Abstract
The regulation network consisting of microRNAs (miRNAs) and their target genes remains largely elusive in hepatocellular carcinoma (HCC), especially the reciprocal loop between specific miRNAs and the miRNA processing machinery. In this study, we found that miR-99a was remarkably decreased in 111 of 152 (73.03%) primary HCC tissues and low-level expression of miR-99a was correlated with low tumor differentiation (P=0.001), liver cirrhosis (P=0.015), poor tumor-free survival (P=0.004) and overall survival (P=0.006) for HCC patients. By restoration of miR-99a, the HCC growth could be considerably inhibited both in vitro and in vivo. Subsequently, Argonaute-2 (Ago2), a central component of RNA-induced silencing complex, was found to be directly regulated by miR-99a via translational repression. Overexpression of Ago2 could partly impair the inhibitory effect of miR-99a on HCC cells in vitro. Then, we demonstrated that Ago2 was upregulated in HCC tissues at both RNA and protein levels and the expression of AGO2 protein and miR-99a was negatively correlated within detected HCC tissues (r=−0.727, P=0.004). Interestingly, the tumorigenicity of Ago2-knockdown HCC cells was severely impaired (4/10 vs 10/10, P<0.05), and this was in contrast to the miR-99a-overexpressing HCC cells. Functionally, the increased AGO2 protein could specifically facilitate oncogenic miR-21 to repress its targeted gene phosphatase and tensin homolog (Pten) in HCC, whereas leave the regulatory capacity of let-7a on its targeted oncogenes almost unaltered. In summary, our study has revealed a novel pathway for the tumor suppressor miR-99a to control tumor growth in HCC, via its downstream signaling of AGO2/miR-21/PTEN. In addition, this study provides potential strategies for HCC therapy by reintroduction of miRNA suppressors.
Keywords: Hepatocellular carcinoma, miR-99a, AGO2, PTEN
Introduction
Hepatocellular carcinoma (HCC), the primary cancer of liver, ranks as the fifth most prevalent malignancy worldwide and accounts for ∼700 000 deaths per year.1 In the past decades, great advances have been made in the treatment of this disease; however, relapse and metastasis occurred frequently and the 5-year survival rate remains quite low among patients with HCC.2 Although it has been identified that infection of hepatitis B virus or hepatitis C virus, intake of aflatoxin B1, alcohol abuse or obesity are the major causes of HCC, the precise mechanisms of HCC oncogenesis remain elusive.3, 4
MicroRNAs (miRNAs) are small endogenous noncoding RNA molecules that have been identified as posttranscriptional regulators of gene expression.5 The mature miRNAs can be loaded onto RNA-induced silencing complex and guided to their mRNA targets through interactions with Argonaute proteins (including Ago subfamily and Piwi subfamily in mammals), resulting in mRNA degradation or blockade of mRNA translation.6 In recent years, the aberrant expression pattern of specific miRNAs has been repeatedly reported and this phenomenon is linked to carcinogenesis and progression in a wide range of cancers, including HCC.7, 8, 9 Recently, the miRNomes of human normal liver and HCC tissues have been successfully identified by using full-scale analysis based on deep sequencing, through which the downregulation of let-7, miR-99a, miR-122, miR-101 and miR-199-3p, as well as the upregulation of miR-21 in HCC are largely verified.10
In this study, we report that miR-99a, a miRNA abundantly expressed in normal liver,10 is downregulated in HCC tissues compared with the matched noncancerous tissues. Deregulation of miR-99a can serve as an independent risk factor for HCC patients' poor survival. Furthermore, miR-99a suppresses the growth of HCCs by inhibiting Ago2. In addition, we found that Argonaute-2 (Ago2) is increased in HCC tissues and can promote oncogenic miR-21 to negatively regulate its target—tumor suppressor phosphatase and tensin homolog (PTEN). Therefore, our study has revealed a novel pathway for HCC suppressor miR-99a (that is, miR-99a/AGO2/miR-21/PTEN) that may facilitate both diagnosis and therapy of this malignancy.
Results
MiR-99a is deregulated in HCC and correlates with the prognosis of HCC patients
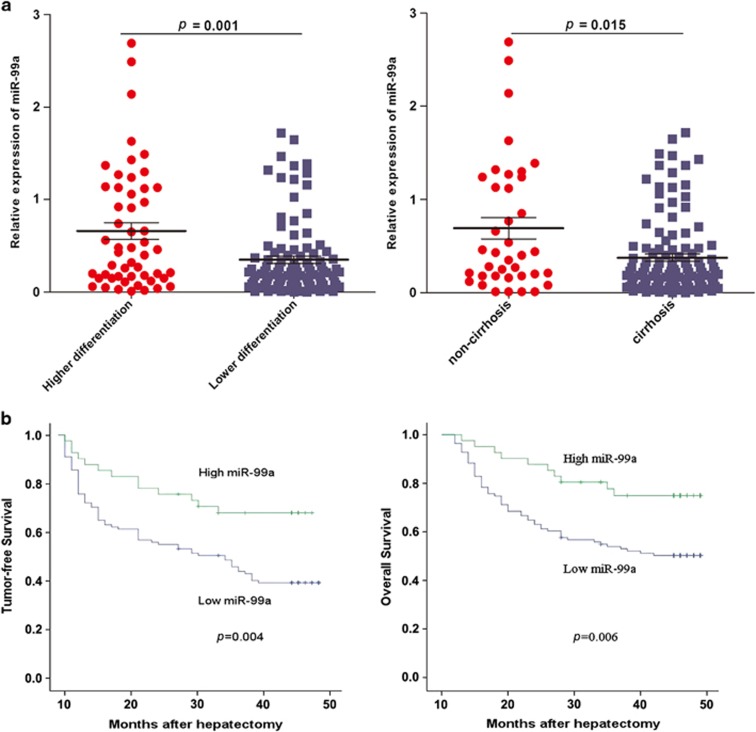
We previously revealed that miR-99a was downregulated in a small amount of HCC tissue samples.10 To further validate this finding, the expression profile of miR-99a in 152 pairs of primary HCC tissues and their matched noncancerous tissues was detected using quantitative reverse transcriptase–PCR (RT–PCR). As a result, we found that miR-99a was significantly decreased (⩾2-fold) in 111 of 152 (73.03%) primary HCC tissues, whose average relative quantity of tumor/normal was 0.19±0.14.
