Abstract
Novel culture-independent techniques have recently demonstrated that the lower respiratory tract, historically considered sterile in health, contains diverse communities of microbes: the lung microbiome. A growing literature has demonstrated that a distinct microbiota of the lower respiratory tract is present both in health and in various respiratory diseases, though the biological and clinical significance of these findings remains undetermined. In this article, we review and synthesize published reports of the lung microbiota of healthy and diseased subjects, discuss trends of microbial diversity and constitution across disease states, and look to the extra-pulmonary microbiome for hypotheses and future directions for study.
Keywords: lung microbiome, pulmonary disease, bacteria, microbial diversity, infection, pathogenesis, 16S, pyrosequencing
Introduction
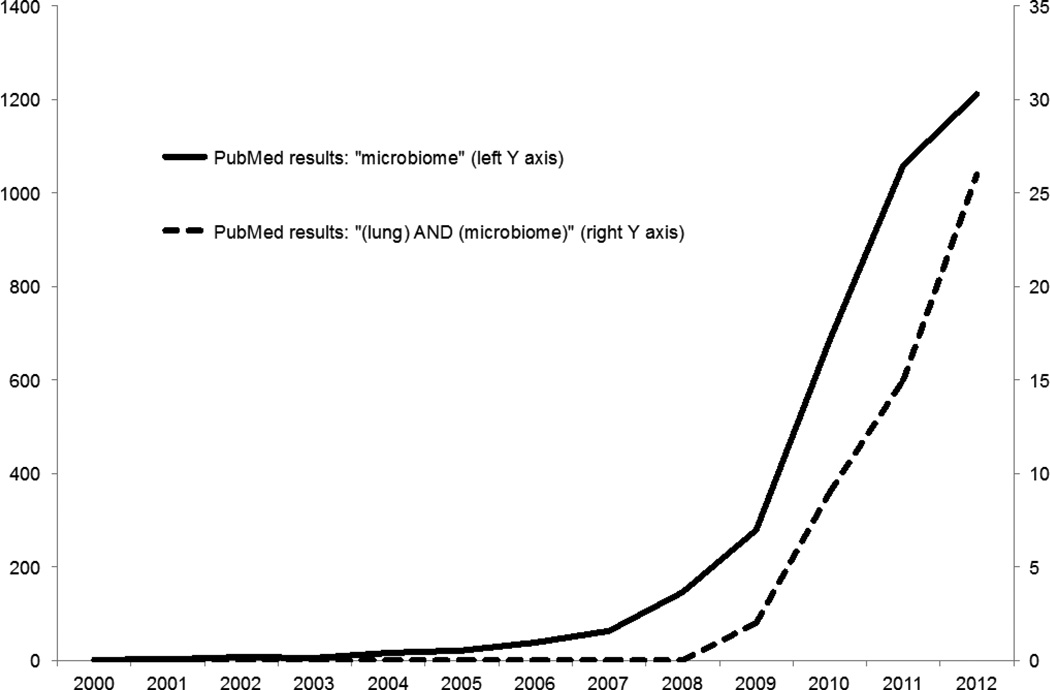
While the lungs have historically been considered sterile in health and were initially omitted from the list of priority organ systems in the Human Microbiome Project (HMP), subsequent studies have since demonstrated that the lower respiratory tract is replete with diverse communities of bacteria both in health and in diseased states. Despite its late start relative to the extra-pulmonary microbiome, the field of lung microbiome studies is rapidly growing and has yielded provocative observations regarding the relationship between lung microbiota and respiratory disease (Figure 1). Study of the lung microbiome may unfold new insights and approaches to the pathogenesis of lung infection, which is consistently a leading cause of worldwide disease burden [1], as well as that of various lung diseases previously thought only indirectly related to microbial pathogenesis.
Figure 1.
PubMed results by year for “microbiome” (left Y axis) and “(lung) AND (microbiome)” (right Y axis)
In the past decade, novel culture-independent techniques of microbial identification have expanded upon our understanding of the abundance and diversity of microbial cells on and within the human body, collectively referred to as the microbiome. A single human subject is host to more than one trillion bacterial cells, and genes present in an individual’s microbiome outnumber his/her own by a factor of 100 [2]. Resident bacteria serve roles in absorption of nutrients, synthesis of vitamins, metabolism of xenobiotics, and regulation of immunity, and derangements in microbiota have been associated with infection, autoimmunity, obesity, cardiovascular disease and malignancy [3]. Recognizing the importance of the microbiome in human health, in 2008 the National Institute for Health (NIH) launched the HMP to characterize the communities of bacteria on and within the human body and to explore their effects on health and disease. This review will discuss our current understanding of (I) the methods used in the analysis of the microbiome, (II) the microbiome of the healthy lung, (III) the microbiome of the diseased lung in specific conditions, (IV) general themes about the lung microbiome observed across disease states, and (V) lessons, implications and hypotheses from studies of the extra-pulmonary microbiome.
I. Methods used in the analysis of the lung microbiome
The sterility of the lower respiratory tract in healthy individuals has never been demonstrated by any study using modern techniques of molecular microbial detection. The conventional wisdom that healthy lungs are sterile was derived from 130 years of defining bacterial presence by the ability to grow organisms from tissue samples on culture media. Bacterial etiologic agents or co-factors have even remained elusive in infectious disease states such as pneumonia, in which 75% of patients admitted with the diagnosis have no pathogen identified [4]. Considered with the observation that 70% of bacterial species within the human body cannot be cultured with standard medical microbiology media [5], these clinical observations alone illustrate deficits in our understanding of host-microbial interactions in the lungs during health and disease.
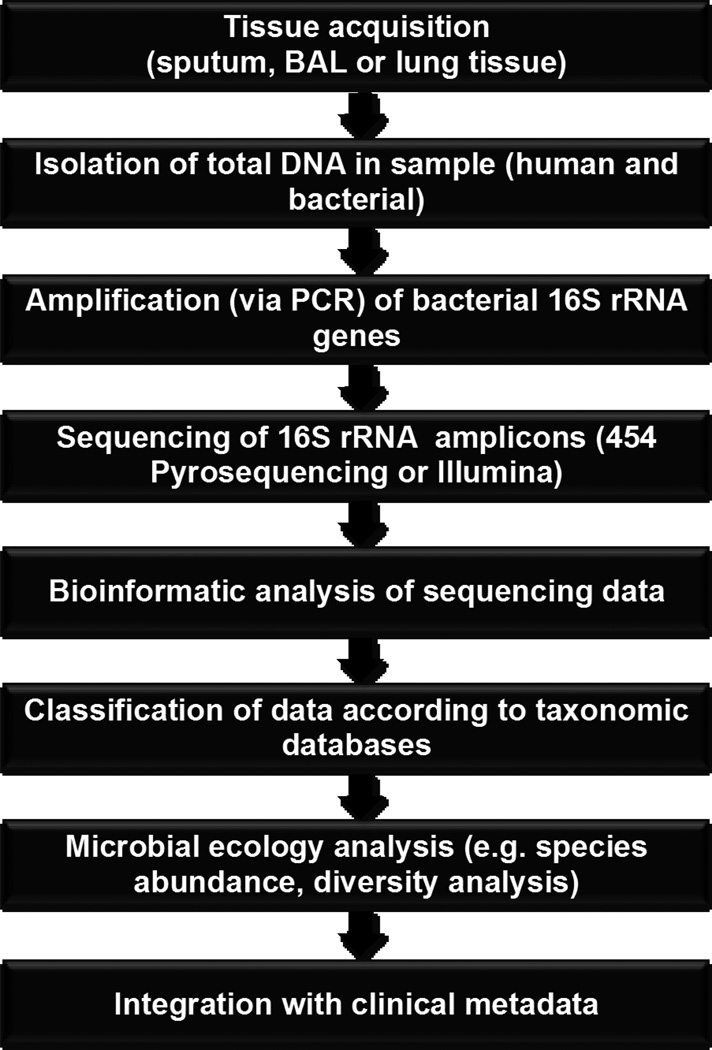
Many modern molecular techniques for bacterial identification exploit variation in the 16S rRNA gene, a small and highly conserved locus of the bacterial genome that permits genus- and species-level identification. High-throughput sequencing of 16S rRNA gene amplicons generated from bacteria-containing biosamples yields a large number of short sequences that can be subsequently aligned and sorted according to a predefined level of homology. These are then classified according to publically available taxonomic databases [6,7]. Using these principles, platforms such as 454 pyrosequencing and Illumina sequencing enable the identification and quantification of entire microbial communities in multiple tissue samples simultaneously [8]. (Figure 2) These sensitive and rapid techniques, often more thorough and accurate than traditional culture-dependent methods, have improved dramatically in efficiency and decreased in cost in the past decade.
