Abstract
Background
Identification of the physiological changes that occur during the early stages of Alzheimer’s disease (AD) may provide critical insights for the diagnosis, prognosis and treatment of disease. Cerebrospinal fluid (CSF) biomarkers are a rich source of information that reflect the brain proteome.
Methods
We applied a novel approach to screen a panel of ~190 CSF analytes quantified by multiplex immunoassay and detected common associations in the Knight- Alzheimer’s Disease Research Center (ADRC;N=311) and the Alzheimer’s Disease Neuroimaging Initiative (ADNI;N=293) cohorts. CSF ptau181-Aβ42 ratio was used as a continuous trait, rather than case control status in these analyses.
Results
We demonstrate the ptau181-Aβ42 ratio has more statistical power than traditional modeling approaches and that the levels of CSF Fatty Acid Binding Protein (H-FABP) and 12 other correlated analytes increase as the disease progresses. These results were validated using the traditional case control status model. Stratification of our dataset demonstrated that increases in these analytes occur very early in the disease course and were apparent even in non-demented individuals with AD pathology (low ptau181, low Aβ42) compared to pathology-negative elderly control subjects (low ptau181, high Aβ42). FABP-Aβ42 ratio demonstrates a similar hazard ratio for disease conversion to ptau181-Aβ42 even though the overlap in classification is incomplete suggesting that FABP contributes independent information as a predictor
Conclusions
Our results clearly indicate that the approach presented here can be employed to correctly identify novel biomarkers for AD, and that CSF H-FABP levels start to increase at very early stages of the disease.
Keywords: Alzheimer’s disease, Biomarkers, cerebrospinal fluid (CSF), Ptau-Aβ42 ratio, Heart Fatty Acid binding protein, Brain Proteome - Rules Based Medicine Discovery Multi-Analyte Profile 1.0
Introduction
There is accumulating evidence that the clinical symptoms of Alzheimer’s disease (AD) are preceded by a long preclinical phase in which pathological protein aggregation occurs in the brain (1–4). β-amyloid plaques are estimated to develop ~15–20 years before the onset of cognitive impairment and neurofibrillary tangles begin to accumulate at least 5 years before symptom onset (2; 3). Furthermore, substantial neurodegeneration is apparent in even mildly symptomatic individuals (1). These observations, together with the failure of clinical trials in symptomatic individuals illustrate the urgent need for additional biomarkers that characterize the preclinical stage of disease and enable treatment before the brain undergoes irreversible neurological damage (2–4).
Analytes in cerebrospinal fluid (CSF) reflect the brain proteome and are a rich source of biomarkers. In fact, CSF levels of Aβ42, tau, and tau phosphorylated at threonine 181 (ptau181) are related to AD by their association with the presence of β-amyloid deposition, neurofibrillary tangles and neuronal cell death (5–8). Similarly, changes in CSF Visinin-like protein-1 (VILIP-1), a neuronal calcium-sensor protein employed as a marker of neuronal injury (2; 9), and and chitinase-3 like-1, and chitinase 3-like 1, cartilage glycoprotein-39 (YKL-40), an inflammatory biomarker (10), have also been associated with AD. Additionally, both ptau181-Aβ42 ratio (ptau181/Aβ42) and YKL-40-Aβ42 ratio (YKL-40/Aβ42) have been shown to be strong predictors of conversion from cognitively normal to very mild/mild cognitive impairment over a 3~4 year period (l; 10; 11). Despite this, there is a need for additional biomarkers to identify and characterize the “preclinical” stage (pathology present with cognition intact) of the disease and to track the effectiveness of therapies. A challenge for clinical trials is to identify a target population of individuals with a high risk for converting from cognitively normal to impaired over a relatively short period of time. Currently elevated ptau181-Aβ42 ratio is used in some clinical trials to select such individuals. Other predictive biomarkers are needed both for the selection of patient cohorts for clinical trials and for monitoring disease progression and the success of disease-modifying treatment outcomes. Ultimately, biomarkers will enable early intervention in presymptomatic individuals with the goal of delaying the onset or preventing the cognitive decline seen in AD before the brain is irreversibly injured (2).
Two features of AD result in misclassification of subjects and severely reduce the power of traditional clinical measures in the search for novel biomarkers: 30% of cognitively normal individuals show Alzheimer’s-type neuropathology by age 75yrs (2; 3; 12; 13) and current clinical diagnostic methods are only 83% accurate as defined by neuropathological confirmation at autopsy (14). To identify new CSF analytes associated with AD we took a novel approach by developing a quantitative measure of “caseness”, employing the CSF ptau181-Aβ42 ratio (ptau181/Aβ42) as an endophenotype for AD. This status independent criterion avoids the misclassification of controls and cases, noted above and is a continuous quantitative trait. Both of these characteristics improve the power of this approach over traditional clinical measures.
Methods and Materials
Subjects
Participants enrolled in the Knight-ADRC (N=311) were diagnosed based on criteria from the National Institute of Neurological and Communicative Diseases and Stroke-Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA) (15). CSF collection, processing and assessment is described elsewhere: (11; 16). Data for subjects included in the ADNI study (N=292) were accessed from the ADNI website (See Supplementary Methods and Table SI in Supplement 1) (8). Processing, aliquoting and storage were performed according to the ADNI Biomarker Core Laboratory Standard Operating Procedures (http://adni.loni.ucla.edu). Additional details for these two studies are provided elsewhere (10; 17) Genotyping of rs7412 and rs429358 which define the APOE ε2/ε3/ε4 isoforms were previously described (17; 18).
Expression studies were carried out using cDNA obtained from the parietal lobe from 82 AD cases (86±7years; 45%male; and 41% APOE ε4 carriers) and 39 cognitively normal individuals (85±9; 41% male; and 23% APOE ε4 carriers) obtained through the Knight-ADRC Neuropathology Core. Real-time data were analyzed using the comparative Ct method (19) (See Supplementary Methods in Supplement 1).
Analyte Measurement
The samples from both the Knight-ADRC and ADNI were evaluated by Rules Based Medicine, Inc. (RBM) (Austin, TX) for levels of 190 analytes using the Human Discovery Multi-Analyte Profile (MAP) 1.0 panel and a Luminex 100 platform. Only analytes with <10% missing values in each study were analyzed. The protocol used to quantify plasma analytes is described elsewhere (20; 21).
