Summary
Unlike humans or mice, some species have limited genome encoded combinatorial diversity potential, yet mount a robust antibody response. Cows are unusual in having exceptionally long CDR H3 loops and few V-regions, but the mechanism for creating diversity is not understood. Deep sequencing revealed that ultralong CDR H3s contain a remarkable complexity of cysteines, suggesting that disulfide-bonded mini-domains may arise during repertoire development. Indeed, crystal structures of two cow antibodies reveal that these CDR H3s form a very unusual architecture composed of a β-strand “stalk” that supports a structurally diverse, disulfide-bonded, “knob” domain. Sequence analysis suggests that diversity arises from somatic hypermutation of an ultralong DH with a severe codon bias towards mutation to cysteine. These unusual antibodies can be elicited to recognize defined antigens through the knob domain. Thus, the bovine immune system produces an antibody repertoire composed of CDR H3s of unprecedented length that fold into a diversity of mini-domains generated through combinations of somatically generated disulfides.
Introduction
Antibodies are quite diverse but this heterogeneity is present within the constraints of the immunoglobulin fold. The most diverse portion of the antibody molecule is the complementarity determining region 3 of the heavy chain (CDR H3), which is derived from DNA rearrangement of variable (V), diversity (D), and junctional (J) gene segments (Fugmann et al., 2000; Kato et al., 2012; Smider and Chu, 1997). Additional point mutations are acquired in the variable regions after antigen exposure through somatic hypermutation (SH) (Di Noia and Neuberger, 2007; Kocks and Rajewsky, 1988). Despite the genetic modifications of gene rearrangement and SH, the overall structure of the antibody is maintained within the immunoglobulin fold and the associated CDR loops of the heavy and light chains. Variations on this theme include VHH antibodies from camelids and the IgNAR of sharks (Decanniere et al., 1999; Stanfield et al., 2004), which contain bivalent heavy chain domains without light chains; however, both of these still utilize their heavy chain CDR loops to bind antigen. The only known exception to this structural paradigm for antigen recognition is the variable lymphocyte receptor of jawless vertebrates, which use a leucine-rich repeat scaffold with variable loops to bind antigen (Alder et al., 2005; Pancer et al., 2004). Interestingly, some vertebrates, such as Bos taurus, have a very limited diversity of V gene segments (Berens et al., 1997; Lopez et al., 1998; Saini et al., 2003; Sinclair et al., 1997; Zhao et al., 2006), yet maintain a perfectly robust adaptive immune response, suggesting unique diversification mechanisms at work to generate a functional antibody repertoire.
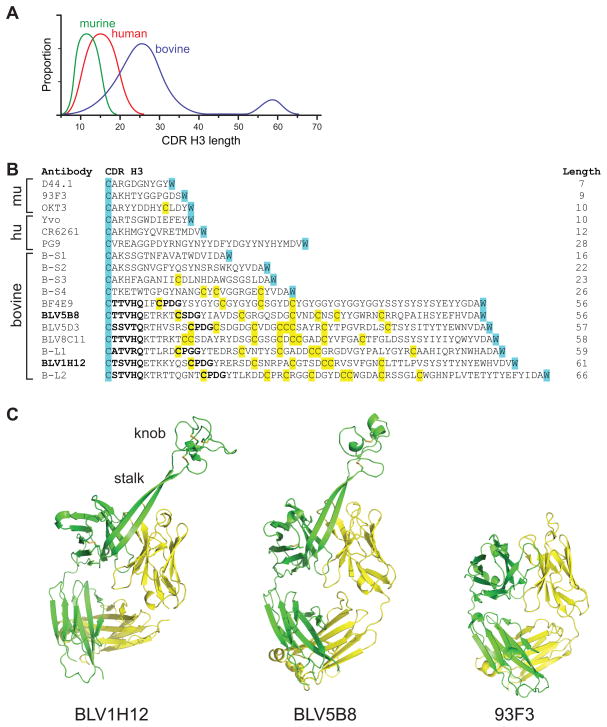
The CDR H3 is typically 8–16 amino acids in length in humans (Figure 1A) and, along with the other CDRs of the heavy and light chain, usually forms a flat or undulating binding surface for antigen recognition. In humans, some longer CDR H3 loops with unusual protruding structures have been described that contribute to important functions such as virus neutralization (Collis et al., 2003; Kwong and Wilson, 2009; Pejchal et al., 2010; Saphire et al., 2001). Different species exhibit a diversity of CDR H3 length; however, bovine antibodies have the longest CDR H3 regions known, with an ultralong subset that ranges in length from 50 to 61 amino acids (Berens et al., 1997; Lopez et al., 1998; Saini et al., 1999; Saini et al., 2003; Zhao et al., 2006) (Figure 1A). These heavy chains pair with a restricted set of lambda light chains (Saini et al., 2003), and have multiple but an even number of cysteines, suggesting they participate in disulfide bonds (Saini et al., 1999) (Figure 1B). The restricted VH-VL pairing, potential for multiple disulfide bonds, and unusually long length suggests that these bovine CDR H3s might not be loops or β-hairpins, but have a unique and well-defined structural fold. Although they represent over 10% of the bovine repertoire, the structure, function, and underlying genetic mechanisms resulting in ultralong CDR H3 formation and diversity generation have not been elucidated.
Figure 1. Identification of a new structural domain in bovine antibodies.
(A) Comparison of CDR H3 length amongst murine, human, and bovine repertoires. An ultralong subset of over 60 amino acids is uniquely found in bovine heavy chains (blue). (B) Sequences of representative CDR H3 from murine (mu), human (hu), or bovine sequences from the literature along with six bovine sequences (B-S1 to B-S4, and B-L1 and B-L2) from our sequencing results. The conserved cysteine of framework 3 and tryptophan of framework 4 that define CDR H3 boundaries in all antibody variable regions are highlighted in cyan for reference, and cysteines are yellow. The lengths of the CDR H3s are indicated at the right. The murine antibodies include D44.1, an anti-HEL antibody, 93F3, an aldolase, and OKT3, a therapeutic antibody targeting human CD3. This antibody is unusual in having a free cysteine in CDR H3. The human antibodies include Yvo, a cryoglobulin, CR6261, an anti-influenza A hemaglutinin, and PG9, an anti-HIV antibody which has one of the longest human CDR H3s. The bovine antibodies represent the ultralong sequences in the literature, and short sequences for comparison. BLV5B8 and BLV1H12 (indicated in bold) were used in our structure determinations. Relatively conserved TTVHQ and CPDG motifs are in bold. (C) Crystal structures of BLV1H12 (left) and BLV5B8 (middle) Fabs compared to the 93F3 Fab with a “normal” CDR H3 (right). A superlong, two β-stranded stalk protrudes from each bovine VH immunoglobulin domain and terminates in an unusual three disulfide-linked knob domain. See also Figure S1 and Table S1.
Results
A unique antibody structure in cattle
To delineate the architecture of bovine antibodies containing ultralong CDR H3s, we determined crystal structures of two Fab fragments: BLV1H12 and BLV5B8 (Table S1 available online). Each of these antibodies was originally cloned from a fetal calf infected with bovine leukemia virus (which transforms B-cells), however the original antigens illiciting these antibodies are unknown (Saini et al., 1999; Saini et al., 2003). The CDR H3s of BLV5B8 and BLV1H12 are 56 and 61 amino acids, respectively (Figure 1B). The overall structure of the BLV1H12 variable region core is very similar to other antibodies except for the CDRs of the heavy and light chains (Figure 1C). The 61-residue CDR H3 forms an unprecedented structure where a subdomain with an unusual architecture is formed from a “stalk”, composed of two 12-residue, anti-parallel β-strands, and a 39-residue, disulfide-rich “knob” that sits atop the stalk far from the canonical antibody paratope (Figure 1C, left). The long anti-parallel β-ribbon serves as a bridge to link the “knob” domain with the main antibody scaffold, and is rigidified using eight standard β-sheet hydrogen bonds. The CDR H3 of a second antibody, BLV5B8, has little sequence homology to BLV1H12, but the unique “stalk” and “knob” structural features are maintained (Figure 1C, middle). The two bovine antibodies have dramatically different CDR H3 structures compared to a typical CDR H3 in mouse or human antibodies (Figure 1C, right).