Figure 2.
Flow diagram for microbiome analysis of respiratory specimens
While most lung microbiome studies to date have focused on the constitution of bacterial communities in the lung, similar techniques can be used to describe viral and fungal organisms. Both viruses and fungi are likely to be of considerable importance in the pathogenesis of acute and chronic respiratory disease, but to date the literature on this topic is limited and beyond the scope of this review. Targeted PCR technology is already in widespread use in the clinical diagnosis of respiratory viruses and the fungus Pneumocystis jirovecii.
Most lung microbiome analysis to date has been performed on bronchoalveolar lavage (BAL) fluid or, in the case of cystic fibrosis, on spontaneously expectorated sputum. The precise contribution of upper airway microbiota on the microbial content of expectorated sputum or BAL specimens is an issue of active debate. Several studies analyzing lung tissue acquired via sterile surgical explant have been performed, all demonstrating that the lower respiratory tract contains a microbiome that is distinct from but related to that of the upper airways [9–12]. A method has been proposed to account for upper respiratory tract microbiota (acquired via bronchoscope contamination or peri-procedural microaspiration) via subtraction of signal from upper respiratory tract specimens [13,14], but such a protocol has neither been validated nor universally adopted. Some investigators have proposed the adoption of the Neutral Theory of Biodiversity (the “neutral model”) [15,16] as a means of assessing whether microbes detected in the lower respiratory tract are a direct extension of upper airway microbiota (in which case their relative concentrations should mirror those of the upper airways) or whether selective pressures in the lower respiratory factor favor the relative abundance of specific species [17]. Within this conceptual framework, the “null hypothesis” is that no unique selective pressures act on bacteria reproducing in the lower respiratory tract, in which case the relative concentrations of bacterial species in BAL specimens should be comparable to those of upper airway specimens. The presence of species that are disproportionately present in BAL would negate the null hypothesis (and be inconsistent with the “neutral model”), implying a distinct ecological niche for bacterial species in the lower airways.
An accurate comparison of relative bacterial biomass between the lower respiratory tract and other compartments of the body is not possible until these issues are resolved, but the concentrations of bacteria in the alveoli and small airways are likely considerably less than that of the mouth or lower GI tract, at most comparable to that of the stomach or small intestine [18]. These low absolute levels of bacterial biomass introduce specific challenges to the field, namely the risk of false positive signal from the upper airway and accidental amplification of low-level signal present in reagents or the laboratory environment. To address this, recent studies have included control specimens not just of upper airway biosamples but also bronchoscope washings and reagent controls.
An important aspect of the above-described molecular techniques of identifying bacteria typically do not intrinsically discriminate between dead and viable bacteria, instead identifying only the presence of DNA in the analyzed specimen. This is distinct from traditional culture-dependent techniques, which require the presence of viable organisms. This is among the respects in which these novel techniques should be viewed as a complement to, not a replacement for, traditional methods of microbial identification.
II. The microbiome of the healthy lung
Fetal lungs, like the fetal intestines, are presumed to be sterile, and infant lungs likely acquire their microbial communities after birth. In the immediate post-delivery period, infant mucosal surfaces are quickly populated by microbes derived from the mother (vaginal and intestinal microbiota in cases of vaginal delivery, skin microbiota in cases of cesaerean section) [19]. Infant microbiota is initially uniform across various body sites, differentiating in subsequent days and weeks into site-specific communities [20]. The derivation of lung microbiota has not been studied in healthy infants, but a recent longitudinal study of seven infants with cystic fibrosis demonstrated concordance between the microbial communities of the gut and those of the respiratory tract, with evidence of temporal precedence in the gastrointestinal tract [21].
Numerous published studies have characterized the lung microbiome of healthy adult subjects using BAL samples [9,13,14,18,22–24]. When analyzed on the phylum level for relative abundance, the most common phyla consistently observed have been Bacteroides, Firmicutes and Proteobacteria. Described phyla in BAL samples are similar to those seen in concurrently collected upper airway (oropharynx, nasal) samples, but differ in relative abundance (e.g. relative infrequency of Actinobacteria). Prominent genera among healthy controls are Prevotella, Veillonella, Streptococcus and Pseudomonas. Active cigarette smoking appears to alter the microbial constitution of the upper airways [25]; its effects on the lung microbiome are unknown. Differences in the sputum microbiota of cystic fibrosis patients have been observed when comparing samples obtained in England and the United States [26], and profound regional differences have been reported in the gut microbiome in subjects from different geographic origins [27], but the influence of biogeography on the constitution of normal lung microbiota has not been described.
Studies describing the lung microbiome of control patients have been limited by small size and lack of longitudinal studies; the total number of control subjects in published studies by the end of 2012 was 75 [9,13,14,18,22–24], and serial specimens from the same control subject have not been reported. The Lung HIV Microbiome Project (LHMP), an ongoing, multi-center NIH project intended to complement the HMP, aims to address this issue by studying the lung microbiome of a large number of subjects, with and without HIV, free of known lung pathology, at multiple time points.
III. The microbiome of the diseased lung
a. Cystic fibrosis
Since its first description in the 1930’s, cystic fibrosis (CF) has been associated with recurrent respiratory infections [28], and the major cause of morbidity and mortality in patients with this systemic disease remains progressive airway obstruction and associated respiratory infections [29,30]. Common pathogens detected by culture-based techniques include Staphylococcus aureus, Haemophilus influenza and, increasing in frequency with age, Pseudomonas aeruginosa. Intermittent and initially (seemingly) eradicable infections are followed by culture-detectable airway colonization, and patients are susceptible to otherwise uncommon respiratory pathogens such as Stenotrophomonas maltophilia and Burkholderia cepacia complex.
Culture-independent techniques of microbial identification have been used to characterize the airway microbiota of patients with CF for nearly a decade, and the lung microbiome of CF has been more thoroughly reported than that of all of other respiratory diseases combined [10,23,26,31–58]. Numerous molecular techniques have been used, including terminal restriction fragment length polymorphism profiling [23,26,31–35,39,44,47,49,57], clone library sequencing [31,32,36,38,42,45,46], pyrosequencing [10,42,47,48,50–52,58], Phylochip [43] and Illumina [55] technologies. Most studies have been performed on spontaneously expectorated sputum samples; fewer have used BAL [31,35] and explanted lung tissue [10,23,46,48,54,56]. These citation lists are not comprehensive and emphasize studies that have attempted to characterize lower airway microbial communities.
Molecular methods have unveiled a previously unrecognized complexity to the microbiota of CF airways, both during and between active exacerbations. An early and important observation from Rogers et al. was that the sputum of patients with active bacterial respiratory infection contains large quantities of multiple bacterial species [32,33,44]. Subsequent studies have re-demonstrated the polymicrobial nature of airway colonization and infection in CF, including numerous species previously unrecognized via culture-dependent techniques. This insight may partially explain the long-observed discordance between sputum culture speciation and sensitivity results and patients’ response to antimicrobial therapy [59]. The clinical implications of this observation on acute and chronic use of antibiotic therapy are not clear, but it suggests that rather than “clearing” an infection, administered antibiotics instead alter the internal architecture of a dynamic and heterogenous microbial community. A single study has found comparable complexity in the lungs of patients with non-CF bronchiectasis [54], but it has not yet been established how universally applicable these findings are to other forms of lung infection.
Study of the CF lung microbiome has uncovered numerous previously un- or under-recognized bacterial species that are potentially pathogenic in CF exacerbations, inflammation and airway destruction. Special interest has arisen in anaerobic species [32–34], the common presence and viability of which have been subsequently confirmed using culture-dependent techniques [60]. No published clinical trials, prospective or otherwise, have studied whether targeted antibiotic therapy against these microbes is of benefit, and their clinical significance is as of yet undetermined.
Several studies have examined whether the diversity of lung microbiota changes with time or severity of illness in CF. Cox et al. used longitudinal analysis to demonstrate that bacterial community diversity decreases with age and severity of airway obstruction; the same study found that patients with a homozygous ΔF508 mutation had decreased microbiota diversity when compared to heterogenous-ΔF508 patients and non-ΔF508 patients [40]. Klepac-Ceraj et al. also found decreasing microbial diversity with age, and confirmed the negative influence of systemic and inhaled antibiotics on diversity [43]. In the largest and most long-term longitudinal series of CF airways samples published to date, Zhao et al. demonstrated decreasing diversity over time but relative stability among patients with mild airway disease [58]. In this series, recent antibiotic use was by far the strongest predictor of decreased microbial diversity. The immediate influence of antibiotic therapy on lung microbiota was also observed by Stressmann et. al., who observed that pre-antibiotic community structures appeared to return to prominence within one month of antibiotic exposure [57].