CSF Aβ42, and phospho-tau181 (ptau181) levels for the Knight-ADRC participants were obtained in duplicate by the WU-ADRC Biomarker Core by quantitative ELISA (Innotest; Innogenetics, Ghent, Belgium)(16). Quantification of CSF VILIP-1 and YKL-40 is described elsewhere (9; 10). For the ADNI participants the CSF Aβ42, and ptau181 levels were measured using the multiplex xMAP Luminex platform as described (8).
Statistical analysis
The analyses were performed in R (The R Foundation for Statistical Computing v2.14.1). Analytes were log-transformed to approximate a normal distribution, and outliers were removed. The associations with the CSF ptau181-Aβ42 ratio were tested using linear regression models, adjusting for age at LP; sex and APOE genotype, encoded as five levels based on the genetic risk (APOE ε22=0; ε23=l; ε33=2; ε24=3; ε34=4; and ε44=5). For analyses using the combined datasets we included study as a covariate. We used two approaches to correct for the multiple testing associated with examining 64 analytes: Bonferroni correction and SimpleM. For 64 tests the Bonferroni corrected p-value corresponding to p=0.05 is p= 7.81E-04. Because this method is most likely too conservative given the correlation structure of the analyte measurements, we also extended and applied the method termed SimpleM(22) (see Supplementary Methods in Supplement 1), and obtained corrected p-values =1.39E-03 and 1.43E-03 for the Knight-ADRC and ADNI studies respectively. Logistic regression was used to test for association with CDR (CDR=0 vs. CDR>0), adjusting for the covariates mentioned above. Kaplan-Meier survival curves were calculated employing the function survfit, and the Cox proportional hazard regression models were tested using the function coxph (package survival; v2.36–10). The Cox proportional hazard tests were adjusted by age, sex, APOE genotype, and study. Hierarchical clustering was calculated using the complete linkage method provided (function hclust), and heatmaps were plotted using the function heatmap.2 (package gplots v2.10.1). Multivariate stepwise model selection (Table 3AB) was performed using the function stepAIC (package MASS, v7.3–16), optimizing for the Bayesian Information Criterion; and included age, sex and APOE genotype as fixed covariates. The minimal multivariate model shown in Table 3C was obtained by selecting the subset of analytes identified as significant in each study (Table 3AB). Moreover, we constrained this list to the analytes that were also significant (p-value<5.0E-2) when the identified models were applied to the other study. The principal component analysis was performed in R running the method prcomp (package stats v 2.14.1) The random forest results were obtained executing the function randomForest (implemented in the package randomForest, v4.6–6) to grow 1000 trees, and the importance measure was calculated by the mean decrease in accuracy.
Table 3.
Multivariate models. Stepwise optimization in A) the Knight-ADRC study, and B) the ADNI study. C) A minimal model that includes the analytes commonly selected in A and B.
|
|
|
|||
|---|---|---|---|---|
| A) | Inferred Knight-ADRC Model | Replication on ADNI | ||
| p-value | effect | p-value | effect | |
|
|
|
|||
| H-FABP | 3.18E-08 | 0.44 | 1.74E-11 | 0.47 |
| VEGF | 1.01E-05 | −0.63 | 1.85E-04 | −0.66 |
| TNF.RII | 8.56E-04 | 0.63 | 1.08E-01 | 0.22 |
| Thrombomodulin | 2.03E-03 | −0.37 | 1.01E-03 | −0.39 |
| ACE | 2.25E-03 | −0.36 | 8.66E-01 | −0.02 |
| Adiponectin | 2.29E-03 | 0.17 | 7.98E-03 | 0.23 |
| HGF | 2.69E-03 | 0.51 | 5.60E-08 | 0.60 |
| MIF | 3.30E-03 | 0.33 | 5.88E-04 | 0.16 |
| SAP | 4.26E-03 | −0.21 | 9.67E-01 | 0.00 |
| CXCL9 | 8.17E-03 | −0.14 | 9.23E-01 | −0.01 |
| FasL | 1.12E-02 | 0.21 | 2.95E-01 | −0.10 |
| age | 1.33E-01 | 0.01 | 3.17E-01 | −0.01 |
| sex | 4.84E-01 | −0.05 | 3.57E-01 | −0.07 |
| APOE Genotypes | 2.80E-07 | 0.14 | 3.64E-11 | 0.19 |
| R squared/BIC | 0.50/−323.91 | 0.56/−222.42.68 | ||
|
|
|
|||
|---|---|---|---|---|
| B) | Replication on Knight-ADRC | Inferred ADNI Model | ||
| p-value | effect | p-value | effect | |
|
|
|
|||
| H-FABP | 2.50E-09 | 0.44 | 5.06E-16 | 0.50 |
| HGF | 2.39E-02 | 0.37 | 2.19E-11 | 0.68 |
| VEGF | 1.65E-04 | −0.60 | 4.99E-06 | −0.68 |
| Complement 3 | 7.57E-01 | −0.03 | 2.62E-05 | −0.63 |
| ANGPT2 | 4.04E-01 | 0.12 | 2.10E-04 | 0.41 |
| APOA | 1.13E-02 | −0.26 | 2.58E-04 | 0.39 |
| MIF | 2.08E-02 | 0.33 | 6.71E-04 | 0.14 |
| Fibrinogen | 2.02E-01 | 0.09 | 1.73E-03 | −0.12 |
| Prolactin | 6.80E-01 | −0.04 | 9.88E-03 | −0.26 |
| TRAIL.R3 | 8.13E-01 | 0.04 | 1.70E-02 | −0.28 |
| sex | 4.96E-01 | 0.05 | 8.60E-01 | −0.01 |
| age | 7.24E-03 | 0.01 | 9.89E-01 | 0.00 |
| APOE Genotypes | 7.90E-08 | 0.16 | 6.16E-17 | 0.22 |
| R squared/BIC | 0.44/−296.91 | 0.60/−370.60 | ||
|
|
|
|||
|---|---|---|---|---|
| C) | Knight-ADRC | ADNI | ||
| p-value | effect | p-value | effect | |
|
|
|
|||
| H-FABP | 6.44E-10 | 0.45 | 1.87E-14 | 0.50 |
| HGF | 3.25E-02 | 0.33 | 5.93E-09 | 0.51 |
| MIF | 6.27E-04 | 0.40 | 3.11E-04 | 0.16 |
| VEGF | 3.47E-05 | −0.59 | 1.32E-06 | −0.66 |
| age | 6.78E-03 | 0.01 | 3.51E-01 | 0.00 |
| sex | 3.27E-01 | 0.06 | 9.73E-01 | 0.00 |
| APOE Genotypes | 9.46E-09 | 0.16 | 1.44E-14 | 0.21 |
| R squared/BIC | 0.42/−336.92 | 0.53/−360.04 | ||
Results
CSF ptau181/Aβ42 confers more statistical power to identify CSF analytes associated to AD
We first evaluated the statistical power of the CSF ptau181/Aβ42 measure compared to the Clinical Dementia Rating (CDR) at lumbar puncture (LP), using CSF levels of VILIP-1(9) and YKL-40(10), that had previously been measured and shown to be associated with CDR and CDR–sum of boxes in the Knight-ADRC study (11; 20; 23). The CSF ptau181-Aβ42 ratio has previously been shown to be a strong predictor of both the conversion of cognitively normal subjects to very mild or mild dementia (11), and the rate of decline across time in individuals with very mild dementia (24). The CSF ptau181-Aβ42 ratio is strongly associated with CDR at LP (p-value=5.05E-16).