We next sought to explore the association between miR-99a expression and pathological characteristics of HCC patients (Table 1). Results showed that the lower expression level of miR-99a was more frequently observed in tumors from patients with lower tumor differentiation or liver cirrhosis (P=0.001 and P=0.015, respectively, Figure 1a). Furthermore, Kaplan–Meier analysis revealed that low miR-99a level in HCC tissues significantly correlated with the markedly reduced tumor-free survival (P=0.004) and overall survival of HCC patients (P=0.006), suggesting the important roles of miR-99a in the prognosis of HCC patients (Figure 1b). In addition, the result of Cox proportional hazards regression analysis identified that the deregulation of miR-99a was an independent risk factor for reduced tumor-free survival (hazard ratio 0.443, 95% confidence interval 0.233–0.842; P=0.013; Supplementary Table S1) and overall survival of HCC patients (hazard ratio 0.433, 95% confidence interval 0.209–0.897; P=0.024; Supplementary Table S2).
Table 1. Correlation analysis between miR-99a expression and clinical pathological characteristics.
| Variable |
miR-99a subgroupa |
χ2 | P-value | |
|---|---|---|---|---|
| Low | High | |||
| All cases | 111 | 41 | ||
| Age (year), >50:⩽50 | 53:58 | 18:23 | 0.178 | 0.673 |
| Gender, male:female | 90:21 | 33:8 | 0.007 | 0.934 |
| AFP (μg/l), >20:⩽20 | 75:36 | 22:18 | 2.022 | 0.155 |
| Serum bilirubin (μmol/l), >17:⩽17 | 31:80 | 7:34 | 1.881 | 0.17 |
| Serum albumin (g/l), >40:⩽40 | 84:27 | 30:11 | 0.100 | 0.752 |
| ALT (U/l), >40: ⩽40 | 47:64 | 16:25 | 0.136 | 0.712 |
| HBs antigen, positive:negative | 84:27 | 34:7 | 0.907 | 0.341 |
| HBe antigen, positive:negative | 33:78 | 13:28 | 0.055 | 0.814 |
| Tumor size (cm), >5:⩽5 | 53:58 | 22:19 | 0.418 | 0.518 |
| Tumor number, single:multiple | 75:36 | 32:9 | 1.578 | 0.209 |
| Encapsulation, complete:no | 60:51 | 18:23 | 1.235 | 0.266 |
| Liver cirrhosis, with:without | 89:22 | 25:16 | 5.889 | 0.015* |
| Differentiation, I+II:III+IV | 29:82 | 23:18 | 11.950 | 0.001* |
| Microvascular invasion, yes:no | 26:85 | 8:33 | 0.264 | 0.608 |
| TNM stage, I:II:III | 39:31:41 | 21:11:9 | 4.019 | 0.134 |
| BCLC stage, A:B:C | 33:59:19 | 19:19:3 | 4.673 | 0.097 |
Abbreviations: AFP, α-fetoprotein; ALT, alanine aminotransferase; BCLC, Barcelona Clinic Liver Cancer; HBe, hepatitis Be; HBs, hepatitis B surface; TNM, tumor node metastasis.
Data are expressed as ratios.
*P<0.05 by χ2 test.
The relative quantitative expression level was used. Low expression of miR-99a in 111 patients was classified as hepatocellular carcinoma (HCC)/nontumor ratio of ⩽0.5. High miR-99a expression in 41 patients was classified as HCC/nontumor ratio of >0.5.
Figure 1.
miR-99a is correlated with tumor differentiation, liver cirrhosis and patients' outcome in HCC. (a) Relative lower expression of miR-99a preferably observed in HCC tumors from patients with lower tumor differentiation or liver cirrhosis. (b) Kaplan–Meier survival curves of tumor-free survival and overall survival according to relative expression level of miR-99a in primary HCC tissues. The median value of this ratio was chosen as the cutoff point.
MiR-99a suppresses HCC cell growth in vitro and in vivo
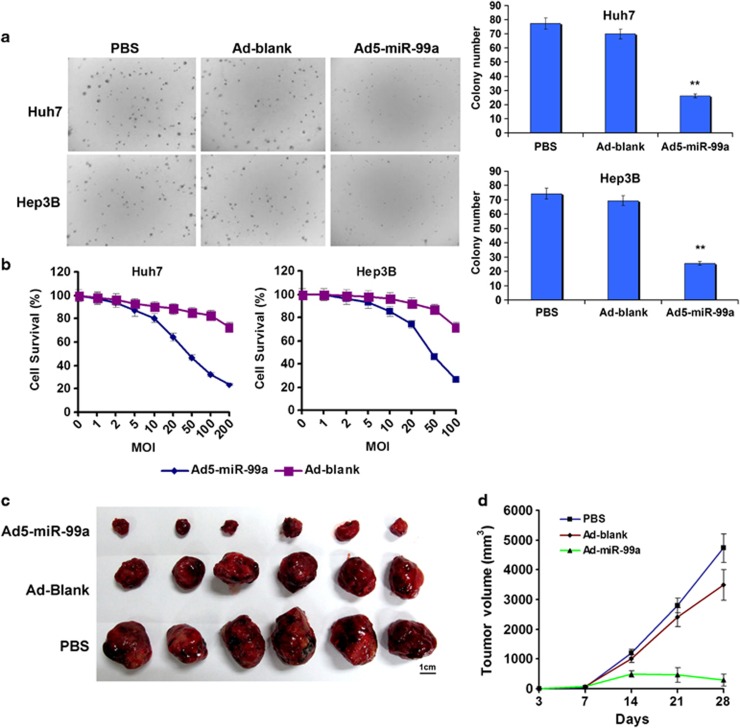
To further investigate the role of miR-99a in HCC, we restored miR-99a expression in Huh7 and Hep3B cells in which miR-99a was greatly reduced (Supplementary Figure S1) via nonreplicative adenoviral vector Ad5-miR-99a that expresses miR-99a (Supplementary Figure S2a). As a result, the growth of transduced cells was significantly decreased and the capacity to form cell colony was remarkably attenuated (Figure 2a) when compared with the cells transduced with the control adenovirus (that is, Ad-Blank). Furthermore, a significant impairment on cell growth was observed in Huh7 and Hep3B cells with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (Figure 2b). These results demonstrated that miR-99a inhibited tumor growth in vitro.
