Abstract
Aim:
To optimize formulation methods for loading gemcitabine (GEM), the main drug against pancreatic cancer, into albumin nanoparticles for extended blood circulation and improved efficacy.
Methods:
GEM was loaded into two sizes of disolvation-crosslinked bovine serum albumin nanoparticles, with a mean diameter of 109.7 nm and 405.6 nm, respectively, by co-precipitation (the direct method) and follow-up adsorption (the indirect method). The antitumor activities of the two nanoparticulate formulations, were evaluated according to their anti-proliferative effects on the human pancreatic cell line BXPC-3, which were assessed using the MTT assay.
Results:
The two nanoparticulate formulations, created by direct co-precipitation and indirect adsorption, possessed smooth surfaces and high drug loading efficiencies, 83% and 93% at 11% and 13% drug loading, respectively. The two formulations released GEM for 8 and 12 h, respectively, and significantly improved anti-BXPC-3 proliferation effects, as compared with the GEM solution and the drug-free albumin particles.
Conclusion:
Co-precipitating and adsorbing GEM into albumin particles resulted in sustained-release nanoparticulate formulations with improved antitumor cytotoxicity. The result suggests that this is a useful formulation strategy for improving the antitumor efficacy of GEM.
Keywords: gemcitabine, serum albumin, nanoparticles, pancreatic cancer
Introduction
Pancreatic cancer is a serious malignant tumor with poor prognosis. Worldwide, approximately 200 000 patients die of pancreatic cancer every year, and the death rate nearly equals the incidence. The 5-year survival is not more than 3%−5%1, 2. Chemotherapy is an important method of combined therapy for pancreatic cancer3, 4, but the therapeutic effect is poor.
Gemcitabine (2′,2′-difluoro-2′-deoxycytidine, GEM) is the main chemotherapeutic agent in the treatment of pancreatic cancer and non-small-cell lung cancer, which targets specific stages of the cell cycle. Compared with classic 5-FU, GEM has significant clinical benefit (clinical benefit response: 23.8% of GEM -treated patients vs 4.8% of 5-FU-treated patients, P=0.0022)5. However, because of its small molecular weight and high hydrophilicity, GEM has a short plasma half-life (17 min) and is decomposed to inactive products quickly after administration. At the standard intravenous infusion dose of 1000 mg/m2, a patient's plasma GEM concentration drops to only 0.4 μg/mL 1 h after administration, considerably below the 5 μg/mL optimal plasma concentration for cancer cell inhibition6. Thus, much larger doses are necessary to reach effective plasma concentrations, posing a greater risk of side effects.
Nanospheres—spherical nanoparticles with a mean diameter of 10–1000 nm—are widely used as carriers in drug-delivery systems in clinical applications7, 8, 9. However, there are no published studies on GEM-loaded serum albumin nanospheres. Albumin is a safe, nontoxic, biocompatible, and biodegradable water-soluble protein present in human plasma10, 11, 12. No problems with hemolysis or immunogenicity have been associated with albumin, in contrast to artificial materials such as polycaprolactone, polycyanoacrylate, or polybutylcyanoacrylate13, 14, 15, 16, 17, 18, 19. Although these compounds are also biodegradable, their safety following intravenous injection has not been confirmed. Bovine serum albumin (BSA) and human serum albumin (HSA) have 80% sequence homology. The difference in their molecular weights is <1%, and they have the same isoelectric point10, 11, 12. For these reasons, we substituted BSA for HSA in the study of GEM-loaded albumin nanospheres.
GEM-loaded albumin nanospheres have many potential chemotherapeutic advantages for the treatment of tumors, including pancreatic cancer: slow release of GEM, deposition of GEM-loaded albumin nanospheres in tissues, an enhanced targeting effect to primary or metastatic tumors, reduced toxicity to normal tissues owing to enhanced permeability and retention (EPR) effects20, 21, 22, 23 in tumor microcirculation, and high permeability in blood sinuses of liver and spleen. Tumor cells, hepatic Kupffer cells, and cells of the mononuclear phagocyte system have higher phagocytotic rates for uptake of nanoparticles than cells of other tissues, thus increasing the distribution of GEM in tumors, the liver, and spleen24. In addition, the lymphatic system is prone to absorb albumin, thus enhancing the drug's potency in the lymph nodes and lymphangiomas, with the added benefit of preventing lymphatic metastasis. Taken together, these factors account for the potential that GEM-loaded albumin nanospheres have fewer side effects and require lower drug doses.
Investigators have used a variety of techniques to prepare albumin nanoparticles, including the hyper-borderline method, the desolvation-crosslinking method (sedimentation-crosslinking), and the desolvation-heating method25, 26. Desolvation-crosslinking is simple and yields particles with well-distributed particle diameters.
The maximum sizes of permeation particles in the microvasculature or blood sinusoids are 400–600 nm in tumor tissues, 50 nm in normal tissues, and 500 nm in the liver or spleen27, 28, 29, 30. Phagocytosis of nanoparticles by tumor cells or cells of the mononuclear phagocyte system is poor for particles with diameters <200 nm, but rapidly increases for particles with diameters >200 nm31. Accordingly, in this study, two sizes of GEM-loaded BSA nanospheres were prepared: 50–200 nm (mean diameter: 110 nm) and 200–600 nm (mean diameter: 406 nm).