Recently, one study compared culture-independent analysis of explanted lung tissue, throat and sputum samples from patients with severe, end-stage CF and found that expectorated sputum accurately represented the dominant microbes in the relatively homogenous lower respiratory tract, but overrepresented the diversity and representation of atypical bacterial species [10]. Most lower respiratory tract samples were overwhelmingly populated by one to three species of classic respiratory pathogens. The authors proposed the development of methods to “subtract” the influence of upper airway microbiota.
Two clinical trials have examined whether administration of enteric probiotics influences the frequency and severity of pulmonary exacerbations in CF patients [61,62]. These studies, though small, both found significant decreases in the frequency of pulmonary exacerbations when compared to control patients receiving placebo [61] and compared to the study subjects in the periods before and after probiotic administration [62]. These findings must be validated in larger trials before routine clinical use can be recommended, but they raise provocative questions regarding the mechanism of efficacy. Both studies’ authors speculate that benefit may arise via decreasing intestinal wall inflammation with indirect effects on pulmonary inflammation, but it is not known how enteric probiotics influence the composition of lung microbiota.
b. Asthma and Allergic Airways Disease
An association between decreasing frequency of childhood infections and the increasing development of asthma and allergies has been recognized for years, giving rise to the “hygiene hypothesis”: that a decrease in infectious exposures early in life results in deranged mucosal tolerance and increased autoimmune pathology [63]. Multiple studies have observed an association between early childhood antibiotic exposure and subsequent development of asthma and allergies [63,64], prompting speculation that disruption of the normal microbiome may be complicit in the pathogenesis of these conditions.
Two important studies have studied the composition of the lung microbiome in patients with asthma as compared to healthy controls [18,24]. Hilty et al. compared the microbiome of oral, nasal and BAL specimens of patients with asthma to that of patients with Chronic Obstructive Pulmonary Disease and healthy controls [18]. Among asthmatics, the authors found increased frequency of Proteobacteriae and decreased frequency of Bacteroidetes when compared to controls. This relative increase in Proteobacteriae was driven by Haemophilus, Moraxella and Neisseria species.
Subsequently, Huang et al. compared the lung microbiota (obtained bronchscopically using protected specimen brushes) of 65 poorly-controlled asthmatics with that of 10 control subjects and found both increased bacterial burden and bacterial diversity among the asthmatic subjects [24]. They confirmed the increased relative abundance of Proteobacteriae among asthmatics, and found a positive correlation between the presence of numerous species and the severity of bronchial hyperresponsiveness. Greater diversity was seen among patients who derived benefit in bronchial hyperresponsiveness when administered clarithromycin, one of the rare observations reported between lung microbiome constitution and a functional response to a clinical intervention.
In addition to the potential role that the lung microbiome may play in allergic disease of the airways, a complementary hypothesis is that perturbations in gastrointestinal microbiota composition due to antibiotic use and poor diet (low fiber, high sugar) in westernized areas have disrupted gastrointestinal microbiome-mediated mechanisms of mucosal tolerance. Data supporting this hypothesis include the correlation between asthma/allergies and antibiotic use in industrialized countries [65–69] and the correlation between altered fecal microbiota and atopic disease [70–76]. Direct testing of this hypothesis in antibiotic-treated and germ free mice has supported this potential link between changes in the gut microbiome and allergic responses in the lungs [77–79]. A few reports have studied the effect of oral delivery of strains of Lactobacillus in modulating experimental allergic pulmonary inflammation.
c. Chronic Obstructive Pulmonary Disease
Since its early modern descriptions, Chronic Obstructive Pulmonary Disease (COPD) has been speculated to be caused at least in part by inflammation secondary to chronic and recurrent airway infections [80]. Yet the direction of causality between airway destruction and respiratory colonization/infection has remained unsettled despite decades of active research [81,82]. A recent trial demonstrating benefit of chronic azithromycin in patients with frequent exacerbations of COPD has sparked renewed interest in the intersection of lung/airway microbiota, inflammation and the development of irreversible airway obstruction [83]. In the past four years, seven studies have reported the lung microbiota of patients with COPD [9,12,18,22,23,39,84]. All but one have studied patients with no active evidence of airway infection and no recent antibiotics, and specimens used for analysis have included BAL [9,12,18,22], explanted lung tissue and expectorated sputum [39]. These studies reveal that the lower airways and alveoli of patients with COPD contain a distinct microbiome that may prove relevant both to the chronic progression of disease and to the intermittent development of infectious exacerbations.
The early study by Hilty et al. on the microbiome of asthmatics included five patients with COPD [18]. The microbiota of specimens of COPD patients clustered with those of asthmatics and separately from those of healthy controls, and exhibited a similar increase in the relative presence of Proteobacteria as well as a relative decrease in Bacteroides. Of note, compared to controls, COPD specimens contained significantly more Haemophilus spp., which is the most commonly cultured organism in COPD exacerbations.
In a subsequent study, Erb-Downward et al. compared the microbiota found in the BAL and explanted lung tissue of patients with severe COPD with that of smokers without evidence of lung disease or obstructive ventilatory deficits and non-smoking control subjects [9]. They found comparable concentrations of 16S rRNA genes across their groups, implying comparable bacterial burdens. Subjects with the most severe airway obstruction had markedly decreased microbial diversity, and extensive overlap in genus-level membership was found across groups. Common genera among COPD samples were Pseudomonas, Streptococcus, Prevotella and Haemophilus. The authors analyzed tissue from multiple sites within the same lungs (acquired from the explanted native lungs of transplant recipients), revealing marked regional heterogeneity of lung microbiota within the lungs (and even individual lobes) of patients with severe disease.
Surgically acquired lung tissue samples were also studied by Sze et al. [23], who found low and comparable quantities of bacterial cells in COPD lung tissue compared to that of controls with no significant difference in microbial diversity. Both COPD subjects and controls had considerably more diversity than CF patients included in analysis. A distinct microbiome was found among COPD subjects when compared to smoking and non-smoking controls, with comparable abundance of Proteobacteria and Bacteroides but increased Firmicutes compared to control subjects. The increase in Firmicutes was driven by increases in the Lactobacillus genus. An important difference between this study and others is that the bulk of analyzed tissue is likely alveolar parenchyma with relatively little airway inclusion, which may explain some of the differences in the relative abundances found.
Cabrera-Rubio et al. studied the respiratory microbiota of eight patients with COPD via analysis of four specimens each: expectorated sputum, bronchial aspirate, BAL and bronchial mucosal biopsy [12]. They observed the same common phyla as previous studies (Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes), with Streptococcus, Prevotella, Moraxella, Haemophilus, Acinetobacter, Fusobacterium, and Neisseria heavily represented among the genera. They found consistent similarity between the microbiota present in BAL and bronchial mucosa specimens, but decreased microbial diversity in the upper respiratory tract specimens (bronchial aspirate and expectorated sputum) with increased prevalence of Bacteroides and Firmicutes.
Pragman et al. used pyrosequencing to compare the microbiota of BAL specimens from 22 COPD patients to that of 10 controls [22] and found increased microbial diversity among patients with COPD that correlated positively with severity of obstruction. The most common phyla among both COPD and control specimens were Actinobacteria, Firmicutes and Proteobacteria. An important contribution from this study was the recognition that the microbiota of BAL samples clustered not merely by disease status but by subject exposure to inhaled bronchodilators and/or inhaled corticosteroids. This observation, combined with the clinical observation that inhaled corticosteroids are associated with increased rate of serious pneumonia [85], should prompt further study regarding the influence of inhaled medications on lung microbiota.
A single study has explored the microbiota of respiratory specimens in patients with COPD in the context of an acute exacerbation [84]. Huang et al. studied eight patients mechanically ventilated for respiratory failure attributed to COPD exacerbations. Unlike the subjects of other COPD studies, these patients all had received recent antibiotics prior to sample acquisition. Despite this, impressive microbial diversity was found in the analyzed endotracheal aspirates. The constitution of the microbiota identified was influenced by duration of mechanical ventilation, and the authors identified a “core” of 75 bacterial taxa and 27 bacterial families that were present in the microbiota of all specimens, including Pseudomonadaceae, Enterobacteriaceae, Campylobacteraceae, and Helicobacteraceae. An important parameter of this study was that the subjects were drawn from a larger population of intubated patients with culture-demonstrated Pseudomonas aeruginosa in respiratory specimens.
d. Lung Transplantation
Mortality and graft dysfunction among lung transplant recipients remain higher than those of all other solid organ transplants, with most morbidity attributable either to infection or bronchiolitis obliterans (BO) and bronchiolitis obliterans syndrome (BOS): chronic fibroproliferation of the small airways resulting in progressive, irreversible airway obstruction. While the mechanisms of pathogenesis remain unknown, multiple associations between microbial infection/colonization and BOS have been observed [86–88]. Lung transplantation results in numerous changes to the host defenses of the respiratory tract that may alter the microbiota of the lung, including distorted airway architecture, disruption of native lymphatic channels and systemic immunosuppression from anti-rejection medications.
