Abstract
Background:
Circulating microRNAs (miRNAs) have been implicated as novel biomarkers for gastric cancer (GC) diagnosis. However, the mixture of GC subtypes may have led to the inconsistent circulating miRNA profiles, and the clinical performance of circulating miRNAs has not yet been evaluated independently on early detection of GC.
Methods:
A four-phase study was designed with a total of 160 cancer-free controls, 124 patients with gastric non-cardia adenocarcinoma (GNCA) and 36 patients diagnosed gastric cardia adenocarcinoma (GCA). In the discovery phase, we screened the miRNA expression profile in plasma of 40 GNCA patients (stage I) and 40 matched controls by TaqMan low density array (TLDA) chips with pooled samples. Differentially expressed miRNAs were further validated in individual sample using quantitative reverse-transcriptase PCR (qRT–PCR) in the training phase. Subsequently, in an independent validation phase, the identified miRNAs were evaluated in 48 GNCA patients (stage I) and 102 matched controls. Finally, the identified miRNAs were further assessed in an external validation phase including advanced GNCA and GCA patients. Additionally, the expression levels of identified miRNAs were measured in the media of BGC823 and MGC803 cell lines.
Results:
Five miRNAs (miR-16, miR-25, miR-92a, miR-451 and miR-486-5p) showed consistently elevated levels in plasma of the GC patients as compared with controls, and were identified to be potential markers for GNCA with area under the receiver operating characteristic (ROC) curves (AUCs) ranging from 0.850 to 0.925 and 0.694 to 0.790 in the training and validation phases, respectively. The five-miRNA panel presented a high diagnostic accuracy for the early-stage GNCA (AUCs=0.989 and 0.812 for the training and validation phases, respectively). Three miRNAs (miR-16, miR-25 and miR-92a) were excreted into the culture media of GC cell lines.
Conclusions:
The five-miRNA panel in plasma may serve as a potential non-invasive biomarker in detecting the early-stage GC.
Keywords: gastric cancer, miRNA, plasma, biomarker
Gastric cancer (GC) is currently the fourth most common malignancy and the second leading cause of cancer death in the world, with an estimate of 989 000 new cases and 738 000 deaths in 2008 (Ferlay et al, 2010). However, nearly half of global GC cases (464 000) and deaths (352 000) occur in China (Ferlay et al, 2010). Early GC is typically small and asymptomatic, and the high mortality from GC is mainly due to late presentation (Hartgrink et al, 2009). Therefore, early detection and treatment is an important way to reduce death from GC. Several tests have been applied in population screening, such as barium meal and endoscopy, especially in Japan, with an ultimate goal of reducing mortality associated with GC (Leung et al, 2008). However, these approaches may be not eligible for mass screening and early detection of GC in most countries including China for the reasons such as acceptance, availability and cost. In addition, non-invasive serological tests, such as serum pepsinogen and gastrin-17, however, have no sufficient sensitivity and specificity to facilitate the early detection (Leung et al, 2008). Therefore, efforts should be invested in the discovery of novel, reliable and non-invasive biomarkers for mass screening and early detection of GC.
MicroRNAs (miRNAs) are a class of small non-coding RNAs that can repress translation or promote degradation of target mRNAs of complementary sequences. A single miRNA can influence the expression of hundreds of target genes, and miRNAs have been implicated as key molecules in various diseases, including cancer (van Kouwenhove et al, 2011). Although the molecular mechanisms of miRNAs are incompletely understood in the process of carcinogenesis, specific patterns of miRNA expression have been associated with almost every type of tumour (Song and Meltzer, 2012), suggesting diagnostic and prognostic potentials for miRNAs. More importantly, circulating miRNAs can be stably detected in the peripheral blood (Chen et al, 2008; Mitchell et al, 2008), though the exact source of circulating miRNAs is a matter of debate. We and other groups have demonstrated that circulating miRNAs had the potential to be new biomarkers in patients with solid tumours for either early detection or evaluation of therapeutic efficiency (Hu et al, 2010, 2012; Chen et al, 2012).
In recent years, several studies have reported a series of circulating miRNAs differentially expressed in patients with GC, and most of these studies focussed on individual miRNAs selected from previous analyses on tissue samples and few with systematic miRNA profiling (Ichikawa et al, 2012; Song and Ajani, 2013). However, the reported circulating miRNAs are not consistent among studies. Liu et al (2011) performed serum miRNA profiling in 20 GC patients and 20 normal controls using Solexa sequencing and identified a five-miRNA signature (miR-1, miR-20a, miR-27a, miR-34 and miR-423-5p) for GC diagnosis with the area under the receiver operating characteristic (ROC) curve (AUC) of >0.800. Thereafter, another group analysed genome-wide serum miRNAs by miRNA microarray, and reported three miRNAs (miR-187*, miR-371-5p and miR-378) significantly elevated in GC patients with the best AUC of 0.861 for miR-378 (Liu et al, 2012). These two studies reported distinct serum miRNA profiles of GC. The inconsistency may be resulted from inclusion of different gastric tumour subtypes. For example, gastric non-cardia adenocarcinoma (GNCA) and cardia adenocarcinoma (GCA) have distinct clinical and epidemiologic features and each form might be characterised by a distinct expression profile of miRNA (Helicobacter and Cancer Collaborative Group, 2001; Lin et al, 2011; Steevens et al, 2011). In addition, identifying differential circulating miRNAs by contrasting controls with GC patients of all tumour stages as a whole might lead to the risk of overrepresenting the aberrant miRNAs from the early-stage GC patients. Eventually, in spite of the promising discriminative ability of circulating miRNAs in studied samples, none of studies have evaluated the performance of these circulating miRNAs in independent samples. Therefore, large-scale, well-designed studies are needed to verify the clinical utility of circulating miRNAs for early detection of GC.