Materials and methods
Materials
Gemcitabine (hydrochloride) was purchased from Hansen Pharmaceutical Co, Ltd (Jiangsu, China), BSA was from Bo'ao Biological Technology Co, Ltd (Shanghai, China), RPMI1640 and DMSO were from GIBCO Laboratories (USA), and MTT and trypsin were from Sigma Chemical Co (USA). Glutaraldehyde, NaOH, dehydrated alcohol, and double-distilled water were used. All the chemicals were of at least analytical grade.
Quantitative evaluation of gemcitabine
The purity and concentration of GEM were determined by high-performance liquid chromatography (HPLC), with a high-performance liquid chromatograph (10A, Shimadzu, Japan), a CLASSVP workstation (Shimadzu, Japan), and a Diamond C18 (5 μm, ID 4.6 mm×300 mm, USA) chromatographic column. The mobile phase was 0.05 mol/L ammonium acetate buffer and methanol (pH 5.7, 90:10, v/v) at a flow rate of 1 mL/min. The procedure was carried out at room temperature.
Absorbance at 269 nm was measured. The calibration curve of GEM absorbance (A) vs concentration (C) was A=35872C (0.1135 μg/mL–5.675 μg/mL, r=0.9999). The lower limit of quantitation was 11.35 ng/mL. Sensitivity and precision were satisfactory.
Preparation of gemcitabine-loaded albumin nanospheres
GEM-loaded BSA nanospheres (GEM-NSP) were prepared using a modified desolvation-crosslinking method25, 26. The modified method was optimized initially during formulation parameter selection and formulation optimization (Tables 1, 2), with a number of important related factors being investigated, such as the concentrations of the albumin, GEM, ethanol and glutaraldehyde, drug-loading methods, and cross-linking time. During the formulation parameter selection and formulation optimization, the yield of nanospheres was calculated as follows: yield of nanospheres=(weight of BSA in the nanospheres/total weight of administered BSA)×100%.
Table 1. Effect of formulation parameters on nanoparticulate characteristics.
| Formulation parameters selection (blank NP) | |||||
|---|---|---|---|---|---|
| pH | 6.0 | 7.0 | 8.0 | 9.0 | 10.0 |
| Particle diameter (nm) | – | – | 251.7±3.1 | 118.9±3.9 | 115.8±4.8 |
| Yield (%) | – | – | 89.7±3.1 | 86.2±2.2 | 73.2±3.2 |
| Ethanol /BSA (v:v) | 1.5:1 | 2.5:1 | 3:1 | 4:1 | 5:1 |
| Particle diameter (nm) | – | 112.2±3.5 | 251.7±5.1 | 335.5±6.2 | 337.0±7.1 |
| Yield (%) | 57.9±3.5 | 82.5±6.6 | 83.3±4.8 | 85.1±5.9 | 84.2±6.2 |
| Cross-linking time | 4 h | 6 h | 12 h | 16 h | 18 h |
| Particle diameter (nm) | – | 124.5±3.2 | 136.7±5.5 | 165.6±6.9 | – |
| Yield (%) | – | 85.7±2.1 | 82.3±3.1 | 78.6±3.9 | – |
| GEM loading methods selection | |||||
|---|---|---|---|---|---|
| Direct loading | Indirect adsorption | Combination | |||
| Particle diameter (nm) | 312.6±5.4 | 267.6±5.1 | 305.7±7.3 | ||
| Encapsulation rate (%) | 73.2±1.9 | 77.2±2.1 | 85.3±1.7 | ||
| Drug loading (%) | 5.0±0.2 | 7.2±0.3 | 8.2±0.5 | ||
| Release time | 12 h | 3 h | 12 h | ||
| Single factor investigations | |||||
|---|---|---|---|---|---|
| BSA concentration | – | 1% | 2% | 3% | 4% |
| Particle diameter (nm) | – | 132.1±7.1 | 189.7±8.6 | 258.7±17.6 | – |
| GEM concentration | – | 10% | 15% | 20% | 30% |
| Particle diameter (nm) | – | 164.6±8.1 | 189.6±10.1 | 232.8±14.5 | 276.7±8.6 |
| Glutaraldehyde | 50% | 100% | 200% | 300% | 400% |
| Particle diameter (nm) | 190.2±3.2 | 210.3±5.9 | 212.6±7.2 | 219.7±3.7 | – |
| Yield (%) | 82.1±1.2 | 88.7±1.9 | 86.7±0.8 | 78.2±0.9 | 74.1±1.7 |
Table 2. Characteristics of optimized formulation.
| 110 nm-GEM-NSP | 406 nm-GEM-NSP | |
|---|---|---|
| BSA | 2% | 2.5% |
| GEM | 15% | 15% |
| Glutaraldehyde | 100% | 300% |
| pH | 9.0 | 8.0 |
| Ethanol | 2.5:1 | 4:1 |
| Cross-linking time | 6 h | 12 h |
| Mean diameter | 109.7±2.2 nm | 405.6±3.5 nm |
| Range | 50–200 nm | 200–600 nm |
| Encapsulation rate | 82.92% | 92.56% |
| Drug loading | 11.25% | 13.40% |
| Zeta potential | −24.4±1.41 mV | −15.6±1.08 mV |
The modified method included five main procedures. (1) Direct drug-loading: Briefly, 10 mL of aqueous BSA (2%–2.5%, w/v) was incubated with 17–22 mg of GEM at room temperature. The pH was adjusted to 8.0–9.0 with NaOH (1 mol/L). Dehydrated ethanol (25–40 mL) was subsequently added drop-wise into the mixture at a rate of 1 mL/min under magnetic stirring (1000 r/min, magnetic stirrer, Sile, Shanghai, China). (2) Indirect drug-adsorption: when the mixture achieved a blue opalescence, 1.7–2.2 mL of aqueous GEM (10 mg/mL, pH adjusted to 8.5 with 1 mol/L NaOH) was added under magnetic stirring. After another 30 min of continued magnetic stirring, glutaraldehyde was added (acid molar ratio of glutaraldehyde and albumin: 1–3:1) into the mixture. Crosslinking was achieved with continued magnetic stirring for 6–12 h. (3) The mixture was rotary evaporated (ZX-91 rotary evaporator, Institute of Organic Chemistry, Chinese Academy of Sciences, Shanghai, China) at 40 °C to remove the ethanol. (4) The nanospheres were separated by centrifugation in a TCL-16C high-speed centrifuge at 14 000 r/min for 20 min (Shanghai Anting Scientific Instrument Factory, Shanghai, China). (5) The precipitated nanospheres were suspended in purified water and subjected to cryodesiccation (Alpha 2-4, Martin Christ, Germany), yielding a desiccated powder of nanospheres.