Figure 2.
Overexpression of miR-99a inhibits HCC growth in vitro and in vivo. (a) The influence of miR-99a restoration on colony formation of HCC cells. Shown was the image 15 days post plating (left) and the quantitative results (right). (b) The killing effect on HCC cells by reintroduction of miR-99a in vitro as examined by MTT assay. (c, d) The tumor morphology and growth curve of subcutaneous Huh7 xenograft after treatment, respectively. Scale bar=1 cm.
In order to investigate the antitumor effect of miR-99a in vivo, we established a human HCC-bearing mouse model by subcutaneously implanting Huh7 cells to BALB/c nude mice (nu/nu). By intratumoral injection of Ad5-miR-99a, miR-99a expression in tumor tissues was greatly increased (Supplementary Figure S2b), whereas tumor growth was significantly inhibited as compared with the control (P<0.001; Figures 2c and d), and more tumor necrosis was observed (Supplementary Figure S3). Thus, our results demonstrated that the restoration of miR-99a expression could dramatically repress HCC growth, suggesting that Ad5-miR-99a might have considerable potential for HCC gene therapy.
MiR-99a directly targets Ago2 by translational repression
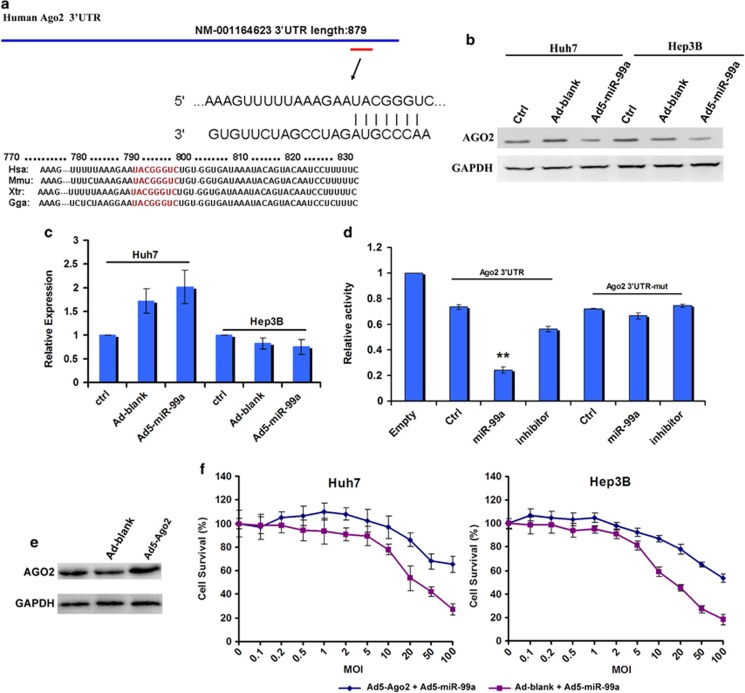
The target genes of miR-99a that mediated its suppressive functions in HCC pathogenesis were further analyzed. By searching predicted target genes of miR-99a overlapped in TargetScan (http://www.targetscan.org) and miRNA database (http://www.microrna.org/microrna/getMirnaForm.do), and subsequently confirmed in RegRNA (http://regrna.mbc.nctu.edu.tw/index1.php), we found that Ago2 (also known as Eif2c2) might be a novel candidate gene targeted by miR-99a (Figure 3a). Interestingly, the putative recognized sites of miR-99 family within the 3′-untranslated region (UTR) of Ago2 were broadly conserved among vertebrates (Figure 3a). When the miR-99a expression was restored by transduction with Ad5-miR-99a in Huh7 and Hep3B cells, the expression level of AGO2 protein was greatly decreased (Figure 3b), whereas the Ago2 mRNA was not significantly reduced (Figure 3c), indicating that miR-99a may inhibit Ago2 via translation repression rather than mRNA degradation.
Figure 3.
Ago2 is directly targeted by miR-99a. (a) Schematic representation of the 3′-UTR of Ago2. Red bars show putative miR-99a target sequences. Sequence alignment of the target site among vertebrate, including amphibians (Xenopus (Xtr)), birds (Gallus (Gga)) and mammals (human (Hsa) and mouse (Mmu)), is shown below. (b, c) The capacity of miR-99a on regulating Ago2 as detected by western blot and qRT–PCR. Ad-blank, the control adenovirus without transgene; Ad5-miR-99a, the recombinant adenovirus expressing miR-99a; Ctrl, untreated HCC cells. (d) Luciferase assay for the regulation capacity of miR-99a on Ago2. Both wild and mutated (mut) Ago2 3′-UTR (the putative seed binding sequence was mutated from TACGGGTC to ATGCCCAG) was used. Empty, Huh7 cells transfected with the empty dual-Luc assay vector, psiCHECK-2; Ctrl, Huh7 cells transfected with psiCHECK-2 containing Ago2 3′-UTR (psiCHECK2-Ago2 UTR) or mutated Ago2 3′-UTR (psiCHECK2-Ago2 mutUTR), respectively. miR-99a, Huh7 cells co-transfected with miR-99a mimic and psiCHECK2-Ago2 UTR/psiCHECK2-Ago2 mutUTR; inhibitor, Huh7 cells co-transfected with miR-99a inhibitor, miR-99a mimic and psiCHECK2-Ago2 UTR/psiCHECK2-Ago2 mutUTR. *P<0.05; **P<0.01. (e) Overexpression of AGO2 protein by transduction with Ad5-Ago2 (MOI=5), a recombinant adenoviral vector expressing Ago2. (f) The counteraction of AGO2 overexpression on miR-99a killing effect on HCC cells as examined by MTT assay.
To verify whether Ago2 is a direct target of miR-99a, a dual-luciferase reporter containing the 3′-UTR of Ago2 was constructed and used to transfect Huh7 cells. As shown in Figure 3d, the luciferase activity (Firefly Luciferase) was significantly repressed by co-transfection with a vector expressing miR-99a, whereas it was recovered by reducing the expression of miR-99a with the miR-99a inhibitor. In contrast, when co-transfection was performed with the miR-99a expression vector and the dual-luciferase reporter containing an artificial 3′-UTR of Ago2, in which the putative miR-99 target sites were mutated, the luciferase activity was not distinctly changed. Together, these results proved that endogenous Ago2 was targeted and regulated by miR-99a, mainly through translation repression.