Charlson et al. recently published a study comparing the microbiota of BAL specimens from 23 lung transplant recipients with those of six non-transplant controls [89]. They found decreased microbial diversity among transplant recipients, and in several cases the overwhelming presence of a single species (e.g. Pseudomonas aeruginosa and Staphylococcus aureus). In these cases the identity of the dominant microbe was usually also found on routine culture. The authors also analyzed for the presence of fungal species and found abundant Candida and Aspergillus species in the BAL of transplant recipients. Significant associations between microbiome constitution and clinical features such as acute or chronic rejection were not reported and remain unknown. A subsequent report by Borewicz et al. found increased diversity among transplant recipients compared to two controls, with transplant phyla dominated by the Burkholderiaceae family of the Proteobacteria phylum [90]. This family was not seen in either of the control specimens. The authors did not find a distinct clustering of the microbiome before or after the development of BOS.
e. Idiopathic Interstitial Pneumonias
A single study has described the microbiota of patients with idiopathic interstitial pneumonia [91], a heterogenous class of chronic diffuse lung diseases. Friaza et al. used denaturing gradient gel electrophoresis to characterize the microbial communities in the BAL of 20 patients with numerous interstitial lung diseases including idiopathic pulmonary fibrosis, non-specific interstitial pneumonia and acute interstitial pneumonia. Both classic respiratory pathogens (e.g. Haemophilus influenza) and a variety of previously un- or under-recognized organisms were identified. Provocatively, a negative association was observed between the presence of Pneumocystis jirovecii and bacterial burden, suggesting a possible in vivo antagonism between Pneumocystis and bacterial species.
f. Mechanical Ventilation
Apart from the study of COPD exacerbations by Huang et al. described above [84], only a single report by Flanagan et al. describes the microbiome of respiratory specimens in patients receiving mechanical ventilation [92]. As in the Huang study, enrolled patients were selected based up the presence of mechanical ventilation and the presence of Pseudomonas aeruginosa in respiratory cultures, limiting generalizability across all ventilated patients. The authors noted a marked decrease in the diversity of respiratory microbiota during the administration of broad-spectrum antibiotics, typically followed by reconstitution with a dominant organism (often P. aeruginosa despite concurrent anti-pseudomonal antibiotics). Further longitudinal analysis in this population may help to unfold the effects of antibiotics and other acute interventions on the dynamic microbial communities of the lung.
IV. General Themes across respiratory disease states
a. Temporal and spatial heterogeneity
No published studies to date describe longitudinal analysis of serial respiratory specimens from healthy controls, so the relative stability or dynamic nature of the “normal” lung microbiome is unknown. Virtually all of the of the longitudinal analysis of lung microbiota specimens to date has been performed on sputum specimens from patients with CF [40,49,50,52,57,58], which may not accurately reflect the microbiota of their lower airways [10]. In the longitudinal studies published, the microbial communities in the sputum of individual patients is relatively stable over time, even despite the development of clinical exacerbations and the administration of antibiotics [52,58]. Because of the limitations of sputum analysis and the unique clinical features of CF, it would be premature to infer from these observations any conclusions regarding microbiome stability in health and other respiratory pathologies. As mentioned above, the ongoing LHMP will include serial sampling of non-diseased patients, and should provide more information regarding the temporal stability/dynamism of lung microbiota. However, the depth/precision of serial sampling of the lung environment (compared to serial sampling of the intestinal environment by serial fecal collection) will always be an issue unless a future technological advance allows for repeated non-invasive microbiome analysis of the lower airways.
In the two studies that have considered spatial variation in microbial communities, impressive spatial heterogeneity has been demonstrated in severe COPD [9] and severe CF [48]. It is unclear whether this finding is specific to the end-stages of these two conditions, which are both characterized by impaired local mucocillary clearance, or if the lungs of other disease states are comparable spatially heterogenous.
Studies that have compared concurrently sampled upper and lower respiratory tract samples have found similar but distinct microbial communities, often with increased relative abundance of Bacteroides and Actinobacter phyla in upper airway specimens. These results suggest that the respiratory tract is not, as traditionally discussed, comprised of discrete and independent compartments. A more accurate model is instead that the respiratory tract is a single but internally heterogenous ecosystem extending from the nares to the alveoli that contains a continuous, and continuously varying, microbial topography. Microbiota from the upper airways are likely constantly introduced to the lower airways via direct mucosal extension and microaspiration during sleep, which is ubiquitous and usually asymptomatic [93]. Microbiota from the lower airways are constantly introduced to the upper airways via cilliary transport and cough, which occurs many times a day even in patients without respiratory complaints [94,95]. The constitution of a microbial community at a specific spot in the respiratory tract is likely an integrated function of these influences as well as local growth conditions such as temperature, oxygen tension, pH, nutrient density, local host defense and intra- and inter-species small molecule signaling.
b. Diversity and severity of illness
An unambiguous trend between microbiome diversity and respiratory disease presence and severity has not emerged in published studies to date. In COPD, the diversity of lung microbiota as compared to controls has been reported as increased [22], decreased [9] and equivalent [23]. Similar analysis performed on BAL samples from lung transplant recipients have found increased [90] and decreased [89] diversity. These conflicting results suggest that the diversity of a single subject’s lung microbiota is likely multifactorial, and cannot be interpreted as a simple function of his/her disease severity. In select disease states, it has been shown that diversity is influenced by administration of antibiotics [43,58,92], age [40,43] and CF genotype [40]. Further study, especially with longitudinal sampling, promises to identify other drivers of diversity and to unfold its clinical significance.
c. Identification of novel pathogens
An immediate and short-term goal of lung microbiome studies is to apply molecular techniques to identify respiratory pathogens previously un- or under-appreciated via culture-based techniques. Our received understanding of the distinctions between “pathogens,” “contaminants” and other identified microbes has been complicated by 1) new recognition of resident bacteria in healthy lungs, 2) identification of concurrent species alongside known pathogens in states of active infection, 3) lack of mechanistic animal modeling and 4) a paucity of longitudinal data or clinical trials. It is likely that a given microbe may have an entirely different relationship with its host depending on context: a host-microbe interaction may be mutually beneficial or neutral in a healthy homeostasis, but pathogenic when homeostasis is disrupted and the microbe overtakes its ecologic niche and causes tissue damage either directly via virulence factors or indirectly via activation of host immunity. The mere identification of microbial DNA in the lungs of patients with symptoms of active infection is only the first step in our evolving understanding of the pathogenesis of lung infection.
With these caveats in mind, and acknowledging that most of the above-reviewed studies excluded patients with clinical evidence of active infection, several studies have identified in the airway specimens of patients with clinical evidence of infection abundant concentrations of possible un- or under-recognized respiratory pathogens. The bulk of these are in the cystic fibrosis literature [35,36], in which most of the sputum collected is either collected context of an active exacerbation or after a patient is “colonized” with known pathogenic organisms. New candidate pathogens include Lysobacter sp., Rickettsiales, I. limosus, D. pneumosintes and D. pigrum, among many others. Particular interest has arisen in anaerobes such as the Prevotella and Veillonella genera, the common presence of which have been confirmed via culture-based techniques [60,96], which have identified these microbes in concentrations equivalent to those of P. aeruginosa and S. aureus. As mentioned above, these findings must be understood to be limited potentially by upper airway contamination [10], and no clinical trials have been published to date examining efficacy of treating these potential pathogens. Their clinical significance is undetermined.
In the few non-CF studies that have included patients with clinical evidence of active infection, culture-independent techniques have confirmed lung microbiota domination by species concurrently detected by culture-based techniques [92] but concurrent presence of many other organisms of uncertain significance [84]. Given the observations described above regarding differences in the constitution of lung microbiota among patients with and without chronic lung disease when not actively infected, it is likely that our understanding of the definition of “pathogen” will require revision and expansion.