At pH 9.0, a volume ratio of albumin and dehydrated ethanol of 1:2.5, an acid molar ratio of glutaraldehyde and albumin of 1:1, and a cross-linking time of 6 h, 110 nm GEM-loaded BSA nanospheres (110 nm-GEM-NSP) were obtained. At pH 8.0, a volume ratio of albumin and dehydrated ethanol of 1:4, an acid molar ratio of glutaraldehyde and albumin of 3:1, and a cross-linking time of 12 h, 406 nm GEM-loaded BSA nanospheres (406 nm-GEM-NSP) resulted. The blank nanospheres (NSP) were prepared using the same procedure as that for the drug-containing nanospheres but without the addition of GEM.
Characterization of GEM-NSPs
The size and zeta potential of the nanospheres were detected by Nicomp™ 380/ZLS electric potential/particle diameter radiometry (Nicomp, USA). Nanospheres were dispersed in isotonic NaCl (1 mg/mL) and examined at 25 °C at a light-scattering angle of 90°. The mean particle size was determined in a condition of scattering intensity as intensity weight. The zeta potential was detected in a 10 V electric field.
The GEM concentration in the supernatant after centrifugation (the fourth procedure in the preparation of nanospheres) was detected by HPLC. The drug encapsulation32, 33 rate of GEM-NSP was equal to (total GEM–GEM in the supernatant)/total GEM×100%.
Drug loading of GEM-NSP=(weight of GEM in nanospheres/total weight of nanospheres)×100%. Some quantitative nanospheres were transferred into a 10-mL graduated flask, with purified water added as needed. The suspension was then processed with a JY92-IIN ultrasonic cell disrupter (Ningbo Scientz Biotechnology Co, Ltd, Ningbo, China) for 30 min, and was filtered through a 0.22-μm membrane. The GEM content in the suspension was then detected by HPLC in order to calculate the weight of GEM in the nanospheres and total weight of the nanospheres32, 33.
The nanosphere suspension was added dropwise onto copper grids, which were then dried at room temperature. Transmission electron micrographs of the nanospheres were acquired with an electron microscope (Hitachi H-600, Japan).
In vitro drug release assay
One milliliter of 1 mg/mL GEM-NSP was sealed in a dialysis bag and immersed in 49 mL of PBS containing 10% (v/v) fetal bovine serum (FBS) at 37 °C with a shaking rate of 100 r/min. A sample (0.5 mL each) was withdrawn from the medium at designated time intervals and the same volume of fresh medium was added. Each sample was mixed with 1.5 mL of acetonitrile, vortexed for 3 min, and centrifuged, and the supernatant was assayed by HPLC.
Cytotoxicity assay
BXPC-3, an ATCC human pancreatic cancer cell line, was purchased from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). The cells were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 50 U penicillin/mL, and 50 μg streptomycin/mL in a humidified atmosphere with 95% O2 and 5% CO2 at 37 °C.
The anti-proliferative effects of pure GEM, 110 nm-GEM-NSPs, 406 nm-GEM-NSPs, and blank NSPs on BXPC-3 cells were detected by the 3-[4,5-dimethylthiazol-2-yl]-3,5-diphenyl tetrazolium bromide (MTT) test. A blank control group without medication was also used. Exponentially growing cells were seeded into 96-well plates and pre-incubated for 24 h. A series of dosages of GEM or GEM-NSPs (containing 0.01, 0.1, 1, 10, and 50 μg/mL of GEM) were added to the cells in the culture medium. Five duplicate wells were used in each set. After 48 and 72 h at 37 °C, 200 μL of MTT (5 mg/mL) was added into each well. The wells were incubated for 4 h, after which the culture medium was removed and the resultant formazan crystals were dissolved in 150 μL extraction solution (DMSO) with shaking for 10 min. The optical density (OD) was measured at 490 nm using a microplate reader (ELx800, BioTek, USA). The inhibitory rate was equal to (1–OD of medication administration group/OD of control group)×100%. The IC50 was calculated with the bliss method34.
Statistical analysis
Data are presented as mean±standard deviation. Statistical analysis was performed using one-way analysis of variance (ANOVA) with SPSS 11.5. P<0.05 was considered significant.
Results
Factors influencing the preparation of GEM-NSPs
During the formulation parameter selection and formulation optimization of GEM-loaded BSA nanospheres (GEM-NSP), a number of factors were investigated (Table 1, 2).