Gastric carcinogenesis is a multistep process that involves many environmental and genetic factors. It has been well demonstrated that the pathogenesis of GNCA initiates with chronic atrophic gastritis, and progresses through intestinal metaplasia and epithelial dysplasia (Shah and Ajani, 2010). A promising diagnostic biomarker may clearly discriminate the GNCA patients from those with precancerous lesions of GC. Herein, we stringently designed a four-phase study to identify a panel of circulating miRNAs as early diagnostic biomarkers of GNCA and evaluated their clinical usefulness in independent samples. In the discovery phase, we systematically screened the miRNA expression profile in plasma of 40 GNCA patients (stage I) and 40 matched controls by TaqMan low density array (TLDA) chips. Differentially expressed miRNAs were further validated in individual sample in the training phase using qRT–PCR assay. Subsequently, in an independent validation phase, the identified miRNAs were evaluated in additional samples, including 48 GNCA patients (stage I), 54 subjects with precancerous lesions of GC (PLGC) and 48 healthy subjects. Finally, the expression levels of the identified miRNAs were assessed in an external validation phase, including 18 stage I and 18 stage III/IV GNCA patients, and 18 stage I and 18 stage III/IV GCA patients.
Materials and methods
Study design and study population
All of the subjects were recruited at the Yangzhou First People's Hospital, the Northern Jiangsu People's Hospital and the First Affiliated Hospital of Nanjing Medical University from January 2007 to November 2011 in Jiangsu province, China. All of the GC patients were confirmed by histopathology of surgical resected tumours and the tumour stage was determined according to the International Union Against Cancer's (UIAC) tumour-node-metastasis (TNM) system. In this study, we restricted the GNCA patients to those with tumour located at the body or antrum of stomach. Each subject was interviewed face-to-face to obtain demographic data and information on related risk factors using a structured questionnaire, including tobacco smoking and alcohol consuming. All of the participants were genetically unrelated, ethnic Han Chinese, and donated 5 ml of venous blood at the recruitment. Exclusion criteria included self-reported previous cancer history, surgical operation or chemo-radiotherapy before blood collection. This study was approved by the institutional review board of Nanjing Medical University, and the written informed consent was obtained from each participant.
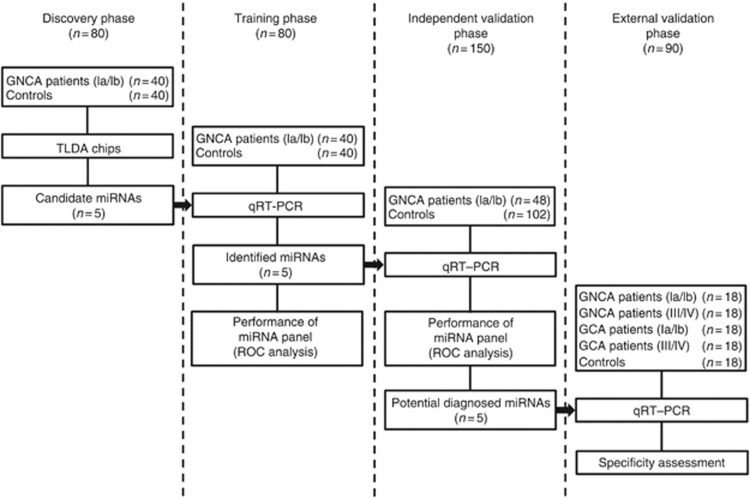
Overall, a total of 320 participants were included in this study, and a four-phase study was designed (Figure 1). In the discovery phase, we performed miRNA profiling in two pooled samples from 40 GNCA patients at stage I and 40 healthy controls, respectively, using TLDA chips (V2.0; Applied Biosystems, Foster city, CA, USA). The controls were frequency matched to the GNCA patients on potential confounding factors including age, gender, smoking and drinking status. In the training phase, the differentially expressed miRNAs discovered via TLDA chips were confirmed by qRT–PCR in the same sample set of the discovery phase individually. These 80 subjects were used to estimate parameters of the identified miRNAs for GNCA diagnosis. In the validation phase, the diagnostic performance of the miRNAs identified by the training phase was independently evaluated in additional 150 subjects (48 GNCA patients at stage I, 54 PLGCs and 48 healthy subjects). The PLGCs included 18 chronic atrophic gastritis, 18 intestinal metaplasia and 18 epithelial dysplasia patients. Both the PLGCs and healthy subjects were also frequency matched to the GNCA patients on the factors of age, gender, smoking and drinking status. In the external validation phase, we enrolled 18 GNCA patients at stage I, 18 GNCA patients at stage III/IV, 18 GCA patients at stage I, 18 GCA patients at stage III/IV and 18 frequency-matched controls to assess the extensionality of the identified miRNAs. In addition, to determine the secretory potential of the identified miRNAs, GC cell lines were cultured and the expression of miRNAs in culture media was measured by qRT–PCR.