Furthermore, we overexpressed AGO2 with Ad5-Ago2, a recombinant adenoviral vector carrying an expression cassette of 3′-UTR-negative Ago2 (Figure 3e). When Ad5-Ago2 was co-transduced at defined multiplicity of infection (MOI) with Ad5-miR-99a at a gradient of MOI into Huh7 and Hep3B cells respectively, the inhibitory effect of Ad-miR-99a could be impaired lower on these HCC cells (Figure 3f). However, the ability of Ago2 overexpression was insufficient to rescue the viability of miR-99a effects when the used MOI of Ad-miR-99a was increased, indicating that miR-99a could also inhibit HCC growth via regulating its other target genes.
Ago2 is upregulated in HCC and negatively correlated with miR-99a
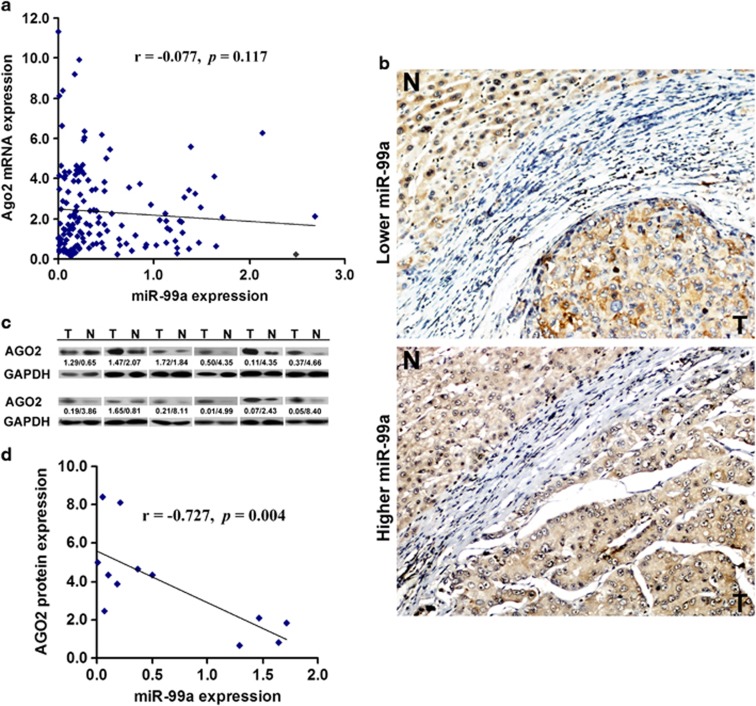
Ago2 plays an important role in miRNA-mediated posttranscriptional regulation, but its expression profile in HCC remains largely unknown. Thus, the expression profile of Ago2 in HCC cell lines and primary HCC tissues was investigated by RT–PCR analysis. As a result, overexpression of Ago2 was found in 5 of 6 detected HCC cells (Supplementary Figure S4) and 105 of 152 cancerous tissues as compared with their paired noncancerous tissues. However, no distinct correlation was revealed between the expression of miR-99a and Ago2 at the RNA level (r=−0.077, P=0.117, Figure 4a).
Figure 4.
Ago2 is upregulated in HCC and negatively correlated with miR-99a. (a) The correlation between relative expression of miR-99a and Ago2 mRNA in HCC tissues as determined by qRT–PCR. (b) Increased AGO2 protein as analyzed by immunohistochemistry in cancerous tissues. The patient serial number was 30 and 6, respectively. (c) Increased AGO2 protein as detected by western blotting. Expression ratio was calculated by normalizing AGO2 to total protein levels in each case. The value of relative expression of miR-99a/AGO2 protein is shown below. The patient serial number was 6, 8, 9, 11, 30, 41, 53, 68, 73, 90, 121, and 132, respectively. N, matched noncancerous control; T, HCC tissue. (d) The correlation between expression of miR-99a and AGO2 protein in 12 pairs of HCC samples mentioned above.
Then, protein expression of Ago2 was examined by immunohistochemistry in 20 pairs of primary HCC tissues with different levels of miR-99a. Among the 10 HCC tissues with lower miR-99a, AGO2 protein was found to be overexpressed (Figure 4b). For the other 10 HCC samples with higher miR-99a, no significant difference of AGO2 expression was detected between the cancerous tissues and the matched noncancerous tissues in 7 samples (Figure 4b). This result was further confirmed by western blotting analysis in which 12 pairs of HCC samples were examined (r=−0. 727, P=0.004, Figures 4c and d). Taken together, these data showed that AGO2 was overexpressed in HCC tissues and the protein expression was negatively correlated with the expression level of miR-99a that validated that Ago2 was a novel target gene directly regulated by miR-99a via translation repression.
Knockdown of Ago2 distinctly inhibits HCC growth in vivo
To identify the role of Ago2 in HCC pathogenesis, the expression of Ago2 was decreased by lentivirus-mediated RNA interference (Supplementary Figure S5). As shown in Figures 5a and b, the abundance of Ago2 was successfully downregulated at both RNA and protein levels in HCC Huh7 cells stably expressing Ago2-specific small interfering RNA (siRNA). However, not distinct alteration on cell growth was observed when Ago2 was downregulated in vitro (Figure 5c).
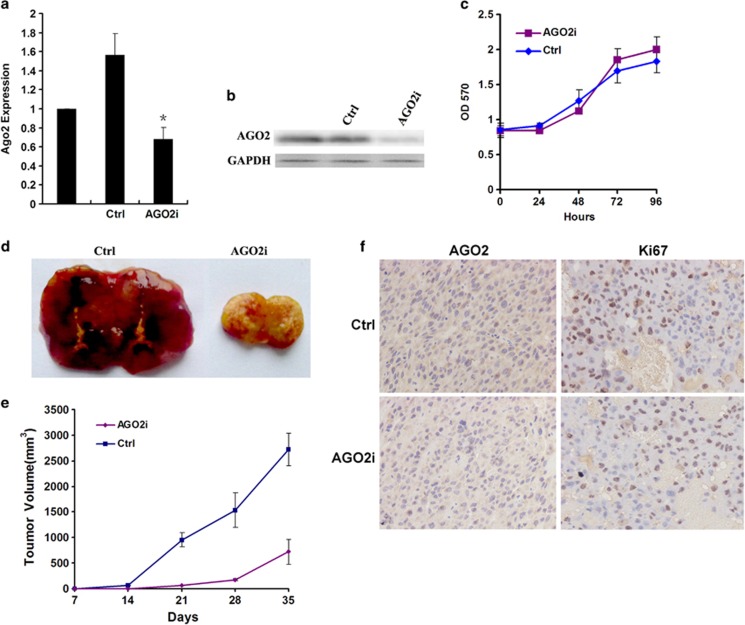
Figure 5.