V. Lessons, implications and hypotheses from study of the extra-pulmonary microbiome
The field of lung microbiome studies enjoys the benefit of concurrent and analogous inquiries into the microbiota of other organ systems, many of which have been studied for years on the human subject, population and animal model level. A review of findings from the extra-pulmonary microbiome reveals potential future directions for study of the lung microbiome.
a. Revision of simplistic and outdated definitions
Investigators of microbiomes of the gut, skin and genitourinary tract have moved past a simplistic designation of all resident microbes as either symbionts (deriving or providing benefit from/to its host) or pathogens. Our evolving understanding of Helicobactor pylori is an informative case study: though once considered sterile (like the lungs), the stomach was shown (via improved diagnostic techniques) in many subjects to contain large numbers of H. pylori, with a strong association with the development of peptic ulcer disease and gastric malignancies [97]. Yet subsequent study has revealed significant negative associations between (and potentially protective effects from) H. pylori and esophageal reflux [98] and asthma [99], as well as mechanistic association between its eradication and obesity [100]. This organism is most accurately considered an “amphibiont,” a resident microbe with a relationship to its host that may be considered symbiotic or pathogenic depending on context [3]. It is unknown whether such “amphibionts” are among the species described in lung microbiota, and what local and systemic factors may influence the nature of their relationship with the host.
As well as refining our understanding of individual microbe designations, study of the microbiome should prompt us to revisit our understanding of what as classically been designated “infection,” “colonization” and “health” (i.e., in the traditional understanding, sterility of the lower respiratory tract). These terms are clearly inadequate to explain lung microbiome findings to date. An improved understanding places active infection and “healthy” lung microbiota diversity at opposite ends of a continuum, separated by varying degrees of “dysbiosis,” as well-described in conditions such as Bacterial Vaginosis and Inflammatory Bowel Disease. This model understands active infection (such as bacterial pneumonia) to be an emergent phenomenon of disruption in the complex homeostasis of the “normal” lung microbiome, rather than the unchecked growth of an invasive organism in a previously sterile compartment. A full account of lung infection will require an understanding of the constitution of a host’s resident lung microbiome and its resilience to perturbation, an element missing from our tradition of Koch’s postulates [101].
b. “Pulmotypes”
An effort has been made in recent years to define clusters of bacterial communities that associate together in the human gut, existing as characteristic “fingerprints” of gut microbiota: “enterotypes.” In an important 2011 study, Arumagam et al. described three distinct enterotypes of fecal microbiota in individuals that had distinct composition and were not geographically specific [102]; a subsequent study demonstrated that gut enterotypes can be influenced by diet [103]. Clinical significance of enterotypes remains uncertain, and subsequent analysis by Arumagam et al. suggests that gut microbiota exist more along spectra and less in distinct enterotypes [104]. But an important and accessible question that may be answered in subsequent studies (especially the LHMP) is whether analogous “pulmotypes” exist among lung microbiota and whether they help in further “phenotyping” complex and heterogenous diseases such as COPD.
c. Mechanisms of host-microbe and microbe-microbe interactions in the lungs
Study of the gut microbiome in humans and animal models has revealed numerous findings regarding the mechanisms by which microbes influence host inflammation and microbe tolerance, host nutrient and drug metabolism, as well as means by which microbial communities communicate with and influence each other. The interface of gut microbiota and mucosal immunity is dynamic with bi-directional communication. Host immune mediators include physical barriers such as mucus layers, secretion of antibacterial peptides by epithelial cells and sophisticated cellular immunity with generation of microbial-specific IgA; conversely, gut microbes prompt lymphatic maturation, mediate epithelial repair via endotoxin signaling, influence Th1/Th2 balance and can promote mucosal tolerance of resident microbes [105]. Most of these interactions have been clarified via what are now well-described mouse models of gut microbiota; the lack of analogous models in the field of lung microbiome studies precludes any such observations regarding lung microbiota. It has, however, been observed that derangements in mouse gut microbiota influence susceptibility to allergic airway disease [106]; it is undetermined how much this is an applied consequence of altered systemic immunity versus more immediate interactions between GI and lung microbiota.
Recent studies have also revealed multiple mechanisms by which bacteria and hosts communicate via small molecules that resemble human hormones and influence bacterial quorum-sensing, host epithelial cell gene expression and activation of microbial virulence factors [107]. It has long been known that many microbes secrete antimicrobial peptides; indeed, most of our clinically available antibiotics were originally derived from bacterial or fungal organisms. These observations suggest that a full understanding of the lung microbiome will require investigation into microbe-microbe interactions, potentially as mediators of the above-discussed “pulmotypes.”
d. Microbiome influences on unexplored disease classes
Associations, some with plausible mechanisms of pathogenic contribution, have been described between microbiome derangements and colorectal cancer [108,109] and rheumatoid arthritis [110], among other pathologies not classically attributed to microbial influence. To date no published study has examined the lung microbiome of patients with primary lung cancer or autoimmune-related lung disease. As we are only beginning to understand the influences of disease states on the microbiome and vice versa, the lung microbiota should not be considered irrelevant a priori to any disease state, however indirectly related it is to our conventional understandings of microbial pathogenesis.
e. The microbiome as a therapeutic target
Should a disordered lung microbiome prove to be involved in the pathogenesis of disease (and not incidentally and secondarily deranged by the disease itself), it will be of immediate interest as a novel target for therapeutic intervention. The lung microbiome, like that of other compartments, may be potentially manipulated with an aim to correct dysbiosis and restore “healthy” microbial communities via use of probiotics (extrinsic microbes administered in the interest of health), prebiotics (non-absorbed molecules that promote specific bacterial growth), antibiotics and quorum-sensing molecule inhibitors. A goal of our more developed understanding of lung microbiota derangements should be to target antibiotics to the narrowest element of the microbial spectrum that is directly pathogenic without disturbing the residual members of the microbial community.
To date, clinical trials investigating the use of orally administered probiotics in gastrointestinal diseases are strongest in the support of use for prevention of antibiotic-associated diarrhea [111] and for the treatment of acute infectious diarrhea [112]; for patients with antibiotic-refractory Clostridium difficile infection, intestinal microbiota transplantation has shown great promise for attaining cure [113]. These suggest that the most appropriate early targets among lung diseases for targeted microbiome therapies may be those with immediate infectious components, such as pneumonia and bronchiectasis.
Indeed, several studies of the pulmonary effects of enterically administered probiotics have already been published, with promising results [114]. In a 2010 study of 146 mechanically ventilated patients, administration of probiotics significantly decreased the rate of development of ventilator-associated pneumonia (40% vs 19%) [115]. Numerous studies have examined the effect of oral probiotics in the prevention of upper respiratory tract infections, most (17/21) with evidence of benefit [116]. Two small randomized controlled trials, discussed above, have demonstrated decreased frequency of CF exacerbations among patients receiving probiotics [61,62]. In none of these studies is it known whether benefit was conveyed via direct alteration of the lung microbiota or indirectly via gut mucosa-mediated effects on systemic immunity.
As illustrated by the case of intestinal microbiota transplantation for C. difficile, route of administration is important in targeted microbiota interventions. Inhalation or direct intrapulmonary instillation of probiotics or prebiotics may have profoundly different effects on lung microbiota when compared to enteric administration.
Beyond strategic manipulation of lung microbiome communities, we may be able to adapt lessons learned from study of the above-mentioned molecular mediators used by microbes to communicate with each other and to host epithelial and inflammatory cells. Resident microbes may be already producing veritable pharmacopoeias of small molecules that can influence local and systemic inflammation, mucosal tolerance, airway reactivity, and microbial cooperation and antagonism.
Expert Commentary
Modern culture-independent techniques of microbial identification have revolutionized our understanding of the bacterial communities that inhabit the respiratory tract both in health and various disease states. Important associations between the constitution of the lung microbiome and key clinical outcomes have already been observed, and further study promises to elucidate novel mechanisms in the pathogenesis of lung infection and chronic lung diseases. This field will prompt researchers to reconsider long-held beliefs regarding the complex and dynamic relationship between microbe and host in the healthy and diseased respiratory tract.
Five-year view
The field of lung microbiome studies is still in relative infancy, with most studies to date limited by small sample sizes, lack of longitudinal samples and control subjects. We expect that in coming years, the following developments will mature our understanding of lung microbiota and their role in human disease:
Standardization of techniques for sample acquisition, quantification of bacterial biomass and data analysis
Ongoing identification of novel pathogens, development of methods for confirming pathogenicity, and development of accurate, rapid and affordable point-of-care diagnostic tools for respiratory infection
Longitudinal studies of samples from individual subjects, both diseased and control, to characterize the stability and robustness of lung microbial communities over time as well as the influence of external factors such as antibiotics, immunosuppression and acute illness
Expansion of tissue acquisition to as-of-yet unstudied respiratory pathologies
Study of non-bacterial lung microbes such as viruses and fungi
Development of animal models of lung microbiome states to test mechanistic hypotheses
Exploration of microbe-microbe and microbe-host signaling mechanisms
Improvement in analytical techniques, including incorporation of analytic tools from other fields such as functional genomics, systems biology and complex systems analysis
Clinical trials of microbiome-targeted interventions
Table 1.