Figure 1.
Overview of the study design. Abbreviations: GCA= gastric cardia adenocarcinoma; GNCA= gastric non-cardia adenocarcinoma; TLDA= TaqMan low density array; qRT–PCR= quantitative reverse-transcriptase PCR assay; ROC= receiver operating characteristic curve.
RNA isolation from plasma and culture medium
The plasma was separated from venous blood within 24 h after sample collection by centrifugation at 4000 r.p.m. for 10 min. Two GC cell lines, BGC823 and MGC803, were cultured and a fraction of the culture media was collected at 0, 24 and 48 h after the initial seeding of cells in 10 cm dishes. In the absence of megascopic haemolysis for plasma samples, the plasma and culture media samples were further resolved by a 15-min high-speed centrifugation at 12 000 r.p.m. at 4 °C to completely remove cell debris. The supernatant samples were stored at −80 °C until analysis. A modified method was used to isolate the total RNAs (Kroh et al, 2010). Following the manufacturer's protocol, the Trizol Reagent (Invitrogen, Carlsbad, CA, USA) was used to denaturise plasma and the Qiagen miRNeasy Mini kit (Qiagen, Valencia, CA, USA) was used to collect and purify RNA. After denaturising, synthetic C. elegans miR-39 (cel-miR-39) (Takara, Japan) was spiked into each sample at a final concentration of 10−4 pmol μl−1 for the purpose of controlling variability in RNA extraction and/or purification procedures.
TLDA chip and qRT–PCR assays
Each plasma sample was pooled equally before RNA isolation with a total amount of 960 μl that was used for the TLDA assay (V2.0; Applied Biosystems). After total RNA isolation, megaplex RT reactions and preamplification reactions were performed according to the manufacturer's protocol. Subsequently, 75 μl 0.1 × TE was added to preamplification product, and 9 μl diluted preamplification product was used to perform the RT–PCRs by dispensing 100 μl of the PCR mix into each port of the TLDA chip. The RQ manager software (Applied Biosystems) was applied in the raw data analysis. ΔCT and ΔΔCT were calculated using the following formulae: ΔCT=CT sample−CT cel-miR-39 and ΔΔCT=ΔCT case−ΔCT control. Differentially expressed miRNAs were chosen as candidates for further confirmation by individual qRT–PCR according to the following two items: (i) having a CT value of <30 in at least one pool to ensure the enough abundance of miRNA for detection; (ii) |ΔΔCT|>2, meaning four-fold altered expression between the GNCA patients and healthy controls to ensure favourable discriminative ability (Supplementary Table 1).
For qRT–PCR, an equal volume (100 μl for plasma and 200 μl for cell-culture media) of sample was processed in RNA isolation for each subject. The total RNA was reverse transcribed to complementary DNA by using the TaqMan miRNA RT Kit and stem-loop RT primers (Applied Biosystems). The quantitative detection of miRNA was performed using the TaqMan PCR kit as implemented in the ABI 7900 Real-Time PCR System (Applied Biosystems). The reactions were initiated with a 384-well optical plate at 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Equal number of GNCA patients and controls was assigned in each plate and the expression levels of target miRNAs and control miRNA (cel-miR-39) were measured simultaneously. All reactions, including no template controls, were performed in triplicate. The relative expression levels of target miRNAs were calculated with 2−ΔΔCT.
Statistical Analysis
Demographic and clinical characteristics among groups were compared by χ2 test or one-way ANOVA. The differences of miRNAs among groups were tested using the Mann–Whitney unpaired test. Receiver operating characteristic curves were constructed in the training phase to obtain the optimal cutoff values for candidate miRNAs. We set the optimal cutoff value as the threshold, denoted as t, to code the expression level of the corresponding miRNA for each sample. The risk score for each miRNA, denoted as s, was calculated as follows:
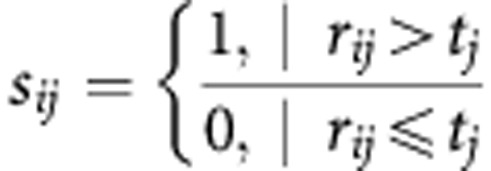 |
Here, we used i to denote the ith sample, j the jth miRNA and r the expression level of miRNA. To additionally investigate the effectiveness of the five-miRNA panel for GNCA diagnosis, a mathematical formula was constructed, taking the contribution of each miRNA into account. Specifically, each subject was assigned a risk score function (RSF) according to a linear combination of the expression level of miRNA, weighted by the regression coefficient developed in the training phase. The RSF for subject i was:
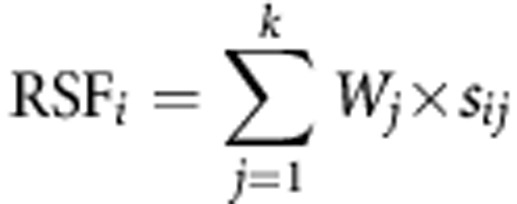 |
Here, sij is the risk score for miRNA j on subject i, and Wj is the weighting for a given miRNA j. The regression coefficient of risk score for each miRNA was estimated by a univariate logistic regression model, and was used as the weighting to indicate the contribution of each miRNA to the RSF. The risk score coefficients developed in the training phase were also applied directly to the validation phase, in which the PLGC subjects and healthy subjects were combined as cancer-free controls to independently evaluate the clinical utility of identified miRNAs individually or in combination. Area under the ROC curve was used as an accuracy index for evaluating the diagnostic performance of the miRNAs. All the statistical analyses were performed with the Statistical Analysis System software (version 9.1.3; SAS Institute, Cary, NC, USA). A P-value of <0.05 was considered as statistically significant.