The initial formulation parameter selection with blank BSA nanospheres indicated that pH and dosage of ethanol significantly influenced preparation of the nanospheres. At pH<8, it was difficult to form albumin nanospheres; at pH 8, nanospheres were reproducibly obtained. With increasing pH, the mean diameter of the nanospheres decreased gradually. A comparison of pH values (6.0, 7.0, 8.0, 9.0, 10.0) showed that pH 8.0–9.0 was optimal. When the ratio of ethanol to 2% BSA (v:v) was greater or equal to 2.5, the yield of nanospheres was greater or equal to 80%; however, with an increasing ratio of ethanol to BSA, the mean diameter of the nanospheres increased significantly (Table 1).
GEM loading method selection indicated that nanospheres prepared by combined methods, including direct drug-loading and indirect drug-adsorption, had the best encapsulation rate, drug loading rate, and release time. Single-factor investigation indicated that particle sizes increased gradually with increasing concentrations of albumin or GEM (Table 1).
After the formulation parameter selection and formulation optimization, a modified desolvation-crosslinking method with optimized formulations for 110 nm and 406 nm GEM-NSPs was chosen (Table 2).
Characterization of GEM-NSPs
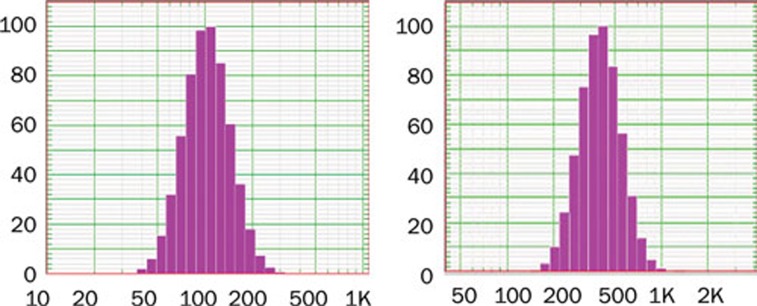
Two sizes of GEM-NSPs with smooth surfaces, good dispersion, and relatively uniform size distributions were obtained. Mean diameters were 109.7±2.2 nm (generally ranging from 50 to 200 nm) and 405.6±3.5 nm (generally ranging from 200 to 600 nm, not exceeding 1000 nm). Drug loadings were 11.25% and 13.40%; drug encapsulation rates were 82.92% and 92.56%, respectively. These parameters were somewhat higher than in other kinds of drug-loaded nanoparticles prepared analogously13, 14, 15, 16, 17, 18, 19. Zeta potentials were -24.4 and -15.6 mV, respectively (Table 2, Figure 1).
Figure 1.
Particle-diameter profiles of two types of GEM-loaded albumin nanospheres.
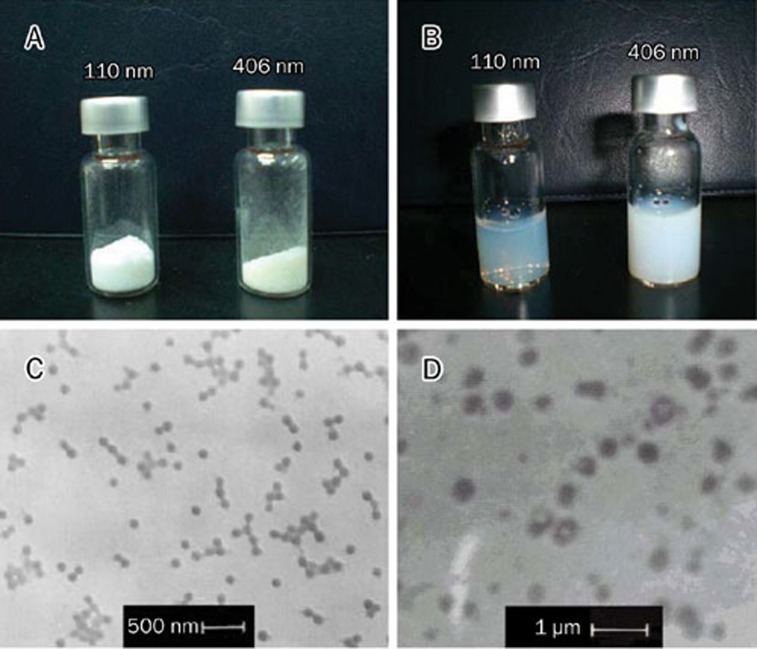
The freeze-dried powder of GEM-NSPs was white and fluffy, whereas that of 406 nm-GEM-NSPs was slightly yellow. Preparation of 110 nm-GEM-NSPs in an aqueous solution yielded a suspension of relative clarity. Transmission electron micrographs of the nanospheres confirmed the smooth surfaces, good dispersion, and moderately uniform size distributions of the particles. Moreover, the drug area in the particle center could be seen in some particles of the 406 nm-GEM-NSPs (Figure 2).
Figure 2.
Photographs of freeze-dried (A), water-soluble (B) GEM-NSP, and transmission electron micrographs of 110 nm-GEM-NSP (C) and 406 nm-GEM-NSP (D). (×10 000 times).