Definitions of key terms
| Term | Definition |
|---|---|
| microbiome | the totality of microorganisms in a given site (such as the human body or respiratory tract) |
| 16S rRNA (ribosomal RNA) | component of the bacterial ribosome, the gene of which is used for sequencing and microbial identification |
| pyrosequencing | method of gene sequencing utilizing serial additions of nucleotides and luminescent enzyme reactions |
| probiotic | microorganisms administered to patients with therapeutic intent |
| prebiotic | administered molecules that promote specific bacterial growth |
| amphibiont | a microbial species that is potentially beneficial or pathogenic depending on context |
| enterotype/pulmotype | communities of distinct bacterial species that associate together in the human gut or lung |
Table 2.
Taxonomy of phyla and genera described in human lung microbiome studies
| Phylum | Genus |
|---|---|
| Bacteroidetes | Prevotella |
| Bacteroides | |
| Firmicutes | Veillonella |
| Streptococcus | |
| Staphylococcus | |
| Proteobacteria | Pseudomonas |
| Haemophilus | |
| Moraxella | |
| Neisseria | |
| Acinetobacter | |
| Escherichia |
Table 3.
Important and unanswered questions regarding the microbiome of the healthy lung
| 1. How are the bacterial communities of the lung established after delivery? |
| 2. Are the bacterial communities of the lung stable or dynamic over time? |
| 3. Are lung microbiota spatial homogenous or heterogenous? |
| 4. Is there geographic variability in the constitution of lung microbial communities? |
| 5. How do microbiota of the lung influence and are they influenced by microbes in other compartments of the body? |
| 6. How are lung microbial communities influenced by clinical factors and exposures such as gender, living environment and vaccination history? |
Key issues.
Recent applications of culture-independent techniques of microbial identification have revealed diverse communities of bacteria in the airways both of diseased patients and healthy control subjects.
These techniques employ amplification of the bacteria-specific 16S rRNA gene, which enables rapid and accurate sequencing of entire bacterial communities in respiratory specimens.
Among healthy subjects, the most commonly reported bacterial phyla in respiratory specimens are Bacteroides, Firmicutes and Proteobacteria. Lower-respiratory tract communities appear related to but distinct from those of the upper airways.
Culture-independent techniques have uncovered un- and underrecognized respiratory pathogens, including a pathogenic role for anaerobic species in polymicrobial CF exacerbations.
Microbiome analysis of various disease states have revealed distinct lung microbiota with altered diversity measures in patients with CF, asthma, COPD, and lung transplantation.
Alterations in lung microbiome constitution and diversity have been associated with clinical parameters such as disease severity, airway obstruction and exposure to antibiotics.
Unanswered questions in this novel field include the temporal stability of lung microbial communities, the contribution of lung microbiota to chronic inflammation and fibrosis, and mechanisms of microbe-microbe and microbe-host signaling.
The few clinical trials of orally-administered probiotics for respiratory diseases to date are small and isolated, but suggest benefit in prevention of ventilator-associated pneumonia, upper respiratory tract infections and cystic fibrosis exacerbations.
References
- 1.Mizgerd JP. Lung infection--a public health priority. PLoS medicine. 2006;3(2):e76. doi: 10.1371/journal.pmed.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science (New York NY) 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature Reviews Genetics. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Restrepo MI, Mortensen EM, Velez JA, Frei C, Anzueto A. A comparative study of communityacquired pneumonia patients admitted to the ward and the ICU. CHEST Journal. 2008;133(3):610–617. doi: 10.1378/chest.07-1456. [DOI] [PubMed] [Google Scholar]
- 5.Suau A, Bonnet R, Sutren M, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Applied and Environmental Microbiology. 1999;65(11):4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z, Desantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic acids research. 2008;36(18):e120. doi: 10.1093/nar/gkn491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and environmental microbiology. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science (New York NY) 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Erb-Downward JR, Thompson DL, Han MK, et al. Analysis of the lung microbiome in the "healthy" smoker and in COPD. PloS one. 2011;6(2):e16384. doi: 10.1371/journal.pone.0016384. Analysis of lung microbiota of patients with severe COPD, comparing BAL and surgically excised tissue; demonstrated spatial heterogeneity of lung microbiota.
- 10. Goddard AF, Staudinger BJ, Dowd SE, et al. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(34):13769–13774. doi: 10.1073/pnas.1107435109. Comparison of microbiota in sputum and surgical lung specimens from patients with cystic fibrosis.
- 11.Rudkjobing VB, Thomsen TR, Alhede M, et al. True microbiota involved in chronic lung infection of cystic fibrosis patients found by culturing and 16S rRNA gene analysis. Journal of clinical microbiology. 2011;49(12):4352–4355. doi: 10.1128/JCM.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabrera-Rubio R, Garcia-Nunez M, Seto L, et al. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. Journal of clinical microbiology. 2012;50(11):3562–3568. doi: 10.1128/JCM.00767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlson ES, Bittinger K, Chen J, et al. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PloS one. 2012;7(9):e42786. doi: 10.1371/journal.pone.0042786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlson ES, Bittinger K, Haas AR, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184(8):957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbell SP. Neutral theory in community ecology and the hypothesis of functional equivalence. Functional ecology. 2005;19(1):166–172. [Google Scholar]
- 16.Hubbell SP. The unified neutral theory of biodiversity and biogeography (MPB-32) Princeton University Press; 2008. [DOI] [PubMed] [Google Scholar]
- 17.Morris A, Beck JM, Schloss PD, et al. Comparison of the Respiratory Microbiome in Healthy Non- Smokers and Smokers. Am J Respir Crit Care Med. 2013 doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PloS one. 2010;5(1):e8578. doi: 10.1371/journal.pone.0008578. Early and important study comparing lung microbiota in the airways of patients with and without asthma.
- 19.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. The Journal of investigative dermatology. 2011;131(10):2026–2032. doi: 10.1038/jid.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madan JC, Koestler DC, Stanton BA, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. mBio. 2012;3(4) doi: 10.1128/mBio.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PloS one. 2012;7(10):e47305. doi: 10.1371/journal.pone.0047305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sze MA, Dimitriu PA, Hayashi S, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185(10):1073–1080. doi: 10.1164/rccm.201111-2075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. The Journal of allergy and clinical immunology. 2011;127(2):372–381. doi: 10.1016/j.jaci.2010.10.048. e371 373. Study identifying association between airway microbiota diversity and clinical response to antibiotic administration in patients with asthma.
- 25.Charlson ES, Chen J, Custers-Allen R, et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PloS one. 2010;5(12):e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stressmann FA, Rogers GB, Klem ER, et al. Analysis of the bacterial communities present in lungs of patients with cystic fibrosis from American and British centers. Journal of clinical microbiology. 2011;49(1):281–291. doi: 10.1128/JCM.01650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen DH. Cystic fibrosis of the pancreas and its relation to celiac diseasea clinical and pathologic study. Archives of Pediatrics & Adolescent Medicine. 1938;56(2):344–399. [Google Scholar]
- 29. Han MLK, Huang YJ, Lipuma JJ, et al. Significance of the microbiome in obstructive lung disease. Thorax. 2012;67(5):456–463. doi: 10.1136/thoraxjnl-2011-201183. Review of lung microbiota and obstructive lung disease with a more detailed discussion of molecular techniques of microbial identification than is contained in this review.
- 30.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clinical microbiology reviews. 2010;23(2):299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers G, Hart C, Mason J, Hughes M, Walshaw M, Bruce K. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. Journal of clinical microbiology. 2003;41(8):3548–3558. doi: 10.1128/JCM.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rogers G, Carroll M, Serisier D, Hockey P, Jones G, Bruce K. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16s ribosomal DNA terminal restriction fragment length polymorphism profiling. Journal of clinical microbiology. 2004;42(11):5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. Early DNA-based analysis of sputum from patients with cystic fibrosis demonstrating complexity of microbial communities.