Structural diversity in bovine CDR H3s
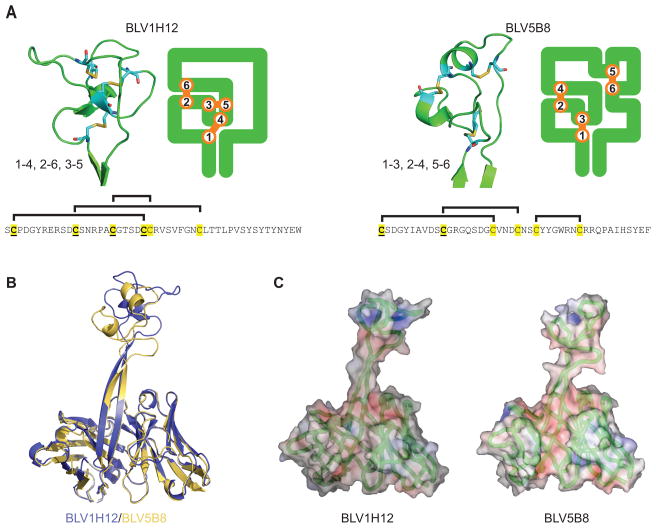
Both BLV1H12 and BLV5B8 have stalk and knob components that share certain features including a “T(T/S)VHQ” motif at the base of the ascending strand, which is connected by a variable number of residues to a “CPDG” motif (CSDG in BLV5B8) that forms a β-turn at the base of each knob (Figure 1B). These motifs are generally conserved in ultralong CDR H3s of bovine antibodies (Figure 1B). Detailed examination, however, reveals that the stalk and knob conformations are otherwise distinct due to different disulfide bond patterns and low amino-acid sequence identity. The 39-residue knob domain of BLV1H12 is composed of two short, anti-parallel β-strands surrounded by three loops and folded such that three disulfide bonds adopt a 1–4, 2–6, 3–5 pattern (Figure 2A, left), which is rarely seen in protein structures. In contrast, the 37-residue knob of BLV5B8 is composed of three loops and two short α-helices and folded such that three disulfide bonds form a 1–3, 2–4, 5–6 pattern (Figure 2A, right). The stalk can be of variable length (Figure 1B); BLV5B8 is two residues shorter than BLV1H12, which reorients the stalk at its distal end and alters the relative position and orientation of the knob domain (Figure 2B). The surface potentials of the two knobs are different, with BLV1H12 generally more positively charged, due to frequent occurrence of arginine (Figure 2C). A search of the Dali protein structure database did not reveal any structurally similar domains to either knob. The ascending β-strand contains mainly hydrophilic side chains, while the descending strand of the stalk is “YTYNY” in BLV1H12 and “HSYEF” in BLV5B8 where the alternating aromatics form a ladder through stacking interactions. Other ultralong sequences (Figure 1B and below) share this motif of alternating aromatics (often YxYxY), suggesting this structural feature is important for integrity of the stalk. This unique amino-acid pattern may contribute to the stability of this long solvent-exposed, two-stranded β-ribbon (Richardson and Richardson, 2002). With the significant CDR H3 amino-acid sequence differences and disulfide patterns, the fold, surface contour, and electrostatic properties of the BLV1H12 and BLV5B8 knob domains are distinct, yet both contain the key structural features of “stalk” and “knob” (Figure 1B and Figure 2).
Figure 2. Structural diversity in ultralong bovine antibodies.
(A)Comparison of the structure of the two knobs showing differences in disulfide patterns. Close up views of the knobs of BLV1H12 (left) and BLV5H8 (right) are shown, in addition to a two-dimensional representation of the knob and its disulfide pattern. Disulfides are in orange. The sequences of the knob regions are shown below, with cysteines in yellow and those conserved with the DH2 germline gene segment underlined. The disulfide pattern is indicated above each sequence. (B) Overlay of the variable regions of BLV1H12 (blue) and BLV5B8 (yellow) shows structural homology in the variable regions except the upper part of the stalk and knob, which are significantly divergent. (C) Surface and charge density representation of BLV1H12 (left) and BLV5B8 (right) showing different shapes and charge in the knob region. The Cα backbone is in green, surface positive charge in blue and negative charge in red.
Genetic basis underlying ultralong CDR H3 structure
The unique disulfide-bonded structures of BLV1H12 and BLV5B8 pose the question as to how such sequences arise in vivo. Antibodies utilize V(D)J recombination and somatic hypermutation (SH) to produce diversity in the antibody repertoire. The VH encodes the majority of the V-region, DH encodes a significant portion of CDR H3, and JH encodes the terminal β-strand. Although CDR H3s can vary in length, they are constrained by the germline-encoded lengths of the DH regions and N or P nucleotide insertions, which usually only account for addition of a few amino acids. Additionally, cysteine residues in CDRs are not common, but when present they are typically conserved between germline and affinity matured sequences (Almagro et al., 2012; Thomson et al., 2008).
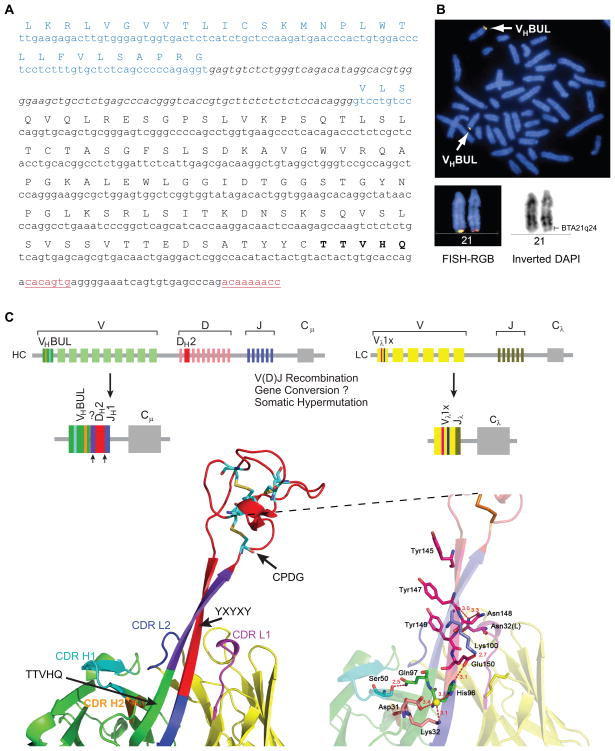
Upon sequencing several bovine V-regions from spleen and lymph node, we found that all sequences with ultralong CDR H3s (>50 amino acids) contained a relatively conserved “T(T/S)VHQ” motif which initiates the ascending strand of BLV1H12 and BLV5B8. This sequence is very unusual, as most human and mouse germline V-regions encode AK or AR in this region which immediately follows the second conserved cysteine in the VH. A search of the bovine genome revealed a single unique germline VH region, which we have termed VHBUL (VH bovine ultralong, Figure 3A) that is present at the immunoglobulin locus on chromosome 21 by FISH analysis, and not at a previously proposed duplicated immunoglobulin locus on chromosome 11 (Hosseini et al., 2004) (Figure 3B). VHBUL contains a functional promoter, leader, intron, and recombination signal sequence and uniquely encodes the terminal “TTVHQ” motif (Figure 3C, left), as well as CDR H1 and H2 motifs that directly interact with the stalk (Figure 3C).
Figure 3. Genetic basis for ultralong antibody formation.