In vitro drug-release characteristics of GEM-NSPs
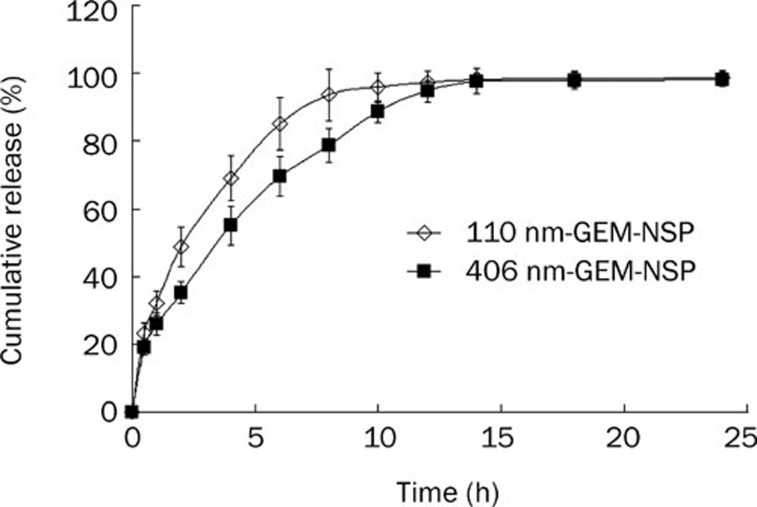
The in vitro drug-release curves for the two types of GEM-NSPs are shown in Figure 3. Burst release occurred for 30 min after administration; then the release gradually became slower. The slow release times were about 8 h (for 110 nm-GEM-NSPs) and 12 h (for 406 nm-GEM-NSPs). The rates of burst release in 30 min were 23.25%±3.05% and 19.13%±2.29%, respectively (Figure 3).
Figure 3.
In vitro release of GEM from GEM-NSP in PBS containing 10% (v/v) fetal bovine serum (FBS) at 37 °C.
Cytotoxicity of GEM-NSPs on BXPC-3 cells in vitro
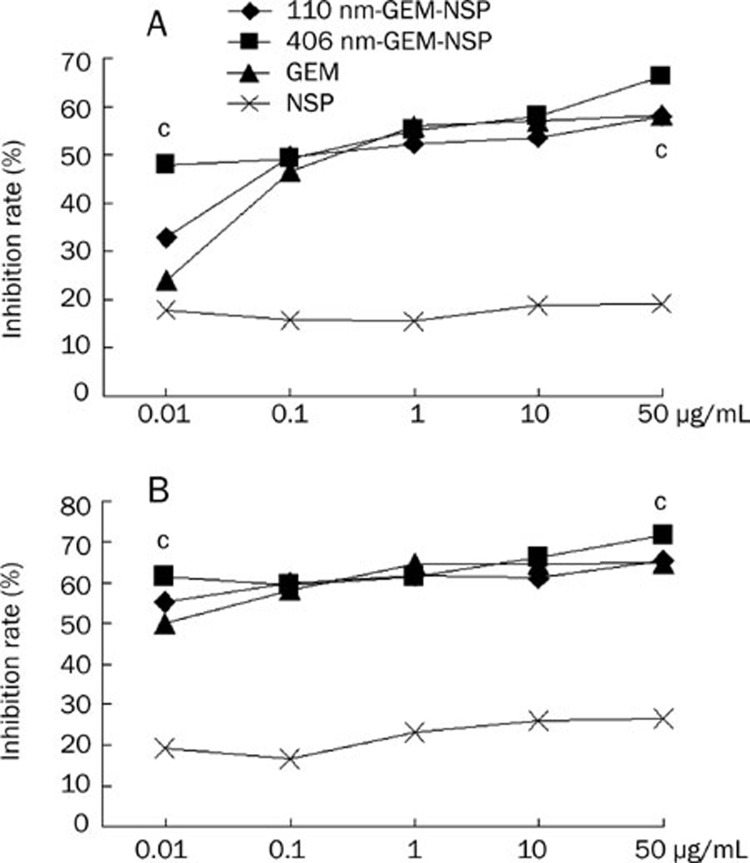
The cytotoxicity of 110 nm-GEM-NSPs and 406 nm-GEM-NSPs on BXPC-3 cells 48 and 72 h after administration was detected by the MTT assay (Figure 4). The inhibition rate-administered concentration profiles of 406 nm-GEM-NSPs, 110 nm-GEM-NSPs, and GEM were all higher than those of NSPs alone (P<0.05). The inhibition rate of blank NSPs was about 20% with good cell morphology, without obvious correlation with time or concentration, which indicated that NSPs had good biocompatibility, without obvious cytotoxicity (grade 1–2), according to the United States Pharmacopeia (USP 28 )35.
Figure 4.
Inhibition rate-GEM concentration profile of 406 nm-GEM-NSP, 110 nm-GEM-NSP, GEM, and NSP on the human pancreatic cancer cell line BXPC-3 48 h (A) and 72 h (B) after administration in vitro. Significance of the difference between 406 nm-GEM-NSP and GEM. cP<0.005.
At 0.01 μg/mL of GEM, the inhibition rates of 406 nm-GEM-NSPs and 110 nm-GEM-NSPs were higher than that of GEM (48 h: 47.88%±1.53% vs 32.99%±0.74% vs 23.99%±1.94%, P<0.05; 72 h: 61.23%±1.43% vs 55.25%±2.07% vs 49.83%±2.53%, P<0.05). At 0.1−10 μg/mL, the differences among inhibition rates of 406 nm-GEM-NSPs, 110 nm-GEM-NSPs and GEM were not significant (P>0.05). At 50 μg/mL, the inhibition rate of 406 nm-GEM-NSPs was higher than those of the 110 nm-GEM-NSPs and GEM alone (P<0.05).
The IC50 values (bliss method) of the GEM-NSPs and GEM are shown in Table 3, which indicates that the IC50 values of 406 nm- and 110 nm-GEM-NSPs were lower than that of GEM.