Ago2 knockdown distinctly inhibits HCC tumor growth in vivo. (a, b) Downregulation of Ago2 through lentivirus-mediated RNA interference (RNAi) as detected by RT–PCR and western blot. Ctrl and AGO2i indicated Huh7 cells stably expressing control siRNA and Ago2-specific siRNA, respectively. (c) The effect on cell proliferation of Ago2-knockdown as measured by MTT assay. Data were obtained from three independent experiments and shown as mean±s.d. (d, e) The influence on tumor growth by Ago2 knockdown in vivo. The growth curve and tumor morphology of Ago2-knockdown Huh7 xenograft. (f) AGO2 and KI67 expression examined by immunohistochemistry ( × 200). Ctrl and AGO2i indicated Huh7 cells stably expressing control siRNA and Ago2-specific siRNA, respectively.
Then, Ago2-knockdown Huh7 cells were subcutaneously injected into BALB/c nude mice to further analyze the influence of Ago2 on HCC growth in vivo. Surprisingly, nearly half of the mice (4 of 10) in Huh7 Ago2-knockdown group failed to generate xenografts, whereas all the mice in control group developed detectable tumors 2 weeks post transplantation (10 of 10), indicating the tumorigenic ability was greatly impaired by Ago2 knockdown in Huh7 cells (P<0.05). Furthermore, the tumors derived from Ago2-knockdown Huh7 cells were much smaller compared with those in the control group transduced with nonsilencing siRNA (P<0.001; Figures 5d and e). The decreased expression of AGO2 and KI67 in tumors derived from Ago2-knockdown Huh7 cells was revealed by immunohistochemistry analyses (Figure 5f). These results indicated that the suppression of Ago2 might greatly inhibit HCC growth in vivo.
Ago2 facilitates miR-21 to repress its targeted gene Pten
The capacity of Ago2 knockdown to inhibit HCC growth in vivo promoted us to further investigate the mechanism of Ago2 in HCC carcinogenesis. Considering Ago2 functions in miRNA-mediated gene silencing, we examined the alteration of miRNA-target gene pairs upon Ago2 knockdown and miR-99a overexpression, including oncomiRs (for example, miR-21), miRNA suppressors (for example, let-7) and their targets. As detected by RT–PCR, miR-21a was upregulated in HCC cells (Huh7 and Hep3B) but decreased in both Ago2-knockdown cells and miR-99a-overexpressing cells, whereas let-7a was almost silenced and this silencing was maintained in both Ago2-knockdown and miR-99a-overexpressing HCC cells (Supplementary Figure S6). The results of western blotting showed that the expression of PTEN (targeted by miR-21) was increased, whereas those of NRAS, HMGA2 and C-MYC (targeted by let-7) were not distinctly altered in either Ago2-knockdown or miR-99a-overexpressing HCC cells (Figure 6a and Supplementary Figure S7), suggesting an indirect regulation of miR-99a on PTEN.
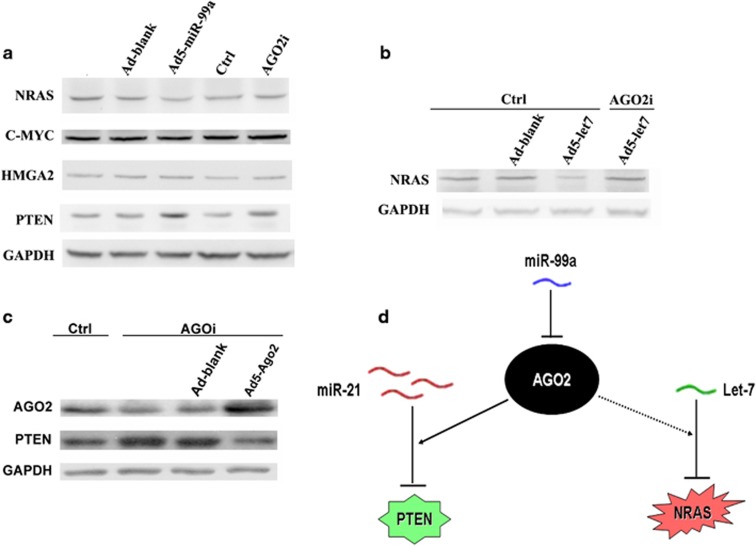
Figure 6.
Ago2 facilitates miR-21 to repress its targeted gene Pten. (a) The expression of a list of cancer-associated proteins as analyzed by western blot. GAPDH served as loading control. Ctrl and AGO2i indicated Huh7 cells stably expressing control siRNA and Ago2-specific siRNA, respectively. Ad-blank, the control adenovirus without transgene; Ad5-miR-99a, the recombinant adenovirus expressing miR-99a. Data are shown as representative of three independent experiments. (b) The expression of NRAS protein under different AGO2 conditions as determined by western blot. Ctrl and AGO2i indicated Huh7 cells stably expressing control siRNA and Ago2-specific siRNA, respectively. Ad5-let7a, the recombinant adenovirus expressing let-7a. (c) The expression of PTEN protein under different AGO2 conditions as determined by western blot. Ctrl and AGO2i indicated Huh7 cells stably expressing control siRNA and Ago2-specific siRNA, respectively. Ad5-Ago2, the recombinant adenovirus expressing Ago2; Ad-blank, the control adenovirus without transgene. (d) A mode of the regulatory network of miR-99a and Ago2. Ago2 is targeted by miR-99a. On the condition that the oncomiRs (for example, miR-21) are upregulated but the tumor suppressors (for example, let-7a) are downregulated, the increased Ago2 can promote miR-21 to repress PTEN, whereas leave the regulation capacity of let-7 on NRAS at low level.