- 33.Rogers GB, Carroll MP, Serisier DJ, et al. Bacterial activity in cystic fibrosis lung infections. Respir Res. 2005;6(1):49. doi: 10.1186/1465-9921-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers G, Carroll M, Serisier D, et al. Use of 16S rRNA gene profiling by terminal restriction fragment length polymorphism analysis to compare bacterial communities in sputum and mouthwash samples from patients with cystic fibrosis. Journal of clinical microbiology. 2006;44(7):2601–2604. doi: 10.1128/JCM.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris JK, De Groote MA, Sagel SD, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proceedings of the National Academy of Sciences. 2007;104(51):20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bittar F, Richet H, Dubus JC, et al. Molecular detection of multiple emerging pathogens in sputa from cystic fibrosis patients. PloS one. 2008;3(8):e2908. doi: 10.1371/journal.pone.0002908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proceedings of the National Academy of Sciences. 2008;105(39):15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armougom F, Bittar F, Stremler N, et al. Microbial diversity in the sputum of a cystic fibrosis patient studied with 16S rDNA pyrosequencing. European journal of clinical microbiology & infectious diseases. 2009;28(9):1151–1154. doi: 10.1007/s10096-009-0749-x. [DOI] [PubMed] [Google Scholar]
- 39.Rogers G, Daniels T, Tuck A, et al. Studying bacteria in respiratory specimens by using conventional and molecular microbiological approaches. BMC pulmonary medicine. 2009;9(1):14. doi: 10.1186/1471-2466-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox MJ, Allgaier M, Taylor B, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PloS one. 2010;5(6):e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doud MS, Light M, Gonzalez G, Narasimhan G, Mathee K. Combination of 16S rRNA variable regions provides a detailed analysis of bacterial community dynamics in the lungs of cystic fibrosis patients. Human genomics. 2010;4(3):147–169. doi: 10.1186/1479-7364-4-3-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guss AM, Roeselers G, Newton ILG, et al. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. The ISME journal. 2010;5(1):20–29. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klepac-Ceraj V, Lemon KP, Martin TR, et al. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environmental microbiology. 2010;12(5):1293–1303. doi: 10.1111/j.1462-2920.2010.02173.x. [DOI] [PubMed] [Google Scholar]
- 44.Rogers GB, Skelton S, Serisier DJ, Van Der Gast CJ, Bruce KD. Determining cystic fibrosisaffected lung microbiology: comparison of spontaneous and serially induced sputum samples by use of terminal restriction fragment length polymorphism profiling. Journal of clinical microbiology. 2010;48(1):78–86. doi: 10.1128/JCM.01324-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Der Gast CJ, Walker AW, Stressmann FA, et al. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. The ISME journal. 2010;5(5):780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudkjøbing VB, Thomsen TR, Alhede M, et al. True microbiota involved in chronic lung infection of cystic fibrosis patients found by culturing and 16S rRNA gene analysis. Journal of clinical microbiology. 2011;49(12):4352–4355. doi: 10.1128/JCM.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sibley CD, Grinwis ME, Field TR, et al. Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PloS one. 2011;6(7):e22702. doi: 10.1371/journal.pone.0022702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willner D, Haynes MR, Furlan M, et al. Spatial distribution of microbial communities in the cystic fibrosis lung. The ISME Journal. 2011 doi: 10.1038/ismej.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniels T, Rogers G, Stressmann F, et al. Impact of antibiotic treatment for pulmonary exacerbations on bacterial diversity in cystic fibrosis. Journal of Cystic Fibrosis. 2012 doi: 10.1016/j.jcf.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Delhaes L, Monchy S, Frealle E, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community--implications for therapeutic management. PloS one. 2012;7(4):e36313. doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Filkins L, Hampton T, Gifford A, et al. Prevalence of Streptococci and Increased Polymicrobial Diversity Associated with Cystic Fibrosis Patient Stability. Journal of Bacteriology. 2012;194(17):4709–4717. doi: 10.1128/JB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fodor AA, Klem ER, Gilpin DF, et al. The Adult Cystic Fibrosis Airway Microbiota Is Stable over Time and Infection Type, and Highly Resilient to Antibiotic Treatment of Exacerbations. PloS one. 2012;7(9):e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goddard AF, Staudinger BJ, Dowd SE, et al. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proceedings of the National Academy of Sciences. 2012;109(34):13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maughan H, Cunningham KS, Wang PW, et al. Pulmonary bacterial communities in surgically resected noncystic fibrosis bronchiectasis lungs are similar to those in cystic fibrosis. Pulmonary medicine. 2012;2012:746358. doi: 10.1155/2012/746358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maughan H, Wang PW, Diaz Caballero J, et al. Analysis of the cystic fibrosis lung microbiota via serial Illumina sequencing of bacterial 16S rRNA hypervariable regions. PloS one. 2012;7(10):e45791. doi: 10.1371/journal.pone.0045791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudkjobing VB, Thomsen TR, Alhede M, et al. The microorganisms in chronically infected endstage and non-end-stage cystic fibrosis patients. FEMS immunology and medical microbiology. 2012;65(2):236–244. doi: 10.1111/j.1574-695X.2011.00925.x. [DOI] [PubMed] [Google Scholar]
- 57.Stressmann FA, Rogers GB, Van Der Gast CJ, et al. Long-term cultivation-independent microbial diversity analysis demonstrates that bacterial communities infecting the adult cystic fibrosis lung show stability and resilience. Thorax. 2012;67(10):867–873. doi: 10.1136/thoraxjnl-2011-200932. [DOI] [PubMed] [Google Scholar]
- 58. Zhao J, Schloss PD, Kalikin LM, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proceedings of the National Academy of Sciences. 2012;109(15):5809–5814. doi: 10.1073/pnas.1120577109. Largest and most thorough longitudinal analysis of changes in sputum microbiota in patients with cystic fibrosis. Clarified influences of disease severity, infection and antibiotics on sputum microbial diversity.
- 59.Smith AL, Fiel SB, Mayer-Hamblett N, Ramsey B, Burns JL. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration Lack of association in cystic fibrosis. CHEST Journal. 2003;123(5):1495–1502. doi: 10.1378/chest.123.5.1495. [DOI] [PubMed] [Google Scholar]
- 60.Tunney MM, Field TR, Moriarty TF, et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med. 2008;177(9):995–1001. doi: 10.1164/rccm.200708-1151OC. [DOI] [PubMed] [Google Scholar]
- 61.Bruzzese E, Raia V, Spagnuolo MI, et al. Effect of Lactobacillus GG supplementation on pulmonary exacerbations in patients with cystic fibrosis: A pilot study. Clinical Nutrition. 2007;26(3):322–328. doi: 10.1016/j.clnu.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 62.Weiss B, Bujanover Y, Yahav Y, Vilozni D, Fireman E, Efrati O. Probiotic supplementation affects pulmonary exacerbations in patients with cystic fibrosis: a pilot study. Pediatric Pulmonology. 2010;45(6):536–540. doi: 10.1002/ppul.21138. [DOI] [PubMed] [Google Scholar]
- 63.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nature reviews. Immunology. 2001;1(1):69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 64.Risnes KR, Belanger K, Murk W, Bracken MB. Antibiotic exposure by 6 months and asthma and allergy at 6 years: findings in a cohort of 1,401 US children. American journal of epidemiology. 2011;173(3):310–318. doi: 10.1093/aje/kwq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alm JS, Swartz J, Lilja G, Scheynius A, Pershagen G. Atopy in children of families with an anthroposophic lifestyle. Lancet. 1999;353(9163):1485–1488. doi: 10.1016/S0140-6736(98)09344-1. [DOI] [PubMed] [Google Scholar]
- 66.Mckeever TM, Lewis SA, Smith C, et al. Early exposure to infections and antibiotics and the incidence of allergic disease: a birth cohort study with the West Midlands General Practice Research Database. J Allergy Clin Immunol. 2002;109(1):43–50. doi: 10.1067/mai.2002.121016. [DOI] [PubMed] [Google Scholar]
- 67.Wjst M, Hoelscher B, Frye C, Wichmann HE, Dold S, Heinrich J. Early antibiotic treatment and later asthma. Eur J Med Res. 2001;6(6):263–271. [PubMed] [Google Scholar]
- 68.Droste JH, Wieringa MH, Weyler JJ, Nelen VJ, Vermeire PA, Van Bever HP. Does the use of antibiotics in early childhood increase the risk of asthma and allergic disease? Clin Exp Allergy. 2000;30(11):1547–1553. doi: 10.1046/j.1365-2222.2000.00939.x. [DOI] [PubMed] [Google Scholar]