Results
Characteristics of study subjects
In the discovery and training phase, the healthy controls (n=40) were adequately frequency matched to the GNCA patients (n=40) with similar distributions of age, gender, smoking and drinking status (all P-values >0.05) (Table 1). All GNCA patients were diagnosed with a stage I tumour at body or antrum of stomach. In the independent validation phase, two groups (n=48 and 102 for GNCAs and controls, respectively) were also well matched (all P-values >0.05). The GNCA patients were also restricted to those with stage I tumour at the site of gastric body or antrum. In the external validation phase, five groups (18 subjects each) with different anatomic locations and clinical stages also showed similar distributions on age, gender, smoking and drinking status (all P-values >0.05) (Table 1).
Table 1. Characteristics of the subjects included in this study.
| |
Discovery/Training phase (n=80) |
Independent validation phase (n=150) |
External validation phase (n=90) |
||||
|---|---|---|---|---|---|---|---|
| Variables | Controls (%) | GNCA (%) | Controls (%) | GNCA (%) | Controls (%) | GNCA (%) | GCA (%) |
| Number |
n=40 |
n=40 |
n=102 |
n=48 |
n=18 |
n=36 |
n=36 |
| Age (mean±s.d.) |
53.55±10.11 |
53.83±10.34 |
54.03±10.45 |
56.63±10.37 |
55.11±11.89 |
55.78±8.77 |
55.36±6.94 |
|
Gender | |||||||
| Male | 29 (72.5) | 29 (72.5) | 72 (70.6) | 35 (72.9) | 13 (72.2) | 27 (75.0) | 26 (72.2) |
| Female |
11 (27.5) |
11 (27.5) |
30 (29.4) |
13 (27.1) |
5 (27.8) |
9 (25.0) |
10 (27.8) |
|
Smoking status | |||||||
| Never | 23 (57.5) | 23 (57.5) | 53 (52.0) | 24 (50.0) | 12 (66.7) | 26 (72.2) | 22 (61.1) |
| Ever |
17 (42.5) |
17 (42.5) |
49 (48.0) |
24 (50.0) |
6 (33.3) |
10 (27.8) |
14 (38.9) |
|
Drinking status | |||||||
| Never | 26 (65.0) | 24 (60.0) | 70 (68.6) | 30 (62.5) | 15 (83.3) | 26 (72.2) | 24 (66.7) |
| Ever |
14 (35.0) |
16 (40.0) |
32 (31.4) |
18 (37.5) |
3 (16.7) |
10 (27.8) |
12 (33.3) |
|
Tumour site | |||||||
| Cardia | 0 | 0 | 0 | 36 (100.0) | |||
| Body | 20 (50.0) | 25 (52.1) | 17 (47.2) | 0 | |||
| Antrum |
|
20 (50.0) |
|
23 (47.9) |
|
19 (52.8) |
0 |
|
Tumour stage | |||||||
| Ia | 20 (50.0) | 32 (66.7) | 11 (30.6) | 6 (16.7) | |||
| Ib | 20 (50.0) | 16 (33.3) | 7 (19.4) | 12 (33.3) | |||
| III | 0 | 0 | 9 (25.0) | 12 (33.3) | |||
| IV | 0 | 0 | 9 (25.0) | 6 (16.7) | |||
Abbreviations: GCA=gastric cardia adenocarcinoma; GNCA=gastric non-cardia adenocarcinoma.