Table 3. IC50 of GEM-NSP on BXPC-3 cells (μg/mL).
| 48 h | 72 h | |
|---|---|---|
| 110 nm-GEM-NP | 1.1368 | 0.0000246 |
| 406 nm-GEM-NP | 0.0674 | 0.0000220 |
| GEM | 1.4740 | 0.0022000 |
| NP | – | – |
Discussion
The aim of this study was to prepare gemcitabine-loaded albumin nanospheres and to investigate their physical characteristics and cytotoxicity. An important feature of the method in this study was that the molecular structure of GEM remained unchanged, as compared with other GEM-loaded particle-delivery systems, in which the particles were prepared with GEM precursors, derivatives, or other modified structures36, 37, 38, 39. Although such changes might improve GEM loading or stability, and might even maintain the drug's cytotoxicity, these modified products introduced new chemical compounds, and their pharmacodynamics, pharmacokinetics, safety, and clinical effects must be re-evaluated. In our study, with the molecular structure of GEM unchanged, high drug loading and improved stability of the GEM-NSP were achieved.
pH is important for the maintenance of the structure and characteristics of albumin. At pH<8, the structure of albumin is “N”, whereas at pH≥8, it is “F”. Albumin at “F” has an augmented viscosity and decreased dissolvability40, which facilitate its preparation into nanospheres. As the amount of ethanol is increased, albumin became sufficiently denatured, and the particle diameters and the yield of nanospheres increased41, 42.
The main drug-loading methods of albumin nanoparticles are direct drug-loading and indirect drug-adsorption25, 26. In direct drug-loading, albumin is prepared and the drug is mixed into the liquor; then an organic solvent is added dropwise, with drug-loading occurring during albumin sedimentation. In drug adsorption, blank albumin nanoparticles are prepared and then added to a solution of the drug under stirring to promote adsorption. Both methods have advantages and disadvantages, as shown by the GEM loading method selection (Table 1). In this study, we combine both techniques, with 50% of the GEM loaded directly and 50% indirectly. This approach, albeit complicated, had a favorable encapsulation rate, drug loading percentage and release time.
In the characterization of GEM-NSPs, the drug loadings and drug encapsulation rates reached relatively high levels with good particle size control and cytotoxicity, as compared with other drug-loading nanoparticles. The time for slow release of GEM from GEM-NSPs observed in this study was not longer than 12 h, probably because of GEM high water solubility, as reported previously13, 14, 15, 16, 17, 18, 19. However, since pure GEM is rapidly metabolized, prolongation of the release time of GEM by GEM-NSPs represents an improvement. The water solubility of GEM can be reduced by molecular modification18, 19, 36, 37, 38, 39, such as modification with hydrophobic stearo-chains, amino-acids, or pteroylglutamic acid. Although these strategies would likely improve drug loading and slow release, the pharmacodynamic action of GEM derivates, ie, new molecules, would require an overall re-evaluation. Therefore, in the present study, we abandoned GEM modification strategies and successfully prepared GEM-NSPs with better effects.
The MTT assay demonstrated that there was no pharmacodynamic loss of GEM during the preparation of GEM-NSPs; both GEM-NSPs, as well as GEM, had a significant inhibition effect against BXPC-3 cells, and blank NSPs had good biocompatibility, without obvious cytotoxicity. It is well known that GEM is highly water-soluble and needs specific transport proteins to transfer it into cancer cells; in the cells, GEM is metabolized by enzymes such as deoxycytidine kinase and deoxycytidine deaminase6, 43. However, GEM-NSPs, particularly particles with diameters >200 nm, could be phagocytized by cancer cells31 and have slow release features, which might cause the differences among the cytotoxicities of 406 nm-, 110 nm-GEM-NSPs and GEM at 0.01 and 50 μg/mL. However, the advantages of the slow-release and anticancer effects of GEM-NSPs in vivo need further evaulation, because the cells cultured in vitro were in a closed system, different from the internal environment in the human body.
Chemotherapeutics loaded in nanoparticles thus far include doxorubicin, camptothecin, and fluorouracil14, 44, 45, 46. This is the first published study on GEM-loaded albumin nanospheres. GEM-NSPs have the potential to be used to treat many kinds of tumors, such as pancreatic and non-small-cell lung cancers. Further improvements are necessary for the GEM-NSP system, such as prolonging the drug release time and enhancing the drug-loading rate, before the system meets clinical requirements. These challenges are the subject of our ongoing experiments.
Author contribution
Xian-jun YU, Hui-min HOU, Wei-yue LU, Quan-xing NI, and De-liang FU designed the research; Jin-ming LI, Wei CHEN, Hao WANG, Long CUI, and Chen JIN performed the research; Jin-ming LI wrote the paper.
Acknowledgments
This work was supported by the Science and Technology Commission of Shanghai Municipality (No 08431902500) and the Shanghai Municipal Economic Commission (No 06-23, 07ZH-028).