We employed linear regression models to analyze the association of CSF VILIP-1 with the log-transformed levels of CSF ptau181-Aβ42 ratio, and observed a much stronger association than with CDR (p-value=3.18E-17 vs. 3.80E-05). The CSF ptau181-Aβ42 ratio remained more powerful than CDR even when converted to a dichotomous variable using two partitioning criteria. First, we split the subjects by the median value of CSF ptau181-Aβ42 ratio and second we compared the subjects from the upper tercile to the lower two terciles (intended to resemble the CDR distribution of the Knight-ADRC series). Both logistic models that included the CSF ratio showed much stronger associations (p-values=7.11E-08 and 4.00E-09 respectively) than the CDR model (p-value=3.80E-05).
Similarly, the association of YKL-40 with CSF ptau181-Aβ42 ratio was stronger than CDR (p-value =8.99E-09 vs. 7.76E-04 respectively). We also tested the normal quantile transformed CSF ratio and the same two dichotomous representations for YKL-40. In every case, the test that included the CSF ratio was more statistically significant than the CDR model (p-value=9.42E-06 and 3.04E-05 partitioning by the median and by the upper tercile respectively). Together these analyses provide a strong rationale for our subsequent use of the CSF ptau181-Aβ42 ratio for novel biomarker discovery.
Correlation structure of the CSF analytes
We applied stringent quality criteria to the CSF analyte measurements, selecting only those that were measurable in >90% of the subjects in each series (n=311 and 293 for the Knight-ADRC and the ADNI series). A total of 64 CSF analytes met this criterion.
Analysis of the correlation matrix for the combined measurements of both datasets showed clear patterns in the analyte levels (Table S2 in Supplement 2). The hierarchical clustering method identified 10 clusters containing 48 of the CSF analytes, with a within-cluster correlation r2>0.50. Six clusters included 2 analytes, and the other 4 clusters included 3, 4, 9 and 20 analytes (Figure S1 in Supplement 1). The remaining 16 CSF analytes showed only weak correlations to other analytes in the dataset (mean r2=0.16, and maximum r2=0.35).
CSF Analytes associated with ptau181-Aβ42 ratio
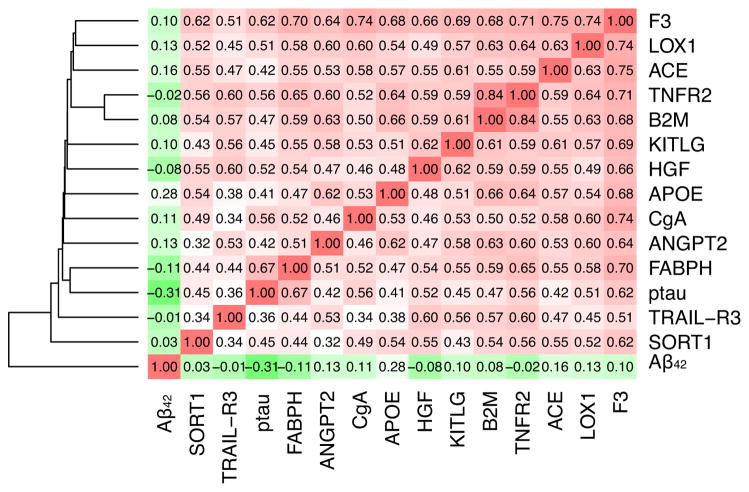
Next we tested for association between each of the CSF analytes and the ptau181-Aβ42 ratio. Thirteen analytes were significantly associated with CSF ptau181-Aβ42 ratio (p-value<1.0E-3) and exhibited the same direction of effect in the Knight-ADRC and the ADNI series (Table 1; and see Table S3 in Supplement 1 for the exhaustive list of the analytes evaluated); Eleven of these CSF analytes showed a mean r2=0.58 with each other (Figure 1), while the other two, Sortilin (S0RT1) and Tumor necrosis factor–Related Apoptosis-Inducing Ligand R3 (TRAIL-R3), exhibited a slightly lower correlation (r2=0.49 and 0.48, respectively) (Figure 1).
Table 1.
