
References
- Taubenberger J. K. & Morens D. M. The pathology of influenza virus infections. Annu. Rev. Pathol. 3, 499–522 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxas M. & Jurenka J. Colds and influenza: a review of diagnosis and conventional, botanical, and nutritional considerations. Altern. Med. Rev. 12, 25–48 (2007). [PubMed] [Google Scholar]
- Torner N., Soldevila N., Martínez A., Pumarola T. & Dominguez A. Timely Prediction of Peak Seasonal Influenza Activity Estimation Using Sentinel Surveillance Data. Public Heal. Res. 2, 53–57 (2012). [Google Scholar]
- Ortiz J. R. et al. Strategy to enhance influenza surveillance worldwide. Emerg. Infect. Dis. 15, 1271–1278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung G. D. Diagnostic virology: from animals to automation. Yale J. Biol. Med. 57, 727–733 (1984). [PMC free article] [PubMed] [Google Scholar]
- Sidwell R. W. & Smee D. F. In vitro and in vivo assay systems for study of influenza virus inhibitors. Antiviral Res. 48, 1–16 (2000). [DOI] [PubMed] [Google Scholar]
- Pascua P. N. Q. et al. Virulence and transmissibility of H1N2 influenza virus in ferrets imply the continuing threat of triple-reassortant swine viruses. Proc. Natl. Acad. Sci. U. S. A. 109, 15900–15905 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya K. et al. Avian flu: influenza virus receptors in the human airway. Nature 440, 435–436 (2006). [DOI] [PubMed] [Google Scholar]
- Amano Y. & Cheng Q. Detection of influenza virus: traditional approaches and development of biosensors. Anal. Bioanal. Chem. 381, 156–164 (2005). [DOI] [PubMed] [Google Scholar]
- Otsubo T. et al. 2-(Benzothiazol-2-yl)-phenyl-β-D-galactopyranoside derivatives as fluorescent pigment dyeing substrates and their application for the assay of β-D-galactosidase activities. Bioorg. Med. Chem. Lett. 23, 2245–2249 (2013). [DOI] [PubMed] [Google Scholar]
- Minami A. et al. Visualization of sialidase activity in Mammalian tissues and cancer detection with a novel fluorescent sialidase substrate. PLoS One 9, e81941 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobasa D., Wells K. & Kawaoka Y. Amino acids responsible for the absolute sialidase activity of the influenza A virus neuraminidase: relationship to growth in the duck intestine. J. Virol. 75, 11773–11780 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 6, 967–974 (2007). [DOI] [PubMed] [Google Scholar]
- Takahashi T. et al. Sulfatide is required for efficient replication of influenza A virus. J. Virol. 82, 5940–5950 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. J. et al. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443, 45–49 (2006). [DOI] [PubMed] [Google Scholar]
- Yang P., Bansal A., Liu C. & Air G. M. Hemagglutinin specificity and neuraminidase coding capacity of neuraminidase-deficient influenza viruses. Virology 229, 155–165 (1997). [DOI] [PubMed] [Google Scholar]
- Nayak D. P. & Reichl U. Neuraminidase activity assays for monitoring MDCK cell culture derived influenza virus. J. Virol. Methods 122, 9–15 (2004). [DOI] [PubMed] [Google Scholar]
- Tomin A., Shkandina T. & Bilyy R. Novel assay for direct fluorescent imaging of sialidase activity. Proc. SPIE. 8087, 80871Z (2011). [Google Scholar]
- Potier M., Mameli L., Bélisle M., Dallaire L. & Melançon S. B. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal. Biochem. 94, 287–296 (1979). [DOI] [PubMed] [Google Scholar]
- Marathe B. M., Lévêque V., Klumpp K., Webster R. G. & Govorkova E. A. Determination of neuraminidase kinetic constants using whole influenza virus preparations and correction for spectroscopic interference by a fluorogenic substrate. PLoS One 8, e71401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton R. C. et al. Development of a sensitive chemiluminescent neuraminidase assay for the determination of influenza virus susceptibility to zanamivir. Anal. Biochem. 280, 291–300 (2000). [DOI] [PubMed] [Google Scholar]
- Tran V., Moser L. A., Poole D. S. & Mehle A. Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. J. Virol. 87, 13321–13329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. et al. Visualizing influenza virus infection in living mice. Nat. Commun. 4, 2369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez J. T., García-Sastre A. & Manicassamy B. Insertion of a GFP reporter gene in influenza virus. Curr. Protoc. Microbiol. 29, 15G.4.1–15G.4.16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T. et al. Sialidase activity of influenza A virus in an endocytic pathway enhances viral replication. J. Virol. 79, 11705–11715 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T. et al. Duck and human pandemic influenza A viruses retain sialidase activity under low pH conditions. J. Biochem. 130, 279–283 (2001). [DOI] [PubMed] [Google Scholar]
- Takahashi T., Suzuki T., Hidari K. I.-P. J., Miyamoto D. & Suzuki Y. A molecular mechanism for the low-pH stability of sialidase activity of influenza A virus N2 neuraminidases. FEBS Lett. 543, 71–75 (2003). [DOI] [PubMed] [Google Scholar]
- Suzuki T. et al. Evolutional analysis of human influenza A virus N2 neuraminidase genes based on the transition of the low-pH stability of sialidase activity. FEBS Lett. 557, 228–232 (2004). [DOI] [PubMed] [Google Scholar]
- Takahashi T. et al. The Low-pH stability discovered in neuraminidase of 1918 pandemic influenza A virus enhances virus replication. PLoS One 5, e15556 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Nidom C. A., Quynh Le M. T., Suzuki T. & Kawaoka Y. Amino acid determinants conferring stable sialidase activity at low pH for H5N1 influenza A virus neuraminidase. FEBS Open Bio 2, 261–266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Song J., Suzuki T. & Kawaoka Y. Mutations in NA that induced low pH-stability and enhanced the replication of pandemic (H1N1) 2009 influenza A virus at an early stage of the pandemic. PLoS One 8, e64439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B. et al. Enhancement of the influenza A hemagglutinin (HA)-mediated cell-cell fusion and virus entry by the viral neuraminidase (NA). PLoS One 4, e8495 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M. N., Matrosovich T. Y., Gray T., Roberts N. A. & Klenk H.-D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 78, 12665–12667 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi M., Asaoka N., Sakai T. & Ohuchi R. Roles of neuraminidase in the initial stage of influenza virus infection. Microbes Infect. 8, 1287–1293 (2006). [DOI] [PubMed] [Google Scholar]
- Suzuki T. et al. Sulphatide binds to human and animal influenza A viruses, and inhibits the viral infection. Biochem. J. 318, 389–93 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Kawagishi S., Masuda M. & Suzuki T. Binding kinetics of sulfatide with influenza A virus hemagglutinin. Glycoconj. J. 30, 709–716 (2013). [DOI] [PubMed] [Google Scholar]
- Matrosovich M., Matrosovich T., Garten W. & Klenk H.-D. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3, 63 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Takaguchi M., Kawakami T. & Suzuki T. Sulfatide regulates caspase-3-independent apoptosis of influenza A virus through viral PB1-F2 protein. PLoS One 8, e61092 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto D. et al. Establishment of a monoclonal antibody directed against Gb3Cer/CD77: a useful immunochemical reagent for a differentiation marker in Burkitt's lymphoma and germinal centre B cells. Glycoconj. J. 14, 379–388 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information