(A) Identification of VHBUL, a germline variable region used in ultralong antibodies. The leader sequence is in blue, coding sequence is indicated with the amino acid translation above, the intron is in italics, and the unique TTVHQ extension, which forms a portion of the ascending strand of the stalk is in bold. The recombination signal sequence heptamer and nonamer are underlined in red. (B) The VHBUL region is found on chromosome 21. Partial cattle metaphase spread (left) and enlarged chromosome 21 (top right) showing the location of VHBUL region in BTA21q24 by two-color FISH with BAC clones 318H2 (green) and 14-74H6 (red). International nomenclature for BTA21 is depicted at the bottom. (C) Schematic of the bovine immunoglobulin loci depicting VHBUL, DH2, and Vλ1x, which are preferentially used in ultralong antibodies. The process of V(D)J recombination assembles the gene segments to form functional ultralong heavy and light chain genes. (bottom left) The V-D-J regions mapped onto the BLV1H12 Fab structure. Colors of the gene segments correlate with the colors of the structure. VHBUL is unique in encoding CDR H1 and CDR H2 residues that interact with the stalk (cyan and orange), as well as a TTVHQ motif that initiates the ascending β-strand. Similarly, the Vλ1x light chain encodes CDR L1 and CDR L2 residues that interact with the stalk (magenta and blue). Arrows indicate areas of potential junctional diversity. Relatively long V-D insertions are indicated in purple. It is unclear whether this sequence results from N-additions, gene conversion, or another mechanism. (bottom right) A detailed depiction of the interactions of CDR H1, H2, L1, and L2 with the stalk of BLV1H12, as well as the location of the YxYxY motif of the descending strand. See also Table S4.
In traditional antibodies, CDRs of the heavy and light chains are normally used for antigen binding. In BLV1H12 and BLV5B8, the CDR H3 stalk is surrounded by the five other CDRs. The base of the stalk interacts with CDRs H1, H2, L1, and L3 (Figure 3C, left). The BLV1H12 “TSVHQ” motif (TTVHQ in the VHBUL germline) at the base of the ascending strand interacts with a “DKAVG” motif in CDR H1 that is also highly conserved in bovine antibodies with ultralong CDR H3s, but divergent from CDR H1 of bovine antibodies with shorter CDR H3s (Figure 1B). The alignment of the crystal structures of BLV1H12 and a typical antibody indicates that this CDR H1 motif is shifted towards the base of the ascending β-strand of the stalk (Figure 3C, bottom). In BLV1H12, Asp31, Lys 32 (CDR H1) and His96 (CDR H3, in TSVHQ in the ascending β-strand) form a hydrogen-bonding network via a water molecule (W286). Ala33 forms a pair of typical β-strand-like hydrogen bonds with His96. The conserved Gln97 (in TSVHQ) forms a close hydrogen-bond interaction (2.5 Å) with Ser50 in CDR H2. The descending β-strand also forms extensive interactions, but with CDRs L1 and L3, which are derived from a lambda light chain, Vλx1. CDR L3 is rotated ~ 90° to accommodate the descending β-strand compared to the search model. Asn32 (CDR L1) hydrogen bonds with the side chain and backbone oxygen of Asn148 and Tyr147, respectively, in the CDR H3 descending strand (Figure 3C, bottom right). These features are not found in the VH regions of conventional antibodies, but are highly conserved between BLV1H12, BLV5B8, and other ultralong sequences (see below), and are encoded in the bovine germline. We speculate that the VHBUL, and the invariant light chain Vλ1x that pairs with ultralong heavy chains, evolved specifically to provide a structural framework to support the stalk and knob, whereas CDR H1 and H2 are not used to bind antigen, but provide structural support for the ultralong CDR H3 stalk. Thus, the germline basis for encoding the base of the stalk structure appears to reside in the VHBUL component of the ultralong CDR H3, with support from CDRs H1 and H2, as well as the CDRs of an invariant lambda light chain Vλx1.
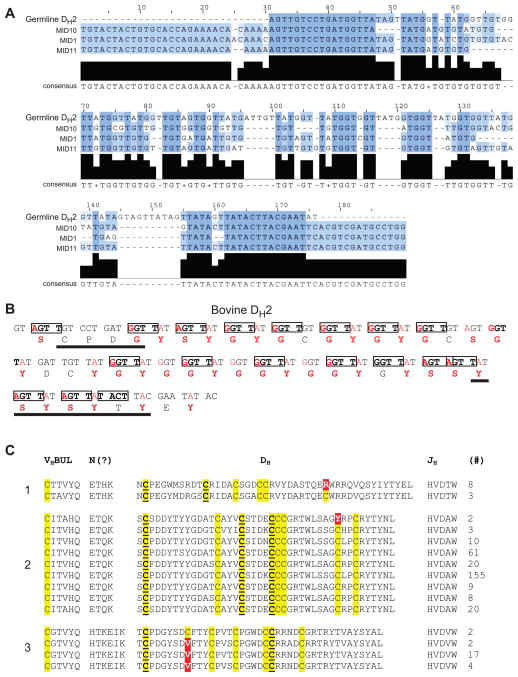
The remaining portion of CDR H3 is comprised of the knob, part of the ascending strand, and the descending strand of the stalk. CDR H3s are typically encoded by the DH region. Cattle have 10 DH regions identified to date (The Bovine Genome Sequencing Analysis Consortium, 2009; Koti et al., 2008; Koti et al., 2010), but only DH2 is long enough to be the genetic basis behind ultralong CDR H3s. Although a draft of the Bos taurus genome is available (The Bovine Genome Sequencing Analysis Consortium, 2009), the assembly of the immunoglobulin heavy chain locus is incomplete, leaving open the possibility of undiscovered ultralong D regions. An initial alignment between DH2, the available literature sequences, and our initial sequences, indicated some limited conservation of the cysteines, but little overall sequence homology within CDR H3s (Figure S1). Nevertheless, the first cysteine in DH2, which is part of the CPDG motif (Figure S1), is highly conserved in ultralong CDR H3s. Additionally, the YxYxY motif forming the descending strand is also encoded by the 3′ portion of DH2 (Figure 3C). Thus, it appears that DH2, (or other similar unidentified DH regions) encodes the “knob” domain and the descending strand of the stalk (Figure 3C, red).
Bovine ultralong CDR H3s are enormously diverse
Despite similar overall “stalk and knob” architectures, BLV1H12 and BLV5B8 have different patterns of disulfide-bonded cysteines that arise from different cysteine sequence positions. The available ultralong CDR H3 sequences are highly diverse, but with limited conservation to the germline DH2, suggesting that they are either derived from different germline DH regions (with cysteines encoded at different positions), or arose through SH or gene conversion from a single DH. In humans, SH is temporally regulated and acts after the naïve B-cell encounters antigen, adding mutations that, through selection, increase the affinity of the antibody. In contrast, ruminants have very limited VH germline diversity, and SH appears to act in the primary repertoire as a mechanism to generate further diversity prior to antigen exposure (Lopez et al., 1998; Zhao et al., 2006). If the cysteines in ultralong CDR H3s are encoded in the germline genome, then the number of different knob minifolds would be limited by the number of ultralong DH regions in the genome. However, if cysteines arise de novo from one or a few D regions through SH or gene conversion, then the knob structural features could form dynamically during B-cell development. These two mechanisms could potentially be distinguished by determining the sequence and cysteine diversity of the bovine ultralong CDR H3 repertoire.