Interestingly, when the let-7 expression was restored by the recombinant adenovirus expressing let-7, NRAS protein could be substantially deregulated, whereas the deregulation was attenuated if Ago2 was knocked down (Figure 6b). In contrast, in Ago2-knockdown Huh7 cells, PTEN protein was upregulated, whereas the upregulation was considerably disrupted when Ago2 was restored via Ad5-Ago2 (Figure 6c). In consistence with previously reported result,10 we found that the tumor-suppressor miRNA let-7a was remarkably downregulated, whereas the oncomiR miR-21 was distinctly upregulated in a list of HCC cell lines (Supplementary Figure S8). Thus, these results had revealed a novel regulation mode of Ago2 in HCC in which upregulation of Ago2 specifically facilitates the increased oncomiRs miR-21 to repress its targeted tumor-suppressors PTEN, whereas it leaves the regulatory capacity of the decreased tumor-suppressor miRNAs let-7 on their oncogenic targets at lower level, thereby promoting HCC progression and carcinogenesis (Figure 6d).
Discussion
We have previously reported that miR-99a is the sixth most abundantly expressed miRNA in human normal liver, but it is deregulated in a high proportion of primary HCC tissues relative to their matched noncancerous tissues via deep sequencing.10 The downregulation of miR-99ab/100 family was also observed in HCC by other groups11, 12, 13, 14 and in other types of cancers,15, 16, 17, 18, 19 including prostate cancer, oral cancer, head and neck squamous cell carcinoma, lung adenocarcinoma and renal cell carcinoma, suggesting an important role of miR-99 in malignant tumors. In this study, using an independent cohort of HCC patients, miR-99a was found to be distinctly decreased in primary HCC tissues and significantly correlated with both tumor-free survival (P=0.004) and overall survival in HCC patients (P=0.006). Furthermore, our result indicated that miR-99a downregulation was more frequently observed in tumors from patients with liver cirrhosis (P=0.015) or lower tumor differentiation (P=0.001), in accord with the finding that miR-99ab/100 dysregulation is an early event maintained along with HCC progression.13 Furthermore, the correlation between miR-99a expression and tumor differentiation may be responsible for the poor survival of HCC patients with lower level of miR-99a.
To date, a number of targets for miRNA-99 have been experimentally validated and most of them are oncogenes. Notable targets include Igf1r (insulin-like growth factor 1 receptor), mTOR (mechanistic target of rapamycin), Mtmr3 (Myotubularin-related protein 3) and two SWI/SNF chromatin remodeling factor Smarca5 and Smarcd1.14, 15, 16, 17, 18, 19, 20, 21 Here, a novel target gene, Ago2, was found to be directly regulated by miR-99a via translational repression and the expression level of AGO2 protein in primary HCC tissue was negatively correlated with miR-99a expression. Thus, a new way that miR-99a functions as a tumor suppressor in HCC has been revealed.
Ago2, also known as Eif2c2, is the central component of RNA-induced silencing complex, the protein complex responsible for gene silencing.22, 23 In addition, Ago2 has been found in Dicer-independent miRNA biogenesis through its catalytic activity.24, 25 The Ago2 gene belongs to an evolutionarily conserved family that is shared among not only eukaryotes, but also archaea and certain bacteria such as Aquifex aeolicus.23 In mice, disruption of Ago2 leads to embryonic lethality early in development after the implantation.26 Despite the fact that Ago2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling at the transcriptional level,27 little is known about the posttranslational regulatory networks of Ago2. Previously, Dicer, another key component of the miRNA processing machinery, has been found to be regulated by miR-103/107 and miRNA-130a,28, 29 forming a regulatory feedback loop that mediated tumor metastasis and survival. In this study, Ago2 is demonstrated to be directly targeted by miR-99a and involved in tumor growth control, and this is the first report revealing the miRNA regulator of Ago2.
Compared with individual miRNA, the deregulation of miRNA processing machinery itself can exert more profound effects on miRNA profile.30 Therefore, to accurately maintain the expression levels of miRNA machinery (for example, Dicer, Drosha and AGO2) is crucial for normal cell activities. In recent years, the dysreglution of Dicer and Drosha has been intensively discussed and linked to the carcinogenesis of several cancers, including HCC.31, 32, 33, 34, 35, 36, 37, 38 Merely, the opposite situations have been revealed between patient's outcome and the expression levels of Dicer and Drosha among different cancer types. As for Ago2, overexpression of AGO2 protein was found within 31 pairs of primary HCC samples during the period that this paper was prepared, and it was found that increased AGO2 can promote tumor growth, migration and metastasis.39 In this study, the upregulation of Ago2 is validated in a large sample of primary HCC tissues (that is, 152) relative to their matched noncancerous tissues at both RNA and protein levels, and this was partly associated with reduced expression of miR-99a. It is noteworthy that the effect of Ago2 knockdown on cell proliferation in vitro and in vivo was varied. A plausible reason for this phenomenon is that Ago2 knockdown can inhibit angiogenesis in vivo that is hardly displayed in vitro (Jin H, et al, unpublished data).
Although several miRNAs are upregulated in specific tumors, a global reduction of miRNA abundance appears a common trait of human cancers.7, 28, 40 Previously, a list of miRNAs suppressors have been found downregulated in HCC, along with the upregulation of some oncoMiRs (for example, miR-21 and miR-221).8, 9, 10 Among them, miR-122, miR-199a-3p, miR-101, let-7 and miR-99a are the first, third, fourth, fifth and sixth most abundant miRNAs in normal human liver, respectively, accounting for ∼71.4% of the miRNome.10 Thus, the global downregulation of miRNA suppressor also applies to HCC. On this basis, the upregulation of Ago2 has aggravated the homeostasis of regulatory networks mediated by miRNA, wherein the capacities of oncoMiRs (for example, miR-21) to regulate tumor suppressors (for example, PTEN) are specifically enhanced, whereas those of miRNA suppressors (for example, let-7) on oncogenes (for example, NRAS) is unaltered. The superposition of these two factors promotes the progression of HCC. In turn, the oncogenic effect exerted by the upregulation of Ago2 in HCC may have also benefitted gene therapy by reintroduction of miR-99a present and miR-199-3p previous.10 Therefore, our study has implied the promising prospect of miRNA therapy for HCC by restoring the expression of miRNA suppressors.