MiRNAs screening and individual qRT–PCR confirmation
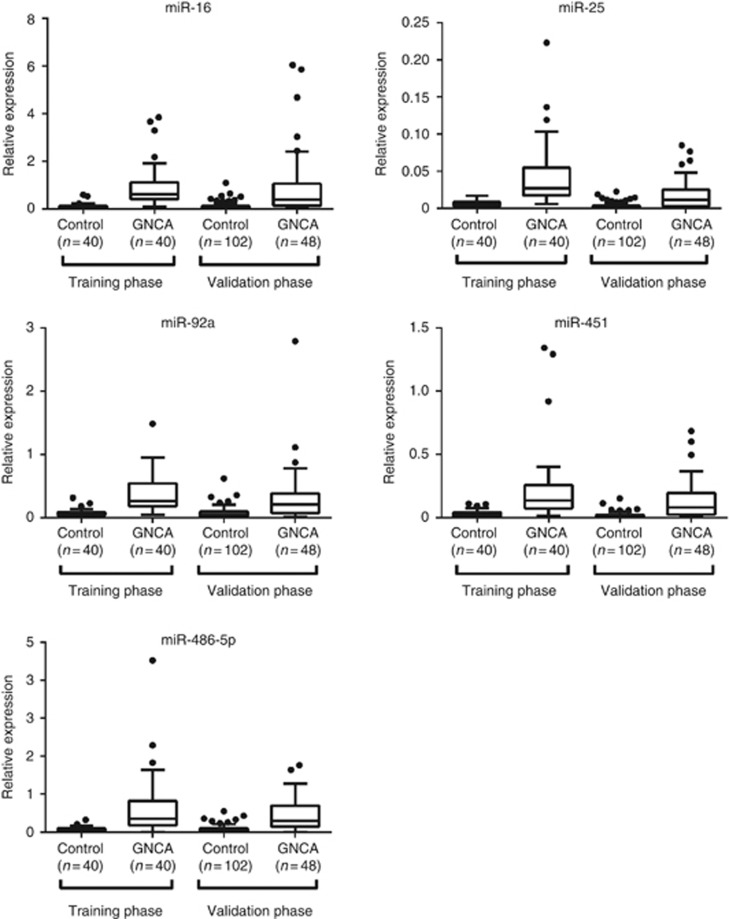
With an effort to identify stably, differentially expressed miRNAs in plasma, we screened a total of 667 human miRNAs by TLDA chips in pooled samples between the GNCA patients and healthy controls (40 subjects for each pool). We further selected candidate miRNAs that had a CT value of <30 in at least one pool and showed more than four-fold altered expression between the two pooled samples. As a result, five upregulated plasma miRNAs, including miR-16, miR-25, miR-92a, miR-451 and miR-486-5p, were subjected to individual qRT–PCR confirmation in the training phase (Supplementary Table 1). Consistent with the results of the discovery phase, all of the five miRNAs had a significantly higher expression level in the GNCA patients as compared with controls (Table 2; Figure 2).
Table 2. Results of five differentially expressed miRNAs between the GNCA patients and controls identified by TLDA chips in the discovery phase and confirmed by individual qRT–PCR assay in the training phase.
| |
Controls (n=40) |
GNCA patients (n=40) | |
|||||
|---|---|---|---|---|---|---|---|---|
| miRNAs | ΔCTa | Mean±s.d. (10−2) | Median (10−2) | ΔCTa | Mean±s.d. (10−2) | Median (10−2) | ΔΔCTb | P-valuec |
| miR-16 | 0.19 | 9.96±11.72 | 6.51 | −2.49 | 93.37±89.26 | 61.39 | −2.68 | 8.71 × 10−13 |
| miR-25 | 6.43 | 0.61±0.39 | 0.58 | 3.51 | 4.17±4.19 | 2.69 | −2.92 | 8.71 × 10−13 |
| miR-92a | 0.13 | 7.10±5.97 | 5.15 | −2.20 | 38.69±30.51 | 26.63 | −2.33 | 3.72 × 10−12 |
| miR-451 | 4.51 | 3.10±2.79 | 2.04 | 1.76 | 22.76±29.78 | 13.57 | −2.75 | 6.42 × 10−11 |
| miR-486-5p | 3.72 | 7.28±6.51 | 5.32 | 0.37 | 66.19±82.92 | 35.63 | −3.35 | 1.26 × 10−9 |
Abbreviations: GNCA=gastric non-cardia adenocarcinoma; microRNA=miRNA; TLDA=TaqMan low density array; qRT–PCR=quantitative reverse-transcriptase PCR.
ΔCT=CT sample−CT cel-miR-39.
ΔΔCT=ΔCT GNCA−ΔCT control.
Mann–Whitney unpaired test for rank sum.
Figure 2.
Expression levels of the five miRNAs in plasma in the training and validation phases. The y axis represents relative expression of miRNAs normalised to cel-miR-39. GNCA, gastric non-cardia adenocarcinoma.
Independent validation of the five identified miRNAs in plasma
Next, we examined the expression levels of the five identified miRNAs in a larger independent sample set comprising of 48 GNCA patients and 102 cancer-free controls in the independent validation phase. As shown in Figure 2 and Supplementary Table 2, the differential expression patterns of the five miRNAs between the GNCA patients and controls were generally concordant between the validation and training phase, suggesting that these five miRNAs (miR-16, miR-25, miR-92a, miR-451and miR-486-5p) in plasma might serve as potential diagnostic markers in the early-stage GNCA.
Performance of the five-miRNA panel for early detection of the GNCA patients
To evaluate the performance of the miRNAs in discriminating the GNCA patients from cancer-free subjects, the discriminatory parameters were first developed in the training phase and were further independently assessed in the validation phase. The optimal cutoff value for individual miRNA was determined according to the ROC curve for each miRNA in the training phase (Supplementary Figure 1). As expected, the performance of the individual miRNA was attractive in the training phase with AUCs being 0.925, 0.925, 0.913, 0.850 and 0.863 for miR-16, miR-25, miR-92a, miR-451 and miR-486-5p, respectively (Table 3). Interestingly, based on the cutoff values developed in the training phase, the AUCs, though reducing, were still very favourable in the validation phase with values varying from 0.694 to 0.790, which may reflect the practical utility of individual miRNA in the diagnosis of the GNCA patients.