CSF analytes significantly associated with CSF ptau181-Aβ42 ratio (p-value<1.0E-03)*
| analyte | Knight-ADRC
|
ADNI
|
||||
|---|---|---|---|---|---|---|
| p-value | Effect | c.i. | p-value | Effect | ci | |
|
|
|
|||||
| Fatty Acid Binding Protein (Heart) - H-FABP | 4.77E-18 | 0.54 | (0.43–0.66) | 7.40E-18 | 0.50 | (0.39–0.61) |
| Tumor Necrosis Factor Receptor 2 - TNFR2 | 2.64E-13 | 0.91 | (0.68–1.15) | 6.11E-07 | 0.55 | (0.34–0.76) |
| Tissue Factor - F3 | 9.18E-13 | 0.54 | (0.40–0.69) | 4.20E-09 | 0.46 | (0.31–0.62) |
| Hepatocyte Growth Factor - HGF | 4.77E-11 | 0.88 | (0.62–1.13) | 5.13E-14 | 0.66 | (0.50–0.82) |
| Chromogranin A-CgA | 1.52E-10 | 1.11 | (0.78–1.44) | 1.27E-07 | 0.95 | (0.60–1.29) |
| Stem Cell Factor - KITLG | 1.08E-08 | 0.64 | (0.42–0.85) | 2.97E-05 | 0.43 | (0.23–0.63) |
| Sortilin - SORT1 | 3.22E-08 | 0.89 | (0.58–1.20) | 9.10E-04 | 0.50 | (0.21–0.79) |
| Lectin Like Oxidized LDL Receptor 1 - LOX1 | 1.54E-07 | 0.51 | (0.32–0.69) | 1.34E-05 | 0.47 | (0.26–0.68) |
| Beta2Microglobulin (B2M) | 3.36E-07 | 0.70 | (0.44–0.97) | 5.07E-05 | 0.53 | (0.28–0.79) |
| TNF-Related Apoptosis-I.L. R3 - TRAIL-R3 | 3.67E-07 | 0.82 | (0.51–1.13) | 4.73E-04 | 0.38 | (0.17–0.58) |
| Angiopoietin 2-ANGPT2 | 2.01E-06 | 0.58 | (0.34–0.81) | 1.90E-04 | 0.36 | (0.17–0.55) |
| Apolipoprotein E - APOE | 4.83E-05 | 0.45 | (0.23–0.66) | 8.26E-04 | 0.40 | (0.17–0.63) |
| Angiotensin-Converting Enzyme - ACE | 6.18E-05 | 0.39 | (0.20–0.58) | 7.86E-04 | 0.34 | (0.14–0.54) |
Only analytes showing similar effect
Figure 1.
Correlation among the analytes associated, CSF Aβ42 and ptau181 in the combined Knight-ADRC and the ADNI studies.
CSF Heart Fatty acid binding protein (H-FABP) was the most significantly associated analyte in both series (p-value= 4.77E-18 and 7.40E-18 for the Knight-ADRC and the ADNI studies, respectively), and was associated with higher levels of H-FABP in individuals with CSF ptau181/Aβ42 ratios indicative of AD (Figure 2a): the regression coefficients for the two studies (Table 1) did not differ significantly (F-Test p-value=0.24).
Figure 2.
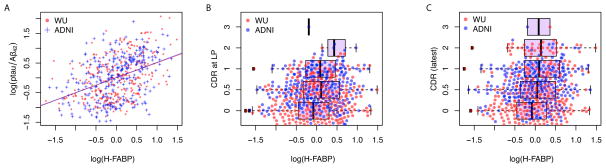
Distribution of CSF H-FABP in the combined Knight-ADRC and the ADNI studies.
A) Linear model of CSF H-FABP compared to CSF ptau/Aβ42; Boxplots for CSF H-FABP stratified by B) CDR at lumbar puncture (LP) and C) latest ascertained CDR.
To investigate whether these associations were apparent in non-demented and demented individuals we combined the subjects from both studies and stratified them by the presence or absence of cognitive impairment. A total of 236 subjects had a CDR>0 and low CSF Aβ42 levels, which we employed as a proxy for brain Aβ42 deposition (16) (cutoff=500 pg/ml and 192 pg/ml for the Knight-ADRC and the ADNI studies, respectively). To evaluate the impact of Aβ42 deposition we further stratified the 300 cognitively normal subjects (CDR=0) by their CSF Aβ42 levels, distinguishing the 192 “clean” controls from the 105 preclinical subjects with lower CSF Aβ42 levels. Each of the 13 CSF analytes was significantly associated (p-value<5.0E-4) in every stratum (Table 2). The ANCOVA analysis revealed that the effect size for CSF H-FABP was significantly lower in controls, compared to the inferred preclinical and clinically assessed cognitively impaired individuals (p-value=4.44E-04 and 5.87E-03 respectively); and that there was no significant difference in the effect observed in the preclinical and symptomatic cases (Figure 3). This is reflected in the different levels of CSF H-FABP among the strata. While H-FABP levels discriminate cognitively impaired individuals from preclinical AD (p-value= 5.05-03), the levels of H-FABP are not significantly different in preclinical individuals from the clean controls, indicating that early amyloid-β plaque deposition does not affect the association of CSF H-FABP with the CSF ptau181-Aβ42 ratio. Importantly, the association of these analytes with the CSF ptau181-Aβ42 ratio is robust to the presence of clinically assessed cognitively impaired individuals with uncharacteristic high levels of CSF Aβ42. (Table S4 in Supplement 1).
Table 2.