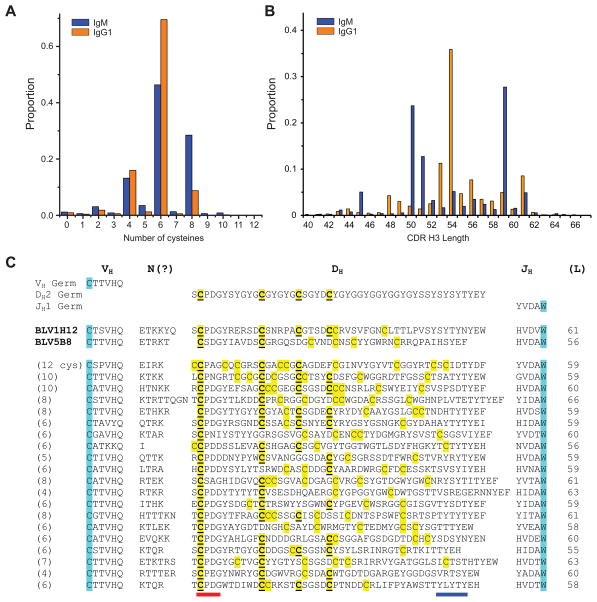
To determine the diversity and content of ultralong bovine CDR H3s, we performed deep sequencing of bovine IgM and IgG variable region genes from two different cows, and analyzed over 10,000 ultralong CDR H3s (Figure 4, Supplemental Information, Table S2 and S3). Sequence analysis showed that an even number of cysteines was strongly preferred, suggesting disulfides were formed in the knob region for nearly all ultralong CDR H3s (Figure 4A). Most sequences had 4, 6, or 8 cysteines, but 33 sequences had 10 and 2 sequences had 12 cysteines (Figure S1). The ultralong CDR H3s ranged in length from 40 to 67 residues (Figure 4B and Figure S1), with the latter being the longest CDR H3 described to date (Figure 4C and Figure S1). Interestingly, the CDR H3 length distribution is distinct between IgM and IgG (Figure 4B). These lengths could be biased due to differential selection of clonally related sequences during an immune response, or alternatively to other selection pressures such as stability or expression (Wang et al., 2013), which may be impacted by CDR H3 length. Several groups of clearly clonally related sequences were found that likely arose during ongoing SH. Amongst non-clonally related sequences, BLAST alignment did not reveal significant sequence conservation throughout the CDRs, or positional conservation of the cysteines. However, when we fixed the first cysteine in each CDR H3 by aligning it with the germline DH2, as in Figure S1, a pattern of conservation for several cysteines emerged that aligned with DH2 (Figure 4C). However, many additional cysteines were not in positions encoded by germline DH2, and did not appear conserved amongst the sequences (Figures 4C and Figure S1). In one sequencing run, 655 out of 5,633 sequences had cysteines in different positions (Table S3), suggesting a significant potential for structural diversity based only on differing disulfide patterns. The sequences that did have a common cysteine pattern were often clearly clonally related, presumably the result of SH and selection in an immune response.
Figure 4. Deep sequence diversity of bovine ultralong VH CDR H3s.
(A) Distribution of the number of cysteines in bovine ultralong CDR H3s of IgM (blue) and IgG (orange). (B) Length distribution of ultralong CDR H3s. Note that clonal sequences selected during an immune response can bias the proportion at any given length. (C)Representative sequences of ultralong bovine VH CDR H3s. The terminal portion of the VHBUL region is shown, along with junctional diversity at the V-D joint, DH2 and JH (top). The sequences of BLVH12 and BLV5B8 are shown for comparison, followed by 20 ultralong CDR H3 sequences (bottom). Cysteines are in yellow, with those conserved with DH2 underlined. The conserved cysteine and tryptophan that define the CDR H3 boundaries in all antibody variable regions are highlighted in cyan for reference. Note that the diversity of many of the cysteines are not conserved between the individual sequences or with DH2. The CPDG motif is underlined in red and the region of the descending strand encoding a possible YxYxY motif is underlined in blue. See also Figure S1, Table S2, S3, and S5 for more sequence information.
Cysteine mutations form diversity in CDR H3
We reasoned that deep sequence analysis would reveal “clusters” of similar sequences if more than one DH region was used to encode the ultralong CDR H3s. However, the sequences of the DH formed only one cluster, without evidence for more than one significantly dissimilar D region (Figure S2), and the consensus sequences of the CDR H3s were highly homologous to DH2, except for a portion at the very N-terminus (Figure 5A). The overall consensus did not encode cysteines in positions divergent from DH2. This result suggested that DH2, or highly related homologues, are the germline precursors of the ultralong repertoire. Indeed, the nucleotide identity of the ultralong sequences to the germline DH2 ranged from 35 to 75% (Supplemental Information and Figure S2). The DH2 region encodes 48 amino acids with four cysteines and a repeating GYG motif (Figure 4C) that leads to a notable sequence bias with 17 tyrosines (35.4%), 14 glycines (31.3%), and 7 serines (14.6%). The limited homology amongst the deep sequences but with conservation of some cysteines (Figure 4C), along with the clustering of nearly 10,000 sequences to a consensus that was highly similar to the germline DH2, suggested that extensive mutation from DH2 could generate the remarkable diversity seen in the bovine repertoire. In this regard, the diversity of cysteines found in bovine ultralong CDR H3s is inconsistent with the known number of DH regions in cattle or any mammalian species, further suggesting that they were somatically generated. Furthermore, the codon usages of the DH2 germline residues are severely biased such that a single nucleotide mutation can produce a cysteine codon (Figure 5B). Indeed, an astonishing 39 of the 48 DH2 residues (81%) can be mutated to cysteine with only one nucleotide change. The DNA sequence of DH2 has numerous RGYW hotspots, which are known to be recognition motifs for the activation-induced (cytidine) deaminase (AID) that produces somatic mutations (Figure 5B). Thus, the DNA sequence of the germline DH2 is primed for mutation to cysteine through SH.
Figure 5. Cysteine mutations contribute to ultralong CDR H3 diversity.
(A) The consensus of ultralong CDR H3 deep sequences aligns with DH2. A consensus sequence for three deep sequencing runs (from two cows) were determined, and aligned with one another and with DH2. The consensus aligns well except for some areas of insertions/deletions. Thus, either a single DH gene, or highly related genes, produce the diversity of sequences in ultralong CDR H3 antibodies. (B) DH2 region analysis showing residues that can readily mutate to cysteine, including SH hotspots. The nucleotide sequence is above and translated amino-acid sequence below. RGYW hotspots, which are recognized by AID for SH and/or gene conversion, are boxed. Red nucleotides indicate positions that can be altered in a single mutation to a cysteine-encoding codon. Red amino acids are the corresponding residues that can be mutated to cysteine in a single step. (C) Affinity maturation groups show mutation to and from cysteine. Several groups of clonally related sequences were identified and analyzed for somatic hypermutation. Three groups are shown as examples (labeled 1 to 3 on the left). Sequence differences from cysteine are highlighted in red. The number of times each sequence is represented in the cluster is shown at the right. See also Figures S1, S2, and S3.
To determine whether the cysteine diversity could be somatically generated, we analyzed clonally related sequences at various stages of somatic hypermutation (Figure 5C and Figure S3). Indeed, we found Arg/Cys, Tyr/Cys, and Cys/Val mutations, directly demonstrating that cysteine patterns can be produced somatically. Since BLV1H12 and BLV5B8 have different disulfide patterns and since an even number of cysteines is strongly favored in our sequences (Figure 4A), the vast diversity of cysteine positions (Table S5) suggests that diverse combinations of disulfide bonds can be formed de novo using residues in DH2, which are primed to mutate to cysteine through SH. Such mutations could occur through base pair changes (Figure 5C) or gene conversion events thought to occur in cattle (Parng et al., 1996), both of which are AID mediated. Irrespective of the mechanism, nucleotide changes resulting in addition or removal of cysteine codons can occur somatically and alter the pattern of cysteines in ultralong CDR H3s.