Materials and methods
Patients and cell lines
A total of 152 pairs of primary HCC tissues and the matched adjacent noncancerous tissues were randomly selected from patients undergone surgical resection at the Eastern Hepatobiliary Surgery Hospital. All samples were collected with the written informed consent of HCC patients and the experiments were approved by the Institutional Review Board of Eastern Hepatobiliary Surgery Hospital Ethics Committee. For the enrolled HCC patients, no local or systemic treatment was conducted before operation. The tumor differentiation was defined according to the Edmondson grading system, in which a higher grade indicated a lower differentiation state.41 The tumor staging was defined as described previously.42 The detailed clinical characteristics of all patients are listed in Table 1. Human HCC cell lines (HepG2, HepG2.15, Hep3B, PLC/PRF/5, Huh7 and SMMC-7721), human embryonic liver cell line (WRL-68) and human embryo kidney cell line (HEK293) were obtained and cultured as described previously.42, 43
Quantitative RT–PCR
Total RNA of all test tissues or cells samples was isolated using a miRNeasy mini kit (Qiagen, Valencia, CA, USA). Then, 1 μg RNA of each sample was reverse transcribed to complementary DNA using a miScript Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. For quantitative PCR analysis, aliquots of complementary DNA were amplified using miScript SYBR Green PCR Kit (Qiagen) and performed on 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The miR-99a expression of all test samples was determined by normalizing to that of U6. The Ago2 expression was measured via similar method except for using Gapdh as an endogenous control. All reactions were performed in triplicate. The sequences of used primers were listed in Supplementary Table S3.
Luciferase assays
Mutagenesis of the putative miR-99-binding sites within the 3′-UTR of human Ago2 was performed using overlapped PCR. Then, the natural and modified 3′-UTRs of Ago2 were cloned into psiCHECK-2 (Promega, Madison, WI, USA), respectively. Cell culture, transfection and luciferase assay were performed as previously described.10, 43 The miR-99a-specific mimic and inhibitor were used to upregulate or suppress the expression of exogenous miR-99a according to the manufacturer's instructions (Life Technologies, Carlsbad, CA, USA).
Virus construction
The replication-defective recombinant adenovirus expressing miR-99a and Ago2 was constructed according to the procedure described previously.43 For lentivirus-based shRNA knockdown systems, five RNA interference candidate target sequences to human Ago2 (Supplementary Table S4) were designed and the oligonucleotides encoding the candidate Ago2-siRNA sequence and a loop sequence separating the complementary domains were synthesized and inserted into the lentiviral transfer vector pGC-LV (Genechem, Shanghai, China), respectively. Among them, Ago2-Si3 had the best interference efficiency in HEK-293T cells cotransfected with Ago2-overexpressing vector and siRNA expression constructs revealed by western blot and was selected to knock down the endogenous Ago2 in HCC cells. The nonsilencing siRNA was used as a control. Then, the obtained lentiviral transfer vector expressing Ago2-specific shRNA was co-transfected with the packaging plasmids pHelper 1.0 and pHelper 2.0 into HEK-293T cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). At 40 h post transfection, the lentiviral particle was harvested from the medium, concentrated by ultracentrifugation and titrated on HEK-293T cells as previously described.44
MTT assay
To examine the therapeutic effect of miR-99a restoration, Huh7 and Hep3B cells were planted at a density of 5 × 103 in 96-well plates and infected with Ad5-miR-99a or Ad-blank at a gradient of MOI. In the rescue experiments, Ad5-Ago2 or Ad-blank at 5 MOI was co-transduced with Ad5-miR-99a at a gradient of MOI into Huh7 and Hep3B cells. On the day of harvest, 100 μl of spent medium was replaced with an equal volume of fresh medium containing MTT 0.5 mg/ml. Plates were incubated at 37 °C for 4 h, and then the medium was replaced by 100 μl of dimethyl sulfoxide (Sigma, St Louis, MO, USA) and plates shaken at room temperature for 10 min. The absorbance was measured at 570 nm.
Proliferation and colony formation assays
Cells (5 × 103) suspended in 1 ml 0.3% top agarose were plated onto 2 ml 0.6% base agarose in six-well plate and maintained for 15 days. Then, complete medium (500 μl) was added every 5 days. On day 15, colonies were counted and photographed by light microscopy. For cell proliferation assay, 1 × 104 Huh7 or Ago2-knockdown Huh7 cells were planted in 96-well plates, and then cells were harvested at a series of time point and detected with MTT assay. Data were obtained from three independent experiments.
Western blot
Protein lysates from cell lines or tissues were prepared in lysis buffer and centrifuged at 12 000 g at 4 °C. The procedure of western blotting was according to the procedure described previously.43 Primary antibodies used were rabbit anti-AGO2 (Abcam, Cambridge, UK), rabbit anti-PTEN (SAB, Pearland, TX, USA), rabbit NRAS (Abgent, San Diego, CA, USA), rabbit HMGA2 (Abgent), rabbit C-MYC (Santa Cruz, CA, USA) and mouse anti-GAPDH (Santa Cruz Ltd.).
In vivo assay
All animal experiments were undertaken in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of Second Military Medical University, Shanghai. Human HCC-bearing male nude mice with Huh7 cells were used for evaluating the antitumor effect of miR-99a in vivo. To prepare the subcutaneous model, mice were injected subcutaneously in the right flank with 1 × 107 Huh7 cells in matrigel (injection volume of 100 μl). At 2 weeks post injection, mice with the xenografts tumor were randomly distributed into three groups (n=6 per group) and injected intratumorally with Ad-blank (at a dose of 5 × 108 plaque-forming units (PFUs)), Ad5-miR-99a (at a dose of 5 × 108 PFUs) or placebo five times (one time every other day), respectively. Tumor volumes (volume=(W2 × L)/2; W, width; L, length, in cubic mm) were measured once weekly after the first adenovirus injection on day 1. To evaluate the effect of Ago2 knockdown in vivo, Huh7 cells stably expressing Ago2-specific siRNA or nonsilencing-siRNA were injected subcutaneously (1 × 107/mouse, n=10 per group). At 2 weeks post injection, tumor volumes were measured once weekly if the xenografts tumors were formed.