Stratified analysis for cognitively normal, preclinical AD and symptomatic AD
| analyte | Cognitive normal (N=195) | Preclinical AD (N=105) | Symptomatic AD (N=236) | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| p-value | effect | c.i. | p-value | effect | c.i. | p-value | effect | c.i. | |
|
|
|||||||||
| Fatty Acid Binding Protein (Heart) - H-FABP | 1.67E-10 | 0.30 | (0.35–0.66) | 8.42E-14 | 0.60 | (0.46–0.74) | 1.59E-22 | 0.49 | (0.40–0.58) |
| Tumor Necrosis Factor Receptor 2 - TNFR2 | 1.41E-09 | 0.51 | (0.30–0.49) | 1.45E-09 | 1.07 | (0.75–1.39) | 1.64E-14 | 0.70 | (0.53–0.87) |
| Tissue Factor - F3 | 7.30E-14 | 0.40 | (0.25–0.59) | 1.49E-13 | 0.72 | (0.55–0.89) | 1.10E-23 | 0.63 | (0.52–0.74) |
| Hepatocyte Growth Factor - HGF | 1.71E-06 | 0.42 | (0.38–0.85) | 8.53E-09 | 1.04 | (0.71–1.37) | 1.86E-16 | 0.66 | (0.51–0.81) |
| Chromogranin A - CgA | 7.74E-07 | 0.61 | (0.32–0.59) | 3.13E-11 | 1.50 | (1.10–1.90) | 4.78E-25 | 1.45 | (1.21–1.70) |
| Stem Cell Factor - KITLG | 1.19E-10 | 0.46 | (0.13–0.57) | 4.59E-10 | 0.99 | (0.71–1.27) | 3.40E-16 | 0.71 | (0.55–0.87) |
| Sortilin - SORT1 | 2.30E-03 | 0.35 | (0.30–0.56) | 1.49E-06 | 1.05 | (0.64–1.45) | 1.88E-07 | 0.67 | (0.42–0.91) |
| Lectin Like Oxidized LDL Receptor 1 - LOX1 | 4.08E-10 | 0.43 | (0.46–0.80) | 1.30E-10 | 0.85 | (0.62–1.09) | 1.67E-11 | 0.57 | (0.41–0.73) |
| Beta2Microglobulin (B2M) | 4.75E-12 | 0.63 | (0.22–0.59) | 5.38E-07 | 0.99 | (0.63–1.36) | 1.73E-09 | 0.64 | (0.44–0.85) |
| TNF-Related Apoptosis-I.L. R3 - TRAIL-R3 | 2.66E-05 | 0.41 | (0.42–0.69) | 1.12E-04 | 0.86 | (0.44–1.29) | 1.58E-03 | 0.32 | (0.12–0.52) |
| Angiopoietin 2 - ANGPT2 | 2.66E-14 | 0.55 | (0.22–0.56) | 2.86E-05 | 0.75 | (0.41–1.08) | 1.13E-08 | 0.49 | (0.33–0.65) |
| Apolipoprotein E - APOE | 6.82E-06 | 0.39 | (0.26–0.52) | 1.29E-06 | 0.76 | (0.47–1.05) | 1.68E-11 | 0.59 | (0.42–0.75) |
| Angiotensin-Converting Enzyme - ACE | 1.53E-08 | 0.39 | (0.46–0.74) | 2.32E-07 | 0.67 | (0.43–0.90) | 3.56E-12 | 0.59 | (0.43–0.75) |
Figure 3.
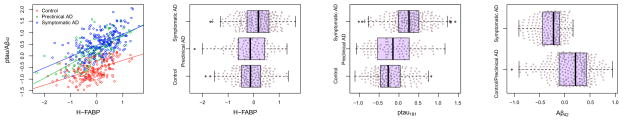
Distribution of CSF H-FABP, ptau181 and Aβ42 stratified by the cognitive status of the subjects (cognitive normal; preclinical; and cognitive decline) in the combined Knight-ADRC and the ADNI studies..
In contrast, the plasma levels of these analytes were not significantly associated with CSF ptau181-Aβ42 ratio (p-values>0.05) in either the Knight-ADRC or the ADNI series (consistent with the null correlation between the CSF and plasma levels of each analyte) (Table S5 in Supplement 1).
CSF heart fatty acid binding protein is associated with ptau181-Aβ42 ratio
To determine whether each of the novel analytes provided independent information to predict the CSF ptaui181-Aβ42 ratio, we combined the subjects from both studies and evaluated joint models that included CSF H-FABP and other CSF analytes from the candidate set, one at a time. Only Hepatocyte Growth Factor (HGF) remained significant after inclusion of CSF H-FABP in the model (p-value=8.56E-06) (Table S6 in Supplement 1).
CSF APOE, which is also associated with the CSF ptaui181-Aβ42 ratio (Table 1) has previously been reported to be associated with AD (17). To determine whether the previously reported association of CSF APOE levels with CSF ptau181-Aβ42 ratio is independent of the other analytes, we extended the models to include CSF APOE. Only CSF Angiopoietin 2 (ANGPT2) and CSF Angiotensin-Converting Enzyme (ACE) show a decrease in the significance of their association (p-values=3.53E-03 and 3.34E-02), while the other CSF analytes remained significant (p-value<1.0E-3) (Table S6 in Supplement 1), indicating they are capturing additional information. Similarly, the joint analysis of CSF H-FABP and VILIP-1(9) or YKL-40(10) demonstrated that CSF H-FABP is providing non-redundant information to the predictive model for CSF ptau181-Aβ42 ratio (See text in Supplement 1).
To determine whether CSF H-FABP levels were a strong predictor of future conversion to CDR>0, we compared the association of CSF H-FABP levels with CDR at LP and CDR after a mean of 4 yrs following LP. We tested the association of CSF H-FABP with CDR at LP and found that it is significantly associated in the ADNI study but marginally associated in the Knight-ADRC study (p-values= 1.55E-04 and 8.66E-02 respectively) (Figure 2b). We also evaluated the association with the latest available evaluation of CDR (mean elapsed years=4.29; and SD=2.30 years) and found it significant (p-value=3.72E-03) for the Knight-ADRC study (36 converted from CDR=0 to CDR>0 and for the ADNI study (p-value=6.17E-05; mean elapsed years=3.90; and SD=1.92 years). We observed a similar trend when we analyzed the CDR sum of boxes (CDR-SB), (25) and the Mini-mental state examination (MMSE) (26). H-FABP levels in the ADNI study were strongly associated with both endophenotypes at LP and the latest available evaluations, while in the Knight-ADRC study H-FABPH levels were significantly associated (p<0.05) only for the latest evaluations (Table S7 in Supplement 1).
To test whether the mRNA expression of H-FABP is also associated with CDR, we measured the mRNA levels in parietal lobe tissue from 112 subjects (73 AD cases and 39 controls) (27). mRNA levels of H-FABP in parietal lobe were not associated with expiration CDR (p-value=0.76) or Braak and Braak staging of the pathology (p-value=0.46) (28).