References
- Michaud. Epidemiology of pancreatic cancer Minerva Chir 20045999–111. [PubMed] [Google Scholar]
- Mitry E, Rachet B, Quinn MJ, Cooper N, Coleman MP. Survival from cancer of the pancreas in England and Wales up to 2001. Br J Cancer. 2008;99:S21–3. doi: 10.1038/sj.bjc.6604576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Ni Q, Yu X, Zhang Q, Hua Y, Zhang Y.et al. Regional intra-arterial infusion chemotherapy for pancreatic cancer: an experimental study Zhonghua Yi Xue Za Zhi 200282371–5.Chinese. [PubMed] [Google Scholar]
- Ishikawa T, Kamimura H, Tsuchiya A, Togashi T, Watanabe K, Seki K.et al. Clinical efficacy of intra-arterial pharmacokinetic chemotherapy with 5-fluorouracil, CDDP, gemcitabine, and angiotensin-II in patients with advanced pancreatic cancer Hepatogastroenterology 2007542378–82. [PubMed] [Google Scholar]
- Burris HA, 3rd, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR.et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial J Clin Oncol 1997152403–13. [DOI] [PubMed] [Google Scholar]
- Eli Lilly Co. Summary of product characteristics: gemcitabine UK prescribing information Indianapolis: Eli Lilly 1997
- Niu L, Xu YC, Xie HY, Dai Z, Tang HQ. Expression of human insulin gene wrapped with chitosan nanoparticles in NIH3T3 cells and diabetic rats. Acta Pharmacol Sin. 2008;29:1342–9. doi: 10.1111/j.1745-7254.2008.00888.x. [DOI] [PubMed] [Google Scholar]
- Meng XX, Wan JQ, Jing M, Zhao SG, Cai W, Liu EZ. Specific targeting of gliomas with multifunctional superparamagnetic iron oxide nanoparticle optical and magnetic resonance imaging contrast agents. Acta Pharmacol Sin. 2007;28:2019–26. doi: 10.1111/j.1745-7254.2007.00661.x. [DOI] [PubMed] [Google Scholar]
- Deng WJ, Yang XQ, Liang YJ, Chen LM, Yan YY, Shuai XT.et al. FG020326-loaded nanoparticle with PEG and PDLLA improved pharmacodynamics of reversing multidrug resistance in vitro and in vivo Acta Pharmacol Sin 200728913–20. [DOI] [PubMed] [Google Scholar]
- Kurrat R, Prenosil JE, Ramsden JJ. Kinetics of human and bovine serum albumin adsorption at silica-titania surfaces. J Colloid Interface Sci. 1997;185:1–8. doi: 10.1006/jcis.1996.4528. [DOI] [PubMed] [Google Scholar]
- Huang BX, Kim HY, Dass C. Probing three-dimensional structure of bovine serum albumin by chemical cross-linking and mass spectrometry. J Am Soc Mass Spectrom. 2004;15:1237–47. doi: 10.1016/j.jasms.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Gelamo EL, Tabak M. Spectroscopic studies on the interaction of bovine (BSA) and human (HSA) serum albumins with ionic surfactants. Spectrochim Acta A Mol Biomol Spectrosc. 2000;56:2255–71. doi: 10.1016/s1386-1425(00)00313-9. [DOI] [PubMed] [Google Scholar]
- Reddy LH, Couvreur P. Novel approaches to deliver gemcitabine to cancers. Curr Pharm Des. 2008;14:1124–37. doi: 10.2174/138161208784246216. [DOI] [PubMed] [Google Scholar]
- Stella B, Arpicco S, Rocco F, Marsaud V, Renoir JM, Cattel L.et al. Encapsulation of gemcitabine lipophilic derivatives into polycyanoacrylate nanospheres and nanocapsules Int J Pharm 200734471–7. [DOI] [PubMed] [Google Scholar]
- Gang J, Park SB, Hyung W, Choi EH, Wen J, Kim HS.et al. Magnetic poly epsilon-caprolactone nanoparticles containing Fe3O4 and gemcitabine enhance anti-tumor effect in pancreatic cancer xenograft mouse model J Drug Target 200715445–53. [DOI] [PubMed] [Google Scholar]
- Yang J, Park SB, Yoon HG, Huh YM, Haam S. Preparation of poly epsilon-caprolactone nanoparticles containing magnetite for magnetic drug carrier. Int J Pharm. 2006;324:185–90. doi: 10.1016/j.ijpharm.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee H, Hyung W, Park SB, Haam S. Magnetic PECA nanoparticles as drug carriers for targeted delivery: synthesis and release characteristics. J Microencapsul. 2006;23:203–12. doi: 10.1080/02652040500435444. [DOI] [PubMed] [Google Scholar]
- Celano M, Calvagno MG, Bulotta S, Paolino D, Arturi F, Rotiroti D.et al. Cytotoxic effects of gemcitabine-loaded liposomes in human anaplastic thyroid carcinoma cells BMC Cancer 2004463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosilio V, Renoir JM, Couvreur P, Rocco F, Cattel L, Stella B. Gemcitabine derivatives nanoparticles. European Patent. p. 1761551.