Immunohistology
Immunohistochemical staining was performed on formalin-fixed, paraffin-embedded tissue sections using ABC (avidin biotin-peroxi-dase complex) method. The used primary antibodies were AGO2 (1:100, Abcam) and KI67 (1:200, Sigma). The quantification of immunohistology for primary tissues was performed according to the procedure previously described with minor modification.45 In brief, positive reaction was determined based on the presence and intensity of cytoplasm immunoreaction deposits. Tissues were graded using the following criteria: 0, low <25% of total cells; 1, 25–50% of total cells; 2, 50–75% of total cells; 3, >75% of total cells. Two pathologists without knowledge regarding patients' clinical characteristics were independently responsible for the scoring. The concordance between scores from different sections of the same tissue was >90%. All discrepancies in scoring were reviewed, and a consensus was reached.
Statistical analysis
All data are presented as mean±s.d. unless otherwise stated. The independent Student's t-test was used to analyze the variation of two selected groups and Pearson's χ2 test was used to analyze the correlation between two clinical pathologic parameters. P<0.05 was considered significant statistically and is marked with an asterisk. P<0.01 was considered highly significant statistically and is marked with a double asterisk. All statistical analyses were performed with SPSS version 18.0 software (IBM, Chicago, IL, USA).
Acknowledgments
This work was supported by National Science Funds for Distinguished Young Scholar (No. 30925037), general project (No. 81001013, 81071681, 81201940) and Creative Research Groups (No. 81221061), Chinese Key Project for Infectious Diseases (No. 2012ZX10002-010) and Major State Basic Research Development Program (No. 2010CB529900-G).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis)
Supplementary Material
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Huang S, He X. The role of microRNAs in liver cancer progression. Br J Cancer. 2011;104:235–240. doi: 10.1038/sj.bjc.6606010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431–1443. doi: 10.1053/j.gastro.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, et al. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856–864. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- Petrelli A, Perra A, Schernhuber K, Cargnelutti M, Salvi A, Migliore C, et al. Sequential analysis of multistage hepatocarcinogenesis reveals that miR-100 and PLK1 dysregulation is an early event maintained along tumor progression. Oncogene. 2012;31:4517–4526. doi: 10.1038/onc.2011.631. [DOI] [PubMed] [Google Scholar]
- Li D, Liu X, Lin L, Hou J, Li N, Wang C, et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem. 2011;286:36677–36685. doi: 10.1074/jbc.M111.270561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Lee YS, Malhotra A, Kim HK, Matecic M, Evans C, et al. miR-99 family of microRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011;71:1313–1324. doi: 10.1158/0008-5472.CAN-10-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Jin Y, Yu D, Wang A, Mahjabeen I, Wang C, et al. Down-regulation of the microRNA-99 family members in head and neck squamous cell carcinoma. Oral Oncol. 2012;48:686–691. doi: 10.1016/j.oraloncology.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Fang S, Gao L, Tan Y, Yang Z. Clinic significance of microRNA-99a expression in human lung adenocarcinoma. J Surg Oncol. 2013;108:248–255. doi: 10.1002/jso.23381. [DOI] [PubMed] [Google Scholar]
- Kuo YZ, Tai YH, Lo HI, Chen YL, Cheng HC, Fang WY, et al. MiR-99a exerts anti-metastasis through inhibiting myotubularin-related protein 3 expression in oral cancer. Oral Dis. 2014;3:e65–75. doi: 10.1111/odi.12133. [DOI] [PubMed] [Google Scholar]
- Cui L, Zhou H, Zhao H, Zhou Y, Xu R, Xu X, et al. MicroRNA-99a induces G1-phase cell cycle arrest and suppresses tumorigenicity in renal cell carcinoma. BMC Cancer. 2012;12:546–556. doi: 10.1186/1471-2407-12-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman G, Avivi C, Mardoukh C, Barzilai A, Tessone A, Gradus B, et al. MiRNA expression in psoriatic skin: reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS ONE. 2011;6:e20916. doi: 10.1371/journal.pone.0020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AC, Sun D, Dutta A. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene. 2013;32:1164–1172. doi: 10.1038/onc.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik ES, Zamore PD. Argonaute proteins. Curr Biol. 2011;21:R446–R449. doi: 10.1016/j.cub.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Adams BD, Claffey KP, White BA. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150:14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Kovaleva V, Mora R, Park YJ, Plass C, Chiramel AI, Bartenschlager R, et al. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012;72:1763–1772. doi: 10.1158/0008-5472.CAN-11-3671. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, et al. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–7850. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis. 2010;31:1805–1812. doi: 10.1093/carcin/bgq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber C, Horst D, Hlubek F, Kirchner T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur J Cancer. 2011;47:1414–1419. doi: 10.1016/j.ejca.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Dedes KJ, Natrajan R, Lambros MB, Geyer FC, Lopez-Garcia MA, Savage K, et al. Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur J Cancer. 2011;47:138–150. doi: 10.1016/j.ejca.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Ravi A, Gurtan AM, Kumar MS, Bhutkar A, Chin C, Lu V, et al. Proliferation and tumorigenesis of a murine sarcoma cell line in the absence of DICER1. Cancer Cell. 2012;21:848–855. doi: 10.1016/j.ccr.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136:2304–2315. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa N, Ojima H, Shirakihara T, Shimizu H, Kokubu A, Urushidate T, et al. Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2013;104:543–551. doi: 10.1111/cas.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N, Li Y, Han ZG. Ago2 promotes tumor metastasis via upregulating FAK expression in hepatocellular carcinoma. Hepatology. 2013;57:1906–1918. doi: 10.1002/hep.26202. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, et al. Long non-coding RNA-MVIH promotes angiogenesis and serves as a predictor for HCC patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- Jin H, Lv S, Yang J, Wang X, Hu H, Su C, et al. Use of microRNA Let-7 to control the replication specificity of oncolytic adenovirus in hepatocellular carcinoma cells. PLoS ONE. 2011;6:e21307. doi: 10.1371/journal.pone.0021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat Methods. 2006;3:109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- Wang K, Liu J, Yan ZL, Li J, Shi LH, Cong WM, et al. Overexpression of aspartyl-(asparaginyl)-beta-hydroxylase in hepatocellular carcinoma is associated with worse surgical outcome. Hepatology. 2010;52:164–173. doi: 10.1002/hep.23650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.