CSF H-FABP as a predictor of developing cognitive impairment
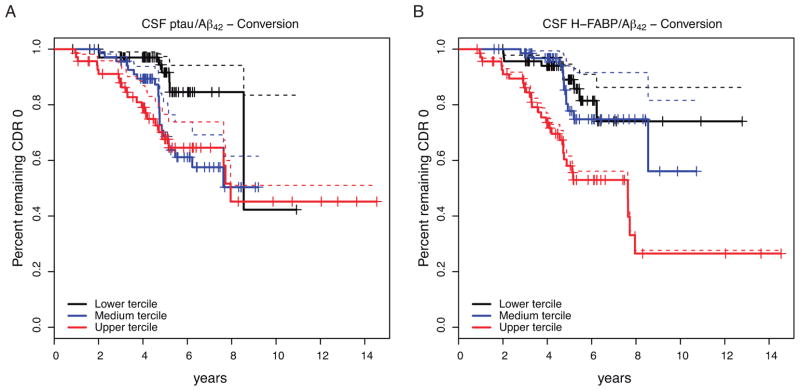
To determine whether higher levels of CSF H-FABP predict conversion from CDR=0 to CDR>0) we performed a survival analysis. The Cox regression analysis showed similar hazard ratios for CSF H-FABP (Hazard ratio = 1.58, 95% CI = 1.02–2.44; p-value = 3.70-02), CSF ptau181 (2.04; 95% CI=1.11–3.71; p-value=1.91E-02) and CSF Aβ42 (2.93; 95% CI = 1.15–7.42; p-value= 2.74E-02). In addition, we compared the survival curves for CSF ptau181-Aβ42 ratio (11) and CSF H-FABP-Aβ42 ratio (29). Both ratios showed similar results (Figure 4) (Cox proportional hazard: ratio=2.05 and 1.85; p-value=2.14E-03 and 3.93E-03; 95% CI= 1.29–3.27 and 1.21–2.79 for CSF ptau181/Aβ42 and CSF H-FABP/Aβ42 respectively). Importantly, only 62% of the subjects were assigned to the same tercile when stratified by these ratios (i.e. P (QCSF ptau/Aβ42 = Q CSF H-FABP/Aβ42)=0.65). Additionally, we observed that the CSF H-FABP-Aβ42 ratio is associated with CDR-SB (Cox proportional hazard: ratio=1.76; p-value=2.83e-05; 95% CI= 1.35–2.29); and it is also associated with the MMSE (Cox proportional hazard: ratio=1.74; p-value=1.47e-04; 95% CI= 1.30–2.32)
Figure 4.
Kaplan-Meyer (solid lines) and Cox survival (dashed lines) curves for the conversion (CDR=0 to CDR>0) for CSF ptau181/Aβ42 and CSF H-FABP/Aβ42. Only 7.46% of the subjects within the lowest tercile for CSF ptau-Aβ42 ratios converted after 5 years (Figure 4a), compared to 23.18% and 27.14% of the subjects in the intermediate and higher terciles. Similarly, 8.69, 10.92 and 34.78% of the subjects within the lower, intermediate and higher terciles of CSF H-FABP/Aβ42 values had converted after 5 years.
CSF Macrophage Migration Inhibitory Factor and Vascular Endothelial Growth Factor are also associated with the CSF ptau181-Aβ42 ratio
We investigated whether multivariate models would detect analytes that did not show strong evidence of association in single analyte models. Our approach included a discovery phase in which we made use of stepwise regression analysis to optimize a multivariate model for the CSF ptau181-Aβ42 ratio in one of the datasets, which was followed by a replication phase, in which the model was applied in the second dataset.
The multivariate model optimized for the Knight-ADRC dataset (Table 3a) showed an increased R2 (0.14 for the discovery set and 0.12 for the ADNI study). It included H-FABP and HGF as well as CSF Vascular Endothelial Growth Factor (VEGF), Macrophage Migration Inhibitory Factor (MIF), Thrombomodulin and Adiponectin (Table 3a). The model optimized for the ADNI dataset (Table 3b) also increased the R2 for both datasets (0.20 and 0.08 for the ADNI and Knight-ADRC dataset respectively). This model also selected H-FABP, HGF, VEGF and MIF. We constructed a multivariate model with these four analytes and observed that all of them remained significantly associated with CSF ptau181-Aβ42 ratio in the two datasets (Table 3c), reflecting an increased goodness of fit for the model (increment of R2=0.08 and 0.09 for the Knight-ADRC and the ADNI datasets, respectively; Table 3c). To evaluate whether the correlation among the analytes produced spurious results, we applying principal component analysis to these selected analytes and evaluated a multivariate model with the values rotated. We found that that all of them remained significant (p-value<0.05), indicating that regardless of their correlation these analytes provide additional information.
CSF MIF levels show a trend toward association with the CSF ptau181-Aβ42 ratio in the single analyte analysis (Table S3 in Supplement 1). Although CSF VEGF is mildly positively associated in the single analyte analysis (Table S3 in Supplement 1), it has a negative effect in the multivariate model. The stratified analysis revealed that the change of direction occurs only in the cases (Figure S2 in Supplement 1).
The random forest method was also used to analyze these datasets and also highlighted the importance of H-FABP, MIF, HGF and VEGF. These analytes are among the top 6 for the Knight-ADRC dataset; and H-FABP, HGF and MIF are among the top 8 analytes for the ADNI dataset (Table S8 in Supplement 1).
Discussion
The goal of this study was to identify novel CSF biomarkers for AD using a discovery dataset collected by the Knight ADRC and a replication cohort ascertained by ADNI. Although the ascertainment and structure of the datasets is quite different, we were able to identify several biomarkers that showed consistent effects across the two datasets. The RBM discovery MAP 1.0 panel includes 64 analytes that are not totally independent of one another. We made use of the correlation structure of these analytes to approximate the number of independent tests, and consequently used this number to adjust our p-values for multiple testing. Moreover, we employed this information to understand the apparent excess of analytes associated with the CSF ptau181-Aβ42 ratio.
A novel feature of our study is the use of the CSF ptau181-Aβ42 ratio as the outcome variable in our analyses rather than comparing the levels of the test analytes in cases versus controls. The CSF ptau181-Aβ42 ratio provides several key advantages over the more traditional approach. It captures the progression of the disease before the onset of clinical symptoms by correcting the misclassification of control subjects with amyloid-β plaques. In addition, this method also correctly assigns clinically demented individuals who do not have Alzheimer’s disease. In a traditional analysis these individuals would be included as cases, but when using the ptau181-Aβ42, ratio would be analyzed as non-AD dementias. We believe that each of these factors contribute to the gain in statistical power of the CSF ptau181-Aβ42 ratio, that we observed comparing its performance to the usual case-control model in the evaluation of the CSF levels of VILIP-1 and YKL-40.