Table 3. Discriminative ability of individual miRNAs between the GNCA patients and controls.
| Stage | miRNAa | Cutoff valueb | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|
| Training phase | miR-16 | 0.255 | 0.900 | 0.950 | 0.925 |
| miR-25 | 0.014 | 0.875 | 0.975 | 0.925 | |
| miR-92a | 0.095 | 0.975 | 0.850 | 0.913 | |
| miR-451 | 0.040 | 0.950 | 0.750 | 0.850 | |
| |
miR-486-5p |
0.178 |
0.775 |
0.950 |
0.863 |
| Validation phase | miR-16 | 0.255 | 0.625 | 0.912 | 0.768 |
| miR-25 | 0.014 | 0.417 | 0.971 | 0.694 | |
| miR-92a | 0.095 | 0.729 | 0.735 | 0.732 | |
| miR-451 | 0.040 | 0.688 | 0.892 | 0.790 | |
| |
miR-486-5p |
0.178 |
0.646 |
0.912 |
0.779 |
| Combined phase | miR-16 | 0.255 | 0.750 | 0.923 | 0.836 |
| miR-25 | 0.014 | 0.625 | 0.972 | 0.798 | |
| miR-92a | 0.095 | 0.841 | 0.768 | 0.804 | |
| miR-451 | 0.040 | 0.807 | 0.852 | 0.829 | |
| miR-486-5p | 0.178 | 0.705 | 0.923 | 0.814 |
Abbreviations: AUC=area under the ROC curve; GNCA=gastric non-cardia adenocarcinoma; microRNA=miRNA; ROC=receiver operating characteristic.
The miRNA expression level was coded as 0 or 1 for each subject according to the optimal cutoff value.
The optimal cutoffs were defined by the ROC curves developed in the training phase.
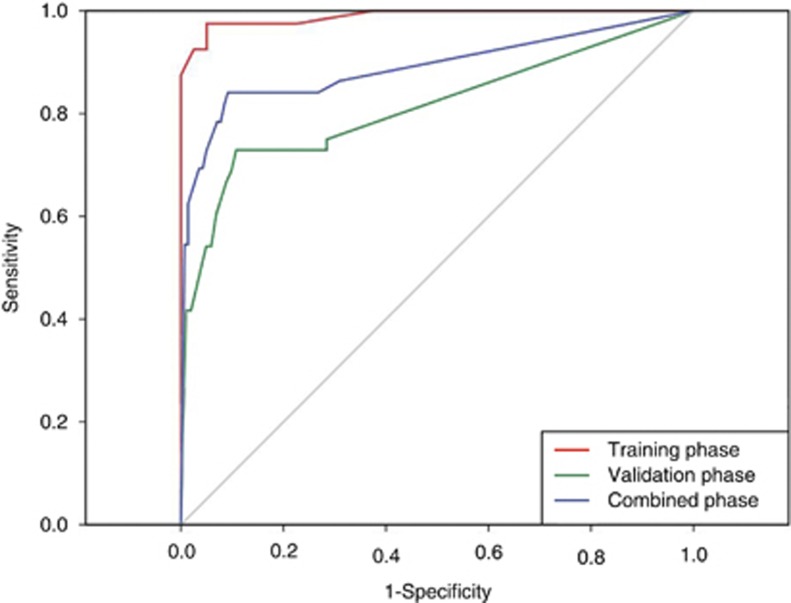
To assess the diagnostic utility of this five-miRNA panel, a linear combination of the risk score of the five miRNAs weighted by the regression coefficient was used to calculate an RSF of the five-miRNA panel for each subject. The RSF was calculated as follows: RSF=(5.142 × risk score of miR-16)+(5.609 × risk score of miR-25)+(5.398 × risk score of miR-92a)+(4.043 × risk score of miR-451)+(4.181 × risk score of miR-486-5p), where the regression coefficients were derived from subjects in the training phase. Similar to but better than the individual miRNA, in the training phase, the AUC of the five-miRNA panel was 0.989 and the diagnostic sensitivity and specificity were 97.5% and 87.5%, respectively (Figure 3). Importantly, in the validation phase, the miRNA panel also showed relatively high ability in discriminating the GNCA patients from the cancer-free controls with an AUC of 0.812 (sensitivity=72.9% and specificity=89.2%) (Figure 3). These results indicate that, although one particular miRNA in plasma may help distinguish different subgroups, a combination of a panel of miRNAs may have a great potential to offer much more sensitive and specific diagnostic test.
Figure 3.
The discriminative ability of the five-miRNA panel between the GNCA patients and cancer-free subjects by receiver operating characteristic curve (ROC) analysis. Red line, the training phase (AUC=0.989, sensitivity=97.5%, specificity=87.5%); green line, the validation phase (AUC=0.812, sensitivity=72.9%, specificity=89.2%); blue line, two phases combined (AUC=0.890, sensitivity=84.1%, specificity=90.8%).
External validation in advanced GNCA and GCA patients
To further assess the extensionality of the identified miRNAs in GC patients at different anatomic sites and clinical stages, we analysed the expression levels of the five identified miRNAs in 18 GNCA patients diagnosed at stage I, 18 GNCA patients at stage III or IV, 18 GCA patients at stage I, 18 GCA patients at stage III or IV and 18 matched controls (Table 1; Figure 1). As compared with the controls, all of the five miRNAs were consistently overexpressed in all GC groups (all P-values <0.05) (Supplementary Figure 2), suggesting that these five miRNAs may be useful in detection of all GC patients.