Antigen binding of ultralong CDR H3 antibodies
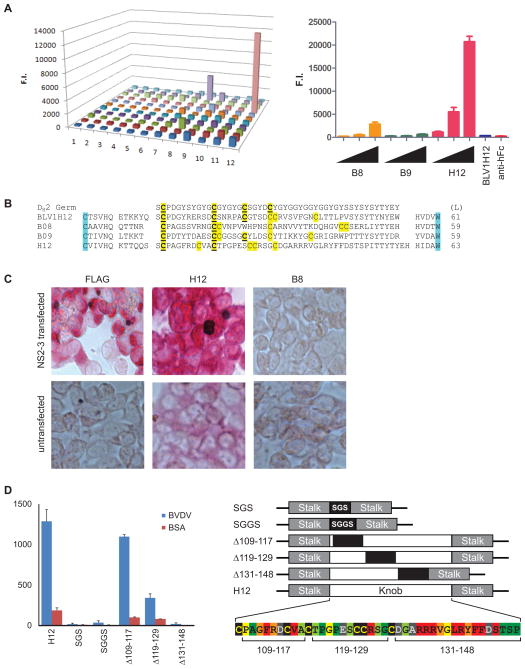
The enormous diversity found in the ultralong repertoire suggested that these ultralong CDR H3 antibodies are a component of the adaptive immune response. To confirm that bovine antibodies utilize their ultralong CDR H3s to bind antigen, we immunized cattle with heat killed bovine viral diarrhea virus (BVDV), a major bovine pathogen of worldwide agricultural economic importance (Figure 6). We collected lymphocyte mRNA, amplified the variable regions, and paired the heavy chain genes with the invariant lambda light chain to produce 132 recombinant bovine-human chimeric IgG (bovine VH with human Fc) in microtiter wells (Mao et al., 2010). These IgGs were screened by ELISA for binding to BVDV and several candidate binders were identified (Figure 6A). The H12 clone has a 63-residue CDR H3 with 6 cysteines (Figure 6B), and could strongly bind virus in a dose-dependent fashion (Figure 6A, right). We then overexpressed BVDV coat Npro, structural (E2), and non-structural (NS2-3) proteins on the surface of HEK293A cells and tested binding of B8 and H12 by immunocytometric analysis. H12 strongly binds HEK293A cells transfected with the NS2-3 non-structural proteins of BVDV, which are required for production of infectious viral particles (Agapov et al., 2004)(Figure 6C), but binds extremely weakly to untransfected cells. As multiple clones derived from BVDV vaccinated cattle had ultralong CDR H3s with the same VHBUL framework (and an identical light chain), the stalk and knob features in the ultralong CDR H3 antibodies appeared to mediate antigen binding.
Figure 6. Bovine antibodies with ultralong CDR H3s bind antigen.
(A) ELISA of 132 ultralong CDR H3 antibodies against BVDV (left), and binding activity of the “hits” B8, B9, and H12 in a titration assay (right). (B) The sequences of B8, H9, and H12 are shown in comparison to BLV1H12 and the germline DH2 region. Lengths (L) of the CDR H3 are indicated at the right. Cysteines conserved with DH2 are underlined. (C) H12 binds NS2-3 on cells. A flag-tagged BVDV NS2-3 protein construct was transfected into HEK293A cells and stained with anti-Flag as a positive control (left), the H12 antibody (middle), and B8 (right). Binding assays with untransfected cells are shown on the bottom. (D) H12 binding to BVDV requires the knob domain. Binding to BVDV (blue) or BSA (red) was assessed by ELISA for knob mutants of H12. Constructs included a total replacement of the knob sequence with a short linker (SGS, or SGGS), partial knob replacements from residues 109–117, 119–129, or 131–148 with an irrelevant sequence (ETYYGSGL). Alanine scan mutants of H12 knob residues were tested for BVDV binding (Figure S4), and the results are summarized in the colored alignment (lower right). Knob point mutant binding to BVDV was compared to that of unmodified H12 (<20%-Red, 20–40%-Orange, 40–60%-Yellow, 60–80% light green, >80%-Green). Some point mutants had greater than 3-fold higher binding to BSA alone, indicating higher non-specific interactions (Grey, Figure S4). All H12 IgGs were normalized to 30 nM (except as indicated in Figure S4 due to poor expression). Data are represented as the mean +/− SEM. See also Figure S4 and Table S7.
To further understand the role of the stalk and knob in the binding mechanism of H12, we deleted the knob domain and replaced it with short SGS or SGGS linkers (Figure 6D). Removal of the knob domain completely abolished binding to BVDV (Figure 6D, left), suggesting that the majority of the antigen binding activity resided in the knob. Next, we replaced approximately each third of the knob domain with the irrelevant sequence ETYYGSGL, and analyzed binding of the resulting mutant antibodies. Replacement of residues 109–117 had a minor impact on binding, whereas replacement of residues 119–129 reduced binding by over 60%, and replacement of the distal residues 131–148 resulted in a complete loss in BVDV binding (Figure 6D, left). While these wholesale swaps of amino acid sequences could result in significant disruptions in folding of the knob, the results suggested that the N-terminal third of the knob is less important to BVDV binding than the C-terminal third. To further define the binding paratope, we generated alanine scan mutants of every residue in the knob domain except the three naturally occurring alanine residues (A110, A117, and A133) which were instead mutated to tyrosine. ELISA analysis of the mutants revealed a substantial decrease in BVDV binding for several residues between 134–145 (Figure 6D and S4), which is consistent with the complete loss of binding activity in the Δ131–148 replacement mutant. Indeed, within this stretch only the relatively conservative G138A mutation retained binding activity. Other point mutations that inhibited binding by over 80% included F112A, V116A, and R127A. Mutations like G111A and R113A in the N-terminal portion of the knob, or V137A and Y141A near the C-terminal portion decreased binding by over 60%. Several mutations had intermediary affects on binding and others had no affect on binding (Figure 6D, right). The heat map in Figure 6D clearly shows a significant impact of mutation of residues between 134–145, with other residues outside this region also playing a role in the binding or structural integrity of the knob domain, which may secondarily affect binding. Thus, in the case of H12, the C-terminal portion of the knob domain appears to mediate significant interaction with the BVDV antigen.
While multiple ultralong CDR H3 sequences have been reported in the literature, the H12 antibody is the first antibody with an ultralong CDR H3 that binds a defined antigen, and we show here that this binding is clearly mediated through the knob domain, with little binding contributed by the stalk or the other five CDRs. Thus the bovine immune system creates a unique repertoire of mega CDR H3s through cysteine diversification, which fold into novel stalk and knob structures that display a unique function in antigen recognition.
Discussion
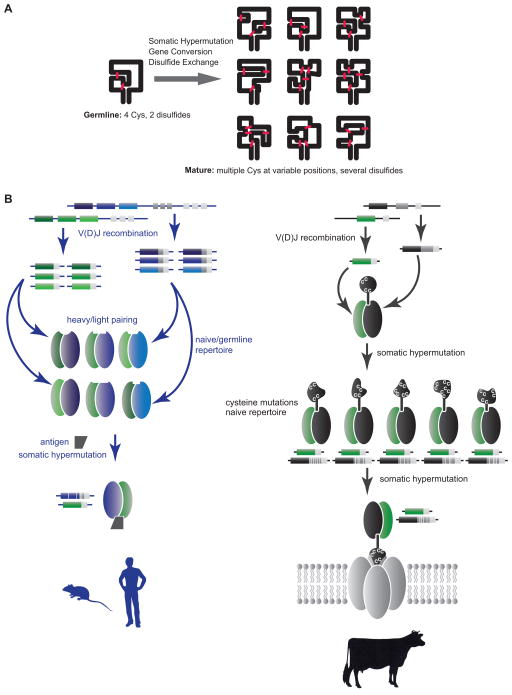
A key component to the clonal selection theory of immune recognition is the generation of a diverse repertoire of antigen receptors. To create this diversity, some species have evolved multiple V, D, and J gene segments, which maximize combinatorial diversity. Other species like chicken and rabbit use a single V(D)J event followed by gene conversion to diversify the repertoire. Cows appear to be unique amongst higher vertebrates in evolving a new domain for antigen recognition, and an unusual mechanism to create diversity in this architecture. Through a single V(D)J event, cows employ cysteine diversification to “reshape” the knob domain in ultralong CDR H3s, creating diverse structures for antigen binding.