The analytes associated with CSF ptau181-Aβ42 ratio were correlated with CSF ptau181 but not Aβ42 (Figure 1), indicating that they do not reflect the very early Aβ – related events in the development of the disease, but they do discriminate between preclinical and symptomatic AD. Query of the Database for Annotation, Visualization and Integrated Discovery (DAVID) (30) failed to identify any pathway that characterized the candidate set of analytes.
CSF levels of H-FABP, the most significant analyte, were consistently associated with the ptau181-Aβ42 ratio in both the Knight-ADRC and ADNI datasets, even when the model included other reported biomarkers such as CSF VILIP-1 and YKL-40. Two previous studies have reported the association of CSF H-FABP with AD in other smaller cohorts (31; 32). In the current study we show that this association is present even at very early stages of the disease, as demonstrated by the analysis of cognitively normal subjects with evidence of amyloid-β plaques. One limitation of our analysis is that although both studies are longitudinal, the novel biomarkers have only been measured in cross sectional data. We are therefore unable to address the role of FABP as a novel AD biomarker across the entire course of disease at the current time. Despite this limitation of our study design, we used the available longitudinal data to show how CSF H-FABP can be employed to predict the conversion from cognitive normality to cognitive impairment. It still remains to be determined whether addition of H-FABP in a model including both CSF ptau181 and Aβ42 levels improves the accuracy of predicting conversion to symptomatic AD. Subjects in different terciles for the CSF H-FABP-Aβ42 and the CSF ptau181-Aβ42 ratios suggest this trend, but the studies included in our analysis did not provide statistical power to test this hypothesis.
In contrast, H-FABP levels do not predict progression from CDR=0.5 to CDR>0.5, but it is noteworthy that the number of subjects in this analysis is small (N=81) compared to the analysis of conversion from CDR=0 to CDR>0 (N=219). Based on these results we believe that H_FABP may be a very useful biomarker in staging preclinical AD
H-FABP is a member of a family of proteins characterized as lipid chaperones (33) that transport lipids to specific compartments in the cell or even outside the cell (33). It has been demonstrated that in vitro fatty acids induce Aβ assembly and modulate the rate of tau polymer assembly (34). Furthermore, genetic studies have implicated several genes involved in lipid metabolism as risk factors for AD, including APOE (35) and ABCA7 (36).
The multivariate analysis of the MAP 1.0 panel of analytes identified distinct complex models for the Knight-ADRC and the ADNI datasets. Nevertheless, these models included a common subset of analytes. CSF H-FABP is part of this subset of selected analytes, as well as HGF, which was reported previously to discriminate AD subjects from other neurodegenerative disorders (32). Similarly VEGF was reported to discriminate controls from moderately-severe AD cases, but not from mild-AD subjects (31). We confirmed the role of these analytes in our dataset by demonstrating an association with CDR at LP.
In summary, the employment of CSF ptau181-Aβ42 ratio as an endophenotype of AD led to the identification of a set of 13 correlated analytes associated with the disease in two independent cohorts. This set of candidate analytes is extended by 2 additional analytes, which are significantly associated only in the context of multivariate models. Despite the obvious differences in our two datasets (the ADNI study has many symptomatic individuals and fewer nondemented controls while the Knight-ADRC study is largely composed of controls and inferred preclinical AD samples), these analytes were consistently associated with the CSF ptau181-Aβ42 ratio and thus AD. Our analysis suggests that these analytes are capturing distinct information from that identified by the traditional analytes (CSF Aβ42 and tau/ptau181) for the disease, and thus describe novel facets of disease pathogenesis. Analysis of the role of these analytes as novel biomarkers of AD pathogenesis in additional datasets will help to determine the specificity of these changes to AD. Longitudinal analyses will also enable characterization of the temporal sequence of analyte changes in the pathological cascade of AD.
Supplementary Material
Acknowledgments
This work was supported by Pfizer, grants from NIH (P30 NS069329-01, R01 AG035083, R01 AG16208, P50 AG05681, P01 AG03991, P01 AG026276, UL1 TR000448), and the Barnes-Jewish Hospital Foundation. The authors thank the supported of the Bright Focus Foundation Alzheimer’s Disease Research Grant A2013359S and Clinical and Genetics Cores of the Knight ADRC at Washington University for clinical and cognitive assessments of the participants and for APOE genotypes. We also thank the Biomarker Core of the Adult Children Study at Washington University for the CSF collection and assays.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Financial Disclosures
The authors have read the journal’s policy and have the following conflicts: K.B. and E.H.P. are employed by Pfizer. A.M.F. served as an advisory board member for Roche and Eli Lilly. D.M.H. reports consulting for Bristol-Myers Squibb, AstraZeneca, and Genentech; is on the scientific advisory board of C2N Diagnostics; and receives research grant support from the NIH, Ellison Medical Foundation, Cure Alzheimer’s Fund, AstraZeneca, C2N Diagnostics, and Integrated Diagnostics. J.C,M has participated or is participating in clinical trials of anti-dementia drugs sponsored by Janssen Alzheimer Immunotherapy Program, Pfizer and Eli Lilly Company; has served as a consultant for Eisai, Esteve, Janssen Alzheimer Immunotherapy Program, GlaxoSmithKline, Novartis, Eli Lily Company and Pfizer; receives research support from Eli Lilly/Avid Radiopharmaceuticals; and is funded by NIH (NIA). A.M.G. reports consulting for Finnegan and Amgen, has received research grant support from Genentech, Pfizer, Astra Zeneca, and royalties from Taconic. O.H., C.C., J.S.K.K., B.J.A., and S.B. report no biomedical financial interests or potential conflicts of interest. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Price JL, McKeel DW, Jr, Buckles VD, Roe CM, Xiong C, Grundman M, et al. Neuropathology of nondemented aging: Presumptive evidence for preclinical Alzheimer disease. Neurobiol Aging. 2009;30:1026–1036. doi: 10.1016/j.neurobiolaging.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holtzman DM, Goate A, Kelly J, Sperling R. Mapping the road forward in Alzheimer’s disease. Sci Transl Med. 2011;3:114ps48. doi: 10.1126/scitranslmed.3003529. Presented at the Science translational medicine. [DOI] [PubMed] [Google Scholar]