Expression of the five identified miRNAs in GC cell-culture medium
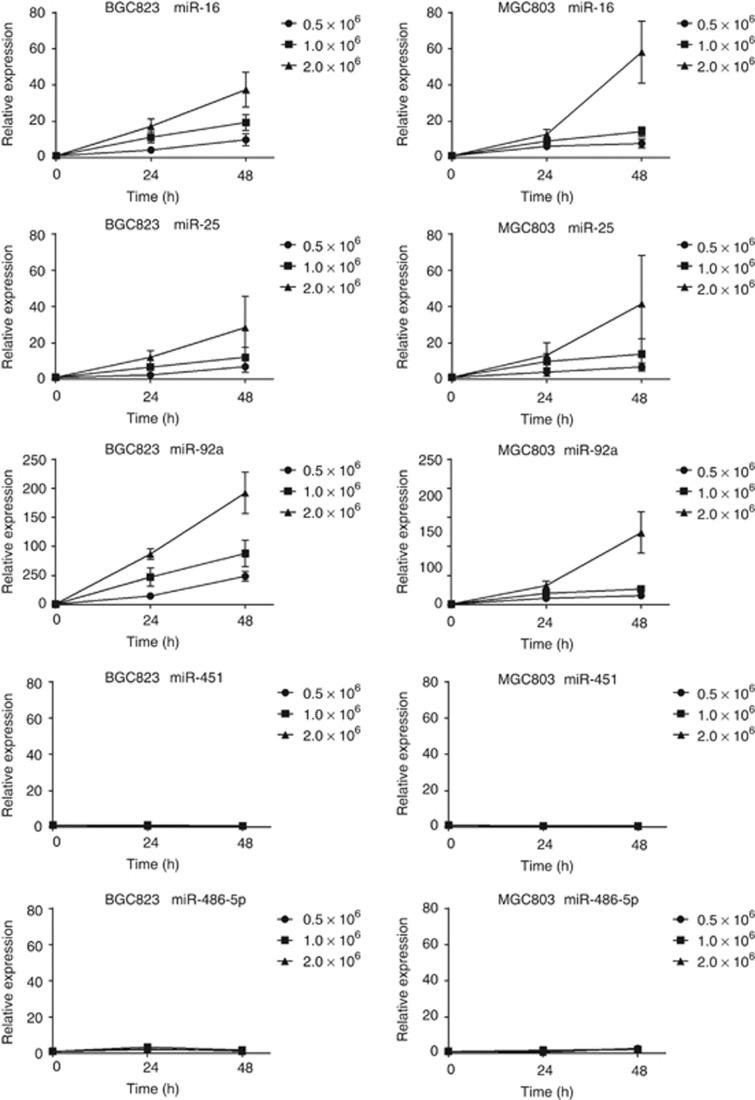
In addition, we determined whether the identified miRNAs (miR-16, miR-25, miR-92a, miR-451 and miR-486-5p) act as secretory miRNAs and are excreted into the culture media by BGC823 and MGC803 cell lines. We found that miR-16, miR-25 and miR-92a levels in media of BGC823 and MGC803 increased with increasing cell counts (0.5–2 × 106 cells per well) and incubation time (24 and 48 h), whereas miR-451 and miR-486-5p levels did not change in either cell line (Figure 4). These data suggest that miR-16, miR-25 and miR-92a but not miR-451 or miR-486-5p may be secretory miRNAs.
Figure 4.
Expression of the five identified miRNAs in culture media of gastric cancer (GC) cell lines (BGC823 and MGC803). MiR-16, miR-25 and miR-92a levels in media of both BGC823 and MGC803 increased with increased cell counts (0.5–2 × 106 cells per well) and longer incubation intervals (24 and 48 h), whereas miR-451 and miR-486-5p levels did not change in either cell line. The y axis represents relative expression of miRNAs normalised to cel-miR-39 and cell-culture media at 0 h.
Discussion
In this study, we systematically screened the miRNA profile in plasma from the GNCA patients and healthy controls, and identified five differentially expressed miRNAs (miR-16, miR-25, miR-92a, miR-451 and miR-486-5p), which were confirmed in independent samples. More importantly, the five-miRNA panel showed high diagnostic performance in discriminating the early-stage GNCA patients from the cancer-free subjects. Our findings highlight that circulating miRNAs may be a class of novel, non-invasive diagnostic biomarkers with sufficient diagnostic accuracy, and a specified miRNA panel can be of great value in mass screening or early detection of GC.
Among the five miRNAs identified in this study, miR-451 and miR-486-5p have been reported to be potentially useful biomarkers for GC (Konishi et al, 2012). In their study, candidate miRNAs were screened by comparing pre- and post-operative paired plasma of GC, and the expression levels of nine miRNAs were markedly decreased in the post-operative plasma. Among these miRNAs, miR-451 and miR-486-5p abundant in plasma were selected and validated in more pre- and post-operative plasma (Konishi et al, 2012). These two miRNAs also showed high AUCs (0.96 and 0.92, respectively) as comparing GC patients (n=56) with healthy controls (n=30) (Konishi et al, 2012). These findings were consistent with our results. Interestingly, the other three miRNAs (miR-16, miR-25 and miR-92a) identified in the current study were also among the nine candidate miRNAs, though not being subject to further study, reported to be markedly decreased in post-operative plasma by Konishi et al (2012). In view of different study designs, populations and platforms between two groups, the consistent findings make us confident that the identified plasma miRNAs in this study may be in great potentials for GC mass screening or early detection, though further perspective clinical trials are warranted to define the usefulness of the five-miRNA panel in clinical application.