Although BLV1H12 and BLV5B8 both contain a stalk with a distal disulfide-bonded knob domain, the ultralong CDR H3s were highly divergent in (i) sequence content, (ii) disulfide bond pattern, and (iii) stalk length. These diversity characteristics were also generally recapitulated in the bovine antibody repertoire sequences (Figure 4C and Figure S1). The first cysteine in CDR H3 forms a disulfide bond at the base of the stalk in both BLV1H12 and BLV5B8 and also is highly conserved in a “CPDG” motif in the ultralong deep sequence data. Thus, we could align all ultralong CDR H3s at this fixed cysteine. This alignment enabled visualization of residues most likely encoded by the VHBUL, DH, JH, and putative N insertions. Notably, the length between the end of the VHBUL and CPDG is variable due to differences in junctional diversity formed through V-D recombination. This region encodes a portion of the β-strand ascending from the VHBUL. Similarly, this change in length is matched through the D-J recombination event, which encodes the descending β-strand of the stalk (Figure 3C). Of note, the YxYxY motif of the descending β-strand is germline encoded in the DH2 region, whereas a portion of the ascending strand does not appear to be encoded in the VHBUL or DH2, and could be the result of random N-insertions, a proposed “oligonucleotide capture” mechanism (Koti et al., 2010), or gene conversion (Parng et al., 1996). Deep sequencing revealed some limited homology within the ascending strand amongst different antibodies; however, evidence for an alternative D-region or D-D fusions has not been found. Although the combinatorial potential is severely limited, the natural diversity mechanism of V(D)J recombination can alter the length and orientation of the stalk, allowing the knob to protrude from the antibody at variable distances and geometries.
The bovine ultralong CDR H3 repertoire represents a new paradigm for the generation of structural diversity by forming a unique architecture distinct from the immunoglobulin domain. Through x-ray crystallography and deep sequencing analysis, we demonstrate that the bovine antibody system utilizes V(D)J recombination and mutational mechanisms to produce CDR H3s with unique “minifolds” comprised of a stalk and a knob, both of which can accommodate significant structural variation including diverse disulfide-bond patterns and loop structures in the knob, as well as differences in length, orientation, and content of the stalk. The codons in the germline D-region encoding ultralong bovine CDR H3s are severely biased towards mutation to cysteine, which may allow new disulfide bonds to be formed or broken in the knob. As both gene conversion and SH utilize AID to create diversity, we suspect that AID produces the remarkable diversity in bovine ultralong CDR H3s through one or both of these mechanisms. With mutation to and from cysteine, the disulfide pattern of germline antibodies, which encodes 4 cysteines in DH2, is distinct from their mature counterparts. Thus, disulfide exchange may occur over time during development of the repertoire (Figure 7A, Table S6). This mechanism suggests ways for rapid minifold evolution in general; a primordial gene with a preferential mutational potential to cysteine could enable new disulfide patterns, which could then be selected and fixed in sequence space based on stability and function. As the number of protein folds in nature is thought to be limited, the bovine antibody repertoire may represent a rich source for discovery of uniquely folded small domains, and provide an unusual opportunity to study protein fold evolution. As antibodies are now a major drug class with alternative scaffolds such as camelid VHHs becoming more important in biomedicine, the bovine structural diversity paradigm could also find utility in drug or diagnostic discovery through further protein engineering efforts.
Figure 7. Model for ultralong CDR H3 diversification into novel minifolds.
(A) A schematic of the DH2 knob with 4 cysteines is shown on the left, with SH and/or gene conversion leading to a multitude of new cysteine patterns and new loops on the right. (B) Mechanisms for generating antibody diversity. In humans and mice (left), combinatorial diversity through V(D)J recombination and VH-VL pairing creates a multitude of different binding sites, which are further optimized following antigen exposure by somatic hypermutation. In cows (right), combinatorial diversity is severely limited; however, somatic mutation to and from cysteines can reshape the “knob” region, creating substantial structural diversity in ultralong CDR H3s. These antibodies may be further optimized through SH and may bind unique targets such as pores or channels. See also Figure S5 and Table S6.
The enormous number of unrelated sequences that we found during deep sequencing suggests that diversity on its own is a major functional driver of the ultralong CDR H3 repertoire. It is curious that cattle have this unique structural repertoire in addition to a more conventional shorter CDR H3 repertoire. Physiologically, cattle are unusual in having a rumen, which functions as a “fermenter” to metabolize feedstuff. Control of the high titer of natural rumen microorganisms is important to inhibit opportunistic digestive tract or serum infections. The added diversity brought about by this unusual antibody structure could serve this purpose, and perhaps be optimized to bind certain antigens like pores, channels, or other receptors that are more difficult to access with typical antibodies (Figure 7B). The rumen biomass includes a substantial portion of eukaryotic microorganisms, which may present different antigen structures than viruses and bacteria, which are the major challenges for other vertebrate immune systems. While we could identify ultralong antibodies against BVDV from immunized cattle, the pressure behind the evolution of “stalk and knob” features may have been by other unknown antigens not easily targeted by the traditional antibody binding scaffold. Several small disulfide-bonded protein families involved in diverse protein-protein interactions have a general shape and dimension similar to the knob of these bovine antibodies, including protease inhibitors, channel blockers, arthropod toxins, and G-protein coupled receptor (GPCR) ligands (Figure S5) (Craik et al., 2001; Silverman et al., 2005; Smith et al., 2011). However, no sequence or structural homology could be found with any of these domains and the BLV1H12 or BLV5B8 knobs. Clearly, small disulfide-bonded protein structures have evolved over time for a multitude of protein-protein interactions of diverse function. Indeed, the “knottin” family of disulfide-bonded proteins have been engineered for a number of different applications using in vitro display technologies (Gracy and Chiche, 2011; Kolmar, 2009; Moore and Cochran, 2012). The bovine antibody system provides an analogous in vivo process for evolution of these small domains, but may also enable unique disulfide pattern diversity and effector functions mediated by the immunoglobulin constant regions.
The propensity for structural and sequence diversity of the stalk and knob motifs could have more general implications. A long stable β-ribbon connecting two unrelated domains is rare. Exposed β-strands can often initiate protein-protein interactions (Richardson and Richardson, 2002). It is interesting to speculate that the significant diversity of the ascending strand provides a nidus for interaction with some antigens, with the diversity of the knob providing additional high affinity contacts through affinity maturation. Also, each ultralong CDR H3 knob has several disulfide-produced loops that could interact with antigen, as we have shown for the H12 antibody. Alternatively, positive charges in the knob could also allow membrane binding or penetration, with the stalk acting to bind surface or membrane proteins. The biophysical and detailed binding properties of this new class of antigen receptor require further investigation.
A significant paradox in adaptive immune evolution is the fact that some species utilize a large number of V, D, and J segments, whereas others have a very limited combinatorial repertoire (Figure 7B, Table S6). For cattle, this limited combinatorial repertoire is expanded enormously by the ability to create structural diversity within ultralong CDR H3s on a scaffold encoded by only a single VHBUL, DH2, and JH paired with a limited number of Vλ light chains (Figures 3 and 7, Table S6). The limitations in VH and VL usage may be due to the structural constraints imposed by the stalk interaction with other CDRs. In the same way that substantial diversity can be produced combinatorially by V(D)J recombination in other species, the bovine mechanism of generating cysteine-mediated hypervariable mini-folds de novo enables a small amount of germline-encoded genetic material to generate substantial sequence and structural diversity, representing a new mechanism for immune receptor repertoire generation.
Experimental Procedures
Crystallization and structure determination of BLV1H12 and BLV5B8
The bovine Fab fragments were cloned and purified as described in the Supplemental Methods. Gel filtration fractions containing the bovine Fabs were concentrated to ~10 mg/mL in 10 mM Tris, pH 8.0 and 50 mM NaCl. Initial crystallization trials were set up using the automated Rigaku Crystalmation robotic system at the Joint Center for Structural Genomics (www.jcsg.org). Several hits were obtained for BLV1H12 and BLV5B8, and crystals used for data collection were grown by the sitting drop vapor diffusion method with a reservoir solution (100 μL) containing 0.27 M potassium citrate and 22% PEG 3350 (BLV1H12) and 0.2 M di-sodium tartrate and 20% PEG 3350 (BLV5B8). Drops consisting of 100 nL protein + 100 nL precipitant were set up at 20 °C, and crystals appeared within 3–7 days. The resulting crystals were cryoprotected using well solution supplemented with 15% ethylene glycol then flash cooled and stored in liquid nitrogen until data collection.