- 3.Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: the challenge of the second century. Sci Transl Med. 2011;3:77sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman RJ, Xiong C, Benzinger TLS, Fagan AM, Goate A, Fox NC, et al. Clinical and Biomarker Changes in Dominantly Inherited Alzheimer’s Disease. N Engl J Med. 2012 doi: 10.1056/NEJMoal202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buerger K, Ewers M, Pirttilä T, Zinkowski R, Alafuzoff I, Teipel SJ, et al. CSF phosphorylated tau protein correlates with neocortical neurofibrillary pathology in Alzheimer’s disease. Brain. 2006;129:3035–3041. doi: 10.1093/brain/awl269. [DOI] [PubMed] [Google Scholar]
- 6.Hardy JJ, Selkoe DJD. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 7.de Leon MJM, DeSanti SS, Zinkowski RR, Mehta PDP, Pratico DD, Segal SS, et al. MRI and CSF studies in the early diagnosis of Alzheimer’s disease. J Intern Med. 2004;256:205–223. doi: 10.1111/j.1365-2796.2004.01381.x. [DOI] [PubMed] [Google Scholar]
- 8.Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarawneh R, Lee J-M, Ladenson JH, Morris JC, Holtzman DM. CSF VILIP-1 predicts rates of cognitive decline in early Alzheimer disease. Neurology. 2012;78:709–719. doi: 10.1212/WNL.0b013e318248e568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig-Schapiro R, Perrin RJ, Roe CM, Xiong C, Carter D, Cairns NJ, et al. YKL-40: a novel prognostic fluid biomarker for preclinical Alzheimer’s disease. Biol Psychiatry. 2010;68:903–912. doi: 10.1016/j.biopsych.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM. Cerebrospinal fluid tau/beta-amyloid (42) ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol. 2007;64:343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- 12.Price JL, Davis PB, Morris JC, White DL. The distribution of tangles, plaques and related immunohistochemical markers in healthy aging and Alzheimer’s disease. Neurobiol Aging. 1991;12:295–312. doi: 10.1016/0197-4580(91)90006-6. [DOI] [PubMed] [Google Scholar]
- 13.Price JL, Morris JC. Tangles and plaques in nondemented aging and “preclinical” Alzheimer’s disease. Ann Neurol. 1999;45:358–368. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 14.Toledo JB, Brettschneider J, Grossman M, Arnold SE, Hu WT, Xie SX, et al. CSF biomarkers cutoffs: the importance of coincident neuropathological diseases. Acta Neuropathol. 2012;124:23–35. doi: 10.1007/s00401-012-0983-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984 Jul; doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 16.Fagan AM, Mintun MA, Mach RH, Lee S-Y, Dence CS, Shah AR, et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006;59:512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 17.Cruchaga C, Kauwe JSK, Nowotny P, Bales K, Pickering EH, Mayo K, et al. Cerebrospinal fluid APOE levels, an endophenotype for genetic studies for Alzheimer’s disease. Human Molecular Genetics. 2012 doi: 10.1093/hmg/dds296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soares HD, Potter WZ, Pickering E, Kuhn M, Immermann FW, Shera DM, et al. Plasma Biomarkers Associated With the Apolipoprotein E Genotype and Alzheimer Disease. Arch Neurol. 2012:1–8. doi: 10.1001/archneurol.2012.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Bio Techniques. 2002;32:1372–4. 1376, 1378–9. [PubMed] [Google Scholar]
- 20.Craig-Schapiro R, Kuhn M, Xiong C, Pickering EH, Liu J, Misko TP, et al. Multiplexed Immunoassay Panel Identifies Novel CSF Biomarkers for Alzheimer’s Disease Diagnosis and Prognosis. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0018850. Public Library of Science e18850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu WT, Holtzman DM, Fagan AM, Shaw LM, Perrin R, Arnold SE, et al. Plasma multianalyte profiling in mild cognitive impairment and Alzheimer disease. Neurology. 2012 doi: 10.1212/WNL.0b013e318266fa70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 23.Johnstone D, Milward EA, Berretta R, Moscato P Alzheimer’s Disease Neuroimaging Initiative. Multivariate protein signatures of pre-clinical Alzheimer’s disease in the Alzheimer’s disease neuroimaging initiative (ADNI) plasma proteome dataset. PLoS ONE. 2012;7:e34341. doi: 10.1371/journal.pone.0034341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snider BJ, Fagan AM, Roe C, Shah AR, Grant EA, Xiong C, et al. Cerebrospinal fluid biomarkers and rate of cognitive decline in very mild dementia of the Alzheimer type. Arch Neurol. 2009;66:638–645. doi: 10.1001/archneurol.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring. Neurology. n.d doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 26.Jefferson AL, Cosentino SA, Ball SK, Bogdanoff B, Leopold N, Kaplan E, Libon DJ. Errors produced on the mini-mental state examination and neuropsychological test performance in Alzheimer’s disease, ischemic vascular dementia, and Parkinson’s disease. J Neuropsychiatry Clin Neurosci. 2002;14:311–320. doi: 10.1176/jnp.14.3.311. [DOI] [PubMed] [Google Scholar]
- 27.Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS ONE. 2012;7:e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 29.Chiasserini D, Parnetti L, Andreasson U, Zetterberg H, Giannandrea D, Calabresi P, Blennow K. CSF levels of heart fatty acid binding protein are altered during early phases of Alzheimer’s disease. J Alzheimers Dis. 2010;22:1281–1288. doi: 10.3233/JAD-2010-101293. [DOI] [PubMed] [Google Scholar]
- 30.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Ohrfelt A, Andreasson U, Simon A, Zetterberg H, Edman A, Potter W, et al. Screening for new biomarkers for subcortical vascular dementia and Alzheimer’s disease. Dement Geriatr Cogn Dis Extra. 2011;1:31–42. doi: 10.1159/000323417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu WT, Chen-Plotkin A, Arnold SE, Grossman M, Clark CM, Shaw LM, et al. Novel CSF biomarkers for Alzheimer’s disease and mild cognitive impairment. Acta Neuropathol. 2010;119:669–678. doi: 10.1007/s00401-010-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson DM, Binder LI. Free fatty acids stimulate the polymerization of tau and amyloid beta peptides. In vitro evidence for a common effector of pathogenesis in Alzheimer’s disease. Am J Pathol. 1997;150:2181–2195. [PMC free article] [PubMed] [Google Scholar]
- 35.Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.