- 69.Wickens K, Pearce N, Crane J, Beasley R. Antibiotic use in early childhood and the development of asthma. Clin Exp Allergy. 1999;29(6):766–771. doi: 10.1046/j.1365-2222.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- 70.Bjorksten B, Naaber P, Sepp E, Mikelsaar M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin Exp Allergy. 1999;29(3):342–346. doi: 10.1046/j.1365-2222.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 71.Bottcher MF, Nordin EK, Sandin A, Midtvedt T, Bjorksten B. Microflora-associated characteristics in faeces from allergic and nonallergic infants. Clin Exp Allergy. 2000;30(11):1590–1596. doi: 10.1046/j.1365-2222.2000.00982.x. [DOI] [PubMed] [Google Scholar]
- 72.Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107(1):129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 73.Kirjavainen PV, Apostolou E, Arvola T, Salminen SJ, Gibson GR, Isolauri E. Characterizing the composition of intestinal microflora as a prospective treatment target in infant allergic disease. FEMS Immunol Med Microbiol. 2001;32(1):1–7. doi: 10.1111/j.1574-695X.2001.tb00526.x. [DOI] [PubMed] [Google Scholar]
- 74.Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001;108(4):516–520. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 75.Kirjavainen PV, Arvola T, Salminen SJ, Isolauri E. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut. 2002;51(1):51–55. doi: 10.1136/gut.51.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adlerberth I, Carlsson B, De Man P, et al. Intestinal colonization with Enterobacteriaceae in Pakistani and Swedish hospital-delivered infants. Acta Paediatr Scand. 1991;80(6–7):602–610. doi: 10.1111/j.1651-2227.1991.tb11917.x. [DOI] [PubMed] [Google Scholar]
- 77.Noverr MC, Falkowski NR, Mcdonald RA, Mckenzie AN, Huffnagle GB. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun. 2005;73(1):30–38. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72(9):4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Herbst T, Sichelstiel A, Schar C, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184(2):198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 80.Stuart-Harris CH, Pownall M, Scothorne CM, Franks Z. The factor of infection in chronic bronchitis. The Quarterly journal of medicine. 1953;22(86):121–132. [PubMed] [Google Scholar]
- 81.Tager I, Speizer FE. Role of infection in chronic bronchitis. The New England journal of medicine. 1975;292(11):563–571. doi: 10.1056/NEJM197503132921105. [DOI] [PubMed] [Google Scholar]
- 82.Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. The New England journal of medicine. 2008;359(22):2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- 83.Albert RK, Connett J, Bailey WC, et al. Azithromycin for prevention of exacerbations of COPD. The New England journal of medicine. 2011;365(8):689–698. doi: 10.1056/NEJMoa1104623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang YJ, Kim E, Cox MJ, et al. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. Omics : a journal of integrative biology. 2010;14(1):9–59. doi: 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh S, Amin AV, Loke YK. Long-term use of inhaled corticosteroids and the risk of pneumonia in chronic obstructive pulmonary disease: a meta-analysis. Archives of internal medicine. 2009;169(3):219. doi: 10.1001/archinternmed.2008.550. [DOI] [PubMed] [Google Scholar]
- 86.Botha P, Archer L, Anderson RL, et al. Pseudomonas aeruginosa colonization of the allograft after lung transplantation and the risk of bronchiolitis obliterans syndrome. Transplantation. 2008;85(5):771–774. doi: 10.1097/TP.0b013e31816651de. [DOI] [PubMed] [Google Scholar]
- 87.Husain S, Singh N. Bronchiolitis obliterans and lung transplantation: evidence for an infectious etiology. Seminars in respiratory infections. 2002;17(4):310–314. doi: 10.1053/srin.2002.36442. [DOI] [PubMed] [Google Scholar]
- 88.Khalifah AP, Hachem RR, Chakinala MM, et al. Respiratory viral infections are a distinct risk for bronchiolitis obliterans syndrome and death. Am J Respir Crit Care Med. 2004;170(2):181–187. doi: 10.1164/rccm.200310-1359OC. [DOI] [PubMed] [Google Scholar]
- 89.Charlson ES, Diamond JM, Bittinger K, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186(6):536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Borewicz K, Pragman AA, Kim HB, Hertz M, Wendt C, Isaacson RE. Longitudinal Analysis of the Lung Microbiome in Lung Transplantation. FEMS Microbiology Letters. 2012 doi: 10.1111/1574-6968.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Friaza V, La Horra C, Rodríguez-Domínguez MJ, et al. Metagenomic analysis of bronchoalveolar lavage samples from patients with idiopathic interstitial pneumonia and its antagonic relation with <i> Pneumocystis jirovecii <i> colonization. Journal of microbiological methods. 2010;82(1):98–101. doi: 10.1016/j.mimet.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 92.Flanagan JL, Brodie EL, Weng L, et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. Journal of clinical microbiology. 2007;45(6):1954–1962. doi: 10.1128/JCM.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gleeson K, Eggli DF, Maxwell SL. Quantitative aspiration during sleep in normal subjects. CHEST Journal. 1997;111(5):1266–1272. doi: 10.1378/chest.111.5.1266. [DOI] [PubMed] [Google Scholar]
- 94.Birring SS, Matos S, Patel RB, Prudon B, Evans DH, Pavord ID. Cough frequency, cough sensitivity and health status in patients with chronic cough. Respiratory medicine. 2006;100(6):1105–1109. doi: 10.1016/j.rmed.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 95.Munyard P, Bush A. How much coughing is normal? Archives of disease in childhood. 1996;74(6):531–534. doi: 10.1136/adc.74.6.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Worlitzsch D, Rintelen C, Böhm K, et al. Antibiotic-resistant obligate anaerobes during exacerbations of cystic fibrosis patients. Clinical Microbiology and Infection. 2009;15(5):454–460. doi: 10.1111/j.1469-0691.2008.02659.x. [DOI] [PubMed] [Google Scholar]
- 97.Mccoll KEL. Helicobacter pylori infection. New England Journal of Medicine. 2010;362(17):1597–1604. doi: 10.1056/NEJMcp1001110. [DOI] [PubMed] [Google Scholar]
- 98.Labenz J, Blum A, Bayerdorffer E, Meining A, Stolte M, Borsch G. Curing Helicobacter pylori infection in patients with duodenal ulcer may provoke reflux esophagitis. Gastroenterology. 1997;112(5):1442–1447. doi: 10.1016/s0016-5085(97)70024-6. [DOI] [PubMed] [Google Scholar]
- 99.Chen Y, Blaser MJ. Inverse associations of Helicobacter pylori with asthma and allergy. Archives of Internal Medicine. 2007;167(8):821. doi: 10.1001/archinte.167.8.821. [DOI] [PubMed] [Google Scholar]
- 100.Nwokolo C, Freshwater D, O’hare P, Randeva H. Plasma ghrelin following cure of Helicobacter pylori. Gut. 2003;52(5):637–640. doi: 10.1136/gut.52.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koch R. Untersuchungen ueber Bakterien V. Die Aetiologie der Milzbrand-Krankheit, begruendent auf die Entwicklungsgeschichte des Bacillus Anthracis. Beitr. Z. Biol. D. Pflanzen. 1876;2:277–310. [Google Scholar]
- 102.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science (New York NY) 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yong E. Gut microbial 'enterotypes' become less clear-cut. Gut microbial 'enterotypes' become less clear-cut. 2013 Jan 3;2013 (2012). [Google Scholar]
- 105.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (New York NY) 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Translational Research. 2012 doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nature Reviews Microbiology. 2008;6(2):111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome research. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nature reviews. Rheumatology. 2011;7(10):569–578. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hempel S, Newberry SJ, Maher AR, et al. Probiotics for the prevention and treatment of antibioticassociated diarrhea: a systematic review and meta-analysis. JAMA : the journal of the American Medical Association. 2012;307(18):1959–1969. doi: 10.1001/jama.2012.3507. [DOI] [PubMed] [Google Scholar]
- 112.Allen SJ, Okoko B, Martinez E, Gregorio G, Dans LF. Probiotics for treating infectious diarrhoea. Cochrane database of systematic reviews (Online) 2004;(2):CD003048. doi: 10.1002/14651858.CD003048.pub2. [DOI] [PubMed] [Google Scholar]
- 113.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2011;53(10):994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 114.Forsythe P. Probiotics and lung diseases. Chest. 2011;139(4):901–908. doi: 10.1378/chest.10-1861. [DOI] [PubMed] [Google Scholar]
- 115.Morrow LE, Kollef MH, Casale TB. Probiotic prophylaxis of ventilator-associated pneumonia: a blinded, randomized, controlled trial. Am J Respir Crit Care Med. 2010;182(8):1058–1064. doi: 10.1164/rccm.200912-1853OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Popova M, Molimard P, Courau S, et al. Beneficial effects of probiotics in upper respiratory tract infections and their mechanical actions to antagonize pathogens. Journal of Applied Microbiology. 2012 doi: 10.1111/j.1365-2672.2012.05394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]