Diffraction data were collected on the GM/CA-CAT 23ID-D beamline at the Advanced Photon Source at Argonne National Laboratory (BLV1H12) and the 11-1 beamline at the Stanford Synchrotron Radiation Lightsource for BLV5B8. Both datasets were indexed in spacegroup P212121, integrated, scaled, and merged using HKL2000 (BLV5B8; HKL Research) or XPREP (BLV1H12; Bruker). The BLV1H12 structure was solved by molecular replacement to 1.88 Å resolution using Phaser (McCoy et al., 2007). Fab variable domains from 1BVK and constant domains from 2FB4 were used as search models and two complete BLV1H12 Fabs were found in the asymmetric unit. The BLV5B8 dataset was also solved by molecular replacement (to 2.20 Å), using the refined BLV1H12 coordinates as a model. Rigid body refinement, simulated annealing and restrained refinement (including TLS refinement, with one group for each Ig domain and one for each CDR H3) were carried out in Phenix (Adams et al., 2010). Riding hydrogens were used during refinement and are included in the final model. Between rounds of refinement, the model was built and adjusted using Coot (Emsley et al., 2010). Waters were built automatically using the “ordered_solvent” modeling function in Phenix (Adams et al., 2010). Structures were validated using the JCSG QC Server (publicly available at http://smb.slac.stanford.edu/jcsg/QC/), which includes Molprobity (Chen et al., 2010). Refinement statistics can be found in Table S1.
Ultralong cDNA generation
Bovine spleen and lymph nodes were obtained from Animal Technologies (Tyler, TX), or from Texas A&M University. Total RNA was isolated from bovine tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol, followed by on column digestion of DNA using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). RNA quantity and quality were assessed with Nanodrop (Thermal Scientific), Qubit RNA and Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA), following the manufacturer’s protocols. Total RNA was used as a template for cDNA synthesis catalyzed by Superscript II (Invitrogen). The antibody variable region was amplified from cDNA using primers 5′-TTGAGCGACAAGGCTGTAGGCTG-3′ and 5′-CTTTCGGGGCTGTGGTGGAGGC-3′.
Deep sequencing
Bar-coded primers (Table S2) were used to amplify VH from bovine spleen cDNA. The amplicons of VH were purified from 2% agarose gels and deep sequenced according to Roche 454 GS FLX instructions. Bioinformatic analysis is described in detail in the Supplemental Methods. CDR H3s were defined by the third residue following the conserved cysteine in framework 3 to the residue immediately preceding the conserved tryptophan in framework 4. This cysteine and tryptophan are highlighted cyan in the figures. VHBUL was identified by BLAST searching the bovine genome (assembly Btau_4.6.1) with multiple ultralong VH sequences identified by deep sequencing. It is unclear whether VHBUL is similar to an uncharacterized partial germline sequence g1.110.10 which has been associated with some ultralong antibodies (Koti et al., 2008; Saini et al., 1999).
FISH analysis
Five sets of primers (Table S3), specific for the VHBUL region exons and flanking sequence were used to screen super-pools and plate-pools of the bovine genomic TAMBET BAC library (Cai et al., 1995) by PCR. Three positive clones, 14–74H6, 318H2, and 7138-19E8 were identified, picked and grown in 2YT with chloramphenicol. BAC DNA was isolated with the Plasmid Midi Kit (Qiagen) according to the manufacturer’s instructions. Physical location of the BACs was determined by fluorescence in situ hybridization (FISH) to cattle metaphase chromosomes as described (Raudsepp and Chowdhary, 2008). Briefly, DNA from individual BAC clones was labeled with biotin-16-dUTP or digoxigenin-11-dUTP, using Biotin- or DIG-Nick Translation Mix (Roche Applied Science), respectively. Differently labeled probes were hybridized in pairs to metaphase chromosomes. Biotin and digoxigenin were detected with avidin-FITC and anti-digoxigenin-Rhodamine, respectively. Images for a minimum of 10 metaphase spreads were captured for each experiment, and analyzed with a Zeiss Axioplan2 fluorescence microscope equipped with Isis V5.2 (MetaSystems GmbH) software. Cattle chromosomes were counterstained with DAPI and identified according to international nomenclature (Cribiu et al., 2001).
Immunization of cattle with whole killed BVDV
A four month-old Holstein steer was immunized by intradermal inoculation of a mixture of heat killed BVDV-1 and BVDV-2 (100 μg of each). The inactivated virus mixture was suspended in 500 μl PBS and emulsified in 500 μl Freund’s Complete Adjuvant by repeated passage through a double barrel needle. The immunogen was inoculated intradermally (200 μl/injection) at the neck region using a 26 × 1½ G needle. The steer was boosted three times at monthly intervals with the same amount of antigen but formulated in Freund’s Incomplete Adjuvant. Sero-conversion was tested by ELISA using plates coated with the inactivated virus and by immunocytometric analysis of MDBK cells infected with either BVDV-1 or BVDV-2. The steer was bled from the jugular vein and blood was collected in heparin. Lymphocytes were purified through Lymphocyte Separation Media (Mediatech) centrifugation and stored in RNAlater.
Anti-BVDV IgG generation
The VH (generated as cDNA, described above) was assembled with bovine CH1 and human IgG Fc and ligated into pFUSE expression vector to afford a full-length heavy chain library. 500 single E. coli transformants were picked and sequenced. 132 clones containing unique heavy chain sequences were selected. The heavy chain library was then co-transfected with pFUSE expression vector encoding the invariant bovine light chain into HEK293T cells using 293Fectin (Life Technologies) to generate a small spatially addressed library (Mao et al., 2010). Antibodies were secreted into culture media and harvested in 96 well format for further testing. The chimeric antibodies were quantified by sandwich ELISA, screened for binding to BVDV by ELISA, and analyzed for cell binding by immunocytometry as described in the Supplemental Procedures.
BDVH12 knob mutation cloning
BsaI restriction sites were engineered into the knob region and used to insert oligonucleotides encoding mutated amino acid residues, as described in detail in the Supplemental Procedures.
Supplementary Material
Research Highlights.
Bovine antibodies form a unique “stalk” and “knob” structure within ultralong CDR H3s
The bovine ultralong CDR H3 repertoire has an enormous diversity of cysteine residues
Cysteine diversity allows different disulfide pattern “knobs” to form in CDR H3
Cows are unusual in binding antigen with a structure distinct from the Ig domain
Acknowledgments
We thank Richard Lerner, Michael McHeyzer-Williams, Jeffery Kelly, and Michael Weiss for helpful discussions, and Cory Bentley, Evan Holmes, James Graziano, Miguel de los Rios and Jocelyn Bray for technical support. This work was supported by American Cancer Society Grant ACS RSG-09-1601 (to V.V.S.), National Institutes of Health Grants R01GM062159 (to P.G.S.) and R01 AI084817 (to I.A.W.), and the Skaggs Institute for Chemical Biology (to P.G.S. and I.A.W.), and the Scripps Translational Sciences Institute Clinical Translational Science Award UL1 RR025774-03 (to A.T.). This is manuscript number 21869 of The Scripps Research Institute.