- Modi S, Prakash Jain J, Domb AJ, Kumar N. Exploiting EPR in polymer drug conjugate delivery for tumor targeting. Curr Pharm Des. 2006;12:4785–96. doi: 10.2174/138161206779026272. [DOI] [PubMed] [Google Scholar]
- Jun YJ, Kim JI, Jun MJ, Sohn YS. Selective tumor targeting by enhanced permeability and retention effect. Synthesis and antitumor activity of polyphosphazene-platinum (II) conjugates. J Inorg Biochem. 2005;99:1593–601. doi: 10.1016/j.jinorgbio.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Greish K. Enhanced permeability and retention of macromolecular drugs in solid tumors: a royal gate for targeted anticancer nanomedicines. J Drug Target. 2007;15:457–64. doi: 10.1080/10611860701539584. [DOI] [PubMed] [Google Scholar]
- Iyer AK, Khaled G, Fang J, Maeda H. Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discov Today. 2006;11:812–8. doi: 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Widder KJ, Marino PA, Morris RM, Howard DP, Poore GA, Senyei AE. Selective targeting of magnetic albumin microspheres to the Yoshida sarcoma: ultrastructural evaluation of microsphere disposition. Eur J Cancer Clin Oncol. 1983;19:141–7. doi: 10.1016/0277-5379(83)90409-1. [DOI] [PubMed] [Google Scholar]
- Weber C, Coester C, Kreuter J, Langer K. Desolvation process and surface characterisation of protein nanoparticles. Int J Pharm. 2000;194:91–102. doi: 10.1016/s0378-5173(99)00370-1. [DOI] [PubMed] [Google Scholar]
- Dreis S, Rothweiler F, Michaelis M, Cinatl J, Jr, Kreuter J, Langer K. Preparation, characterisation and maintenance of drug efficacy of doxorubicin-loaded human serum albumin (HSA) nanoparticles. Int J Pharm. 2007;321:207–14. doi: 10.1016/j.ijpharm.2007.03.036. [DOI] [PubMed] [Google Scholar]
- Hashizume H, Baluk P, Morikawa S, McLean JW, Thurston G, Roberge S.et al. Openings between defective endothelial cells explain tumor vessel leakiness Am J Pathol 20001561363–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs SK, Monsky WL, Yuan F, Roberts WG, Griffith L, Torchilin VP.et al. Regulation of transport pathways in tumor vessels: role of tumor type and microenvironment Proc Natl Acad Sci USA 1998954607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Dellian M, Fukumura D. Vascular permeability in a human tumor xenograft: molecular size dependence and cutoff size. Cancer Res. 1995;55:3752–6. [PubMed] [Google Scholar]
- Kong G, Braun RD, Dewhirst MW. Hyperthermia enables tumor-specific nanoparticle delivery: effect of particle size. Cancer Res. 2000;60:4440–5. [PubMed] [Google Scholar]
- Liu H, Farrell S, Uhrich K. Drug release characteristics of unimolecular polymeric micelles. J Control Release. 2000;10:167–74. doi: 10.1016/s0168-3659(00)00247-9. [DOI] [PubMed] [Google Scholar]
- Chen W, Gu B, Wang H, Pan J, Lu W, Hou H. Development and evaluation of novel itraconazole-loaded intravenous nanoparticles. Int J Pharm. 2008;362:133–40. doi: 10.1016/j.ijpharm.2008.05.039. [DOI] [PubMed] [Google Scholar]
- Sheng Y, Yuan Y, Liu C, Tao X, Shan X, Xu F. In vitro macrophage uptake and in vivo biodistribution of PLA-PEG nanoparticles loaded with hemoglobin as blood substitutes: effect of PEG content. J Mater Sci Mater Med. 2009;20:1881–91. doi: 10.1007/s10856-009-3746-9. [DOI] [PubMed] [Google Scholar]
- Schmidt-Hieber M, Busse A, Reufi B, Knauf W, Thiel E, Blau IW.et al. Bendamustine, but not fludarabine, exhibits a low stem cell toxicity in vitro J Cancer Res Clin Oncol 2009135227–34. [DOI] [PubMed] [Google Scholar]
- The United States Pharmacopeial Convention. USP 28: Biological Reactivity Tests, In-vitro. 2005.
- Arias JL, Reddy LH, Couvreur P. Magnetoresponsive squalenoyl gemcitabine composite nanoparticles for cancer active targeting. Langmuir. 2008;24:7512–9. doi: 10.1021/la800547s. [DOI] [PubMed] [Google Scholar]
- Song X, Lorenzi PL, Landowski CP, Vig BS, Hilfinger JM, Amidon GL. Amino acid ester prodrugs of the anticancer agent gemcitabine: synthesis, bioconversion, metabolic bioevasion, and hPEPT1-mdiated transport. Mol Pharm. 2005;2:157–67. doi: 10.1021/mp049888e. [DOI] [PubMed] [Google Scholar]
- Cavallaro G, Mariano L, Salmaso S, Caliceti P, Gaetano G. Folate-mediated targeting of polymeric conjugates of gemcitabine. Int J Pharm. 2006;307:258–69. doi: 10.1016/j.ijpharm.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Alexander RL, Greene BT, Torti SV, Kucera GL. A novel phospholipid gemcitabine conjugate is able to bypass three drug-resistance mechanisms. Cancer Chemother Pharmacol. 2005;56:15–21. doi: 10.1007/s00280-004-0949-0. [DOI] [PubMed] [Google Scholar]
- Carter DC, Ho JX. Structure of serum albumin. Adv Protein Chem. 1994;45:153–203. doi: 10.1016/s0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- Budavari S, Maryadele J, Smith A. The merck index (An encyclopedia of chemicals, drugs, and biologicals. Twelfth edition. 1996.
- Weber C, Kreuter J, Langer K. Desolvation process and surface characteristics of HSA-nanoparticles. Int J Pharm. 2000;196:197–200. doi: 10.1016/s0378-5173(99)00420-2. [DOI] [PubMed] [Google Scholar]
- Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17:7–12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- Petri B, Bootz A, Khalansky A, Hekmatara T, Müller R, Uhl R.et al. Chemotherapy of brain tumour using doxorubicin bound to surfactant-coated poly(butyl cyanoacrylate) nanoparticles: revisiting the role of surfactants J Control Release 200711751–8. [DOI] [PubMed] [Google Scholar]
- Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW.et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo Proc Natl Acad Sci USA 20061036315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZR, Hua SC, Yang YL, Fang JY. Development and evaluation of lipid nanoparticles for camptothecin delivery: a comparison of solid lipid nanoparticles, nanostructured lipid carriers, and lipid emulsion. Acta Pharmacol Sin. 2008;29:1094–102. doi: 10.1111/j.1745-7254.2008.00829.x. [DOI] [PubMed] [Google Scholar]