Footnotes
Accession Numbers
Crystallographic coordinates and structure factors have been deposited in the Protein Data Bank with the following PDB codes: 4K3D (antibody BLV1H12 Fab) and 4K3E (antibody BLV5B8 Fab).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agapov EV, Murray CL, Frolov I, Qu L, Myers TM, Rice CM. Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus. J Virol. 2004;78:2414–2425. doi: 10.1128/JVI.78.5.2414-2425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- Almagro JC, Raghunathan G, Beil E, Janecki DJ, Chen Q, Dinh T, LaCombe A, Connor J, Ware M, Kim PH, et al. Characterization of a high-affinity human antibody with a disulfide bridge in the third complementarity-determining region of the heavy chain. J Mol Recognit. 2012;25:125–135. doi: 10.1002/jmr.1168. [DOI] [PubMed] [Google Scholar]
- Berens SJ, Wylie DE, Lopez OJ. Use of a single VH family and long CDR3s in the variable region of cattle Ig heavy chains. Int Immunol. 1997;9:189–199. doi: 10.1093/intimm/9.1.189. [DOI] [PubMed] [Google Scholar]
- Cai L, Taylor JF, Wing RA, Gallagher DS, Woo SS, Davis SK. Construction and characterization of a bovine bacterial artificial chromosome library. Genomics. 1995;29:413–425. doi: 10.1006/geno.1995.9986. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis AVJ, Brouwer AP, Martin ACR. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J Mol Biol. 2003;325:337–354. doi: 10.1016/s0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
- Craik DJ, Daly NL, Waine C. The cystine knot motif in toxins and implications for drug design. Toxicon. 2001;39:43–60. doi: 10.1016/s0041-0101(00)00160-4. [DOI] [PubMed] [Google Scholar]
- Cribiu EP, Di Berardino D, Di Meo GP, Eggen A, Gallagher DS, Gustavsson I, Hayes H, Iannuzzi L, Popescu CP, Rubes J, et al. International System for Chromosome Nomenclature of Domestic Bovids (ISCNDB 2000) Cytogenet Cell Genet. 2001;92:283–299. doi: 10.1159/000056917. [DOI] [PubMed] [Google Scholar]
- Decanniere K, Desmyter A, Lauwereys M, Ghahroudi MA, Muyldermans S, Wyns L. A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops. Structure. 1999;7:361–370. doi: 10.1016/s0969-2126(99)80049-5. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Gracy J, Chiche L. Structure and modeling of knottins, a promising molecular scaffold for drug discovery. Curr Pharm Des. 2011;17:4337–4350. doi: 10.2174/138161211798999339. [DOI] [PubMed] [Google Scholar]
- Hosseini A, Campbell G, Prorocic M, Aitken R. Duplicated copies of the bovine JH locus contribute to the Ig repertoire. Int Immunol. 2004;16:843–852. doi: 10.1093/intimm/dxh085. [DOI] [PubMed] [Google Scholar]
- Kato L, Stanlie A, Begum NA, Kobayashi M, Aida M, Honjo T. An evolutionary view of the mechanism for immune and genome diversity. J Immunol. 2012;188:3559–3566. doi: 10.4049/jimmunol.1102397. [DOI] [PubMed] [Google Scholar]
- Kocks C, Rajewsky K. Stepwise intraclonal maturation of antibody affinity through somatic hypermutation. Proc Natl Acad Sci U S A. 1988;85:8206–8210. doi: 10.1073/pnas.85.21.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmar H. Biological diversity and therapeutic potential of natural and engineered cystine knot miniproteins. Curr Opin Pharmacol. 2009;9:608–614. doi: 10.1016/j.coph.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Koti M, Kataeva G, Kaushik A. Organization of DH-gene locus is distinct in cattle. Dev Biol. 2008;132:307–313. doi: 10.1159/000317176. [DOI] [PubMed] [Google Scholar]
- Koti M, Kataeva G, Kaushik AK. Novel atypical nucleotide insertions specifically at VH-DH junction generate exceptionally long CDR3H in cattle antibodies. Mol Immunol. 2010;47:2119–2128. doi: 10.1016/j.molimm.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wilson IA. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat Immunol. 2009;10:573–578. doi: 10.1038/ni.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez O, Perez C, Wylie D. A single VH family and long CDR3s are the targets for hypermutation in bovine immunoglobulin heavy chains. Immunol Rev. 1998;162:55–66. doi: 10.1111/j.1600-065x.1998.tb01429.x. [DOI] [PubMed] [Google Scholar]
- Mao H, Graziano JJ, Chase TM, Bentley CA, Bazirgan OA, Reddy NP, Song BD, Smider VV. Spatially addressed combinatorial protein libraries for recombinant antibody discovery and optimization. Nat Biotech. 2010;28:1195–1202. doi: 10.1038/nbt.1694. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SJ, Cochran JR. Chapter nine - Engineering Knottins as Novel Binding Agents. In: Wittrup KD, Gregory LV, editors. Methods in Enzymology. Academic Press; 2012. pp. 223–251. [DOI] [PubMed] [Google Scholar]
- Njongmeta LM, Bray J, Davies CJ, Davis WC, Howard CJ, Hope JC, Palmer GH, Brown WC, Mwangi W. CD205 antigen targeting combined with dendritic cell recruitment factors and antigen-linked CD40L activation primes and expands significant antigen-specific antibody and CD4(+) T cell responses following DNA vaccination of outbred animals. Vaccine. 2012;30:1624–1635. doi: 10.1016/j.vaccine.2011.12.110. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Amemiya CT, Ehrhardt GR, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- Parng C, Hansal S, Goldsby R, Osborne B. Gene conversion contributes to Ig light chain diversity in cattle. J Immunol. 1996;157:5478–5486. [PubMed] [Google Scholar]
- Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci U S A. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudsepp T, Chowdhary BP. FISH for mapping single copy genes. Methods Mol Biol. 2008;422:31–49. doi: 10.1007/978-1-59745-581-7_3. [DOI] [PubMed] [Google Scholar]
- Richardson JS, Richardson DC. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci U S A. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini SS, Allore B, Jacobs RM, Kaushik A. Exceptionally long CDR3H region with multiple cysteine residues in functional bovine IgM antibodies. Eur J Immunol. 1999;29:2420–2426. doi: 10.1002/(SICI)1521-4141(199908)29:08<2420::AID-IMMU2420>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Saini SS, Farrugia W, Ramsland PA, Kaushik AK. Bovine IgM antibodies with exceptionally long complementarity-determining region 3 of the heavy chain share unique structural properties conferring restricted VH + Vlambda pairings. Int Immunol. 2003;15:845–853. doi: 10.1093/intimm/dxg083. [DOI] [PubMed] [Google Scholar]
- Saphire EO, Parren PWHI, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- Silverman J, Lu Q, Bakker A, To W, Duguay A, Alba BM, Smith R, Rivas A, Li P, Le H, et al. Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nat Biotech. 2005;23:1556–1561. doi: 10.1038/nbt1166. [DOI] [PubMed] [Google Scholar]
- Sinclair MC, Gilchrist J, Aitken R. Bovine IgG repertoire is dominated by a single diversified VH gene family. J Immunol. 1997;159:3883–3889. [PubMed] [Google Scholar]
- Smider V, Chu G. The end-joining reaction in V(D)J recombination. Semin Immunol. 1997;9:189–197. doi: 10.1006/smim.1997.0070. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Hill JM, Little MJ, Nicholson GM, King GF, Alewood PF. Unique scorpion toxin with a putative ancestral fold provides insight into evolution of the inhibitor cystine knot motif. Proc Natl Acad Sci U S A. 2011;108:10478–10483. doi: 10.1073/pnas.1103501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal Structure of a Shark Single-Domain Antibody V Region in Complex with Lysozyme. Science. 2004;305:1770–1773. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- The Bovine Genome Sequencing Analysis Consortium. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson CA, Bryson S, McLean GR, Creagh AL, Pai EF, Schrader JW. Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus. EMBO J. 2008;27:2592–2602. doi: 10.1038/emboj.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Sen S, Zhang Y, Ahmad I, Zhu X, Wilson IA, Smider VV, Magliery TJ, Schultz PG. Somatic hypermutation maintains antibody thermodynamic stability during affinity maturation. Proc Natl Acad Sci U S A. 2013;110:4261–4266. doi: 10.1073/pnas.1301810110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao Y, Jackson SM, Aitken R. The bovine antibody repertoire. Dev Comp Immunol. 2006;30:175–186. doi: 10.1016/j.dci.2005.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.