Emerging evidence suggests that circulating miRNAs are cancer related, but the releasing mechanism is still an open topic (Valadi et al, 2007; Wang et al, 2009; Arroyo et al, 2011). Some researchers believe they are produced from tumour tissues, which leak or actively transport them into the bloodstream (Wang et al, 2009). This hypothesis appears to be reasonable as interpreting our results in part. The levels of miR-16, miR-25 and miR-92a upregulated in GC plasma were also reported to be increased in GC tissue samples (Supplementary Table 3). In the current study, our results suggested that these three miRNAs were excreted into the culture media by BGC823 and MGC803 cell lines and might act as secretory miRNAs. However, there is contradiction of miR-451 and miR-486-5p expression in plasma and GC tissues (Supplementary Table 3). The downregulation of miR-451 was significantly associated with gastric adenocarcinomas as compared with normal gastric mucosa (Bandres et al, 2009; Ueda et al, 2010; Konishi et al, 2012). Similarly, Oh et al (2011) identified miR-486 as a significantly downregulated miRNA in primary GC tumours and GC cell lines, and similar trend was also reported by Konishi et al (2012) and Su et al (2012). Of interest, in the current study, the expression levels of miR-451 and miR-486-5p did not change in culture medium of GC cell lines as cell counts and incubation time increased. The selective release of specific cellular miRNAs from the tumour cells themselves or from normal cells might explain these apparently paradoxical findings (Pigati et al, 2010). Nevertheless, further experimental studies are required to elucidate the releasing mechanism in specific to these five miRNAs, although this may have no substantial influence on the clinical application for GNCA diagnosis as biomarkers.
Moreover, understanding the miRNA targets and the molecular mechanisms by which the miRNAs regulate GC development may promote their clinical application. Zhang et al (2010) reported that miR-16 had a role in the regulation of Wip1 phosphatase in the DNA damage response. Kim et al (2009) showed that miR-25 targets p57 and ectopic expression of miR-25 in GC may result in activation of Cdk2 and facilitation of G1/S phase transition. MiR-92a, a member of miR-17-92 cluster, seems to be tightly linked to the functions of the E2F family of transcription factors, which are critical regulators of the cell cycle and the apoptosis (Petrocca et al, 2008). Microarray and bioinformatic analysis identified that the oncogene macrophage migration inhibitory factor (MIF) may be the potential target of miR-451, indicating that miR-451 may act as a regulator of cancer proliferation (Bandres et al, 2009). As for miR-486-5p, it may direct targeting and inhibition of OLFM4 expression, and function as a tumour-suppressor miRNA in GC (Oh et al, 2011). Nevertheless, additional investigation of the regulatory mechanism of these miRNAs may improve our understanding of the molecular pathogenesis of GC as well as the effectiveness in identifying potential therapy targets and surveillance markers for GC.
This study may have several strengths. First, in the initial screening phase, we restricted GC patients to those with GNCA at stage I, which may exclude the potential confounding due to distinct profiles between subtypes and tumour stages. Our study clearly demonstrated that the five identified miRNAs were consistently elevated in the early-stage GNCA patients. We further explored the extensionality of these miRNAs in GCA and advanced GNCA patients and the results indicated that these five miRNAs may be applicable for the early diagnose of all GC patients, though further large-scale, well-designed studies are warranted to validate these findings. Second, the five-miRNA panel was systematically screened through miRNA profiling on the GNCA patients and healthy controls using TLDA chips, which was validated in both additional platform and independent samples. These may increase the reliability of the findings. Third, in the current study, we selected candidate miRNAs that were abundantly expressed in plasma to ensure the feasibility of examination in clinical practice. Importantly, the clinical utility of identified miRNAs was assessed in independent samples including the early-stage GNCA patients, PLGC subjects and healthy controls. These efforts may offer us an miRNA panel with great potential in early diagnosis of GC.
In summary, we conducted a comprehensive assessment of the diagnostic usefulness of an miRNA panel in plasma for GNCA in a tailored investigation with independent validation. The sensitivity and the specificity were high and the discriminative accuracy was satisfactory for the diagnosis of the early-stage GC. Our work will serve as a basis for further large-scale validation before circulating miRNAs enable their introduction into the clinic setting.
Acknowledgments
This study was funded by the National Basic Research Program (973) (2013CB910304); National Natural Science Foundation of China (81001276, 81373090); Science Foundation for Distinguished Young Scholars in Jiangsu (BK20130042); Jiangsu Natural Science Foundation (BK2011028, BK2012443, BK2012841); Jiangsu Province Clinical Science and Technology Projects (Clinical Research Center, BL2012008); the Fok Ying-Tong Education Foundation for Young Teachers in the Higher Education Institutions (122031) and Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108 (12:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ, Jimenez P, Rodriguez J, Garcia-Foncillas J. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15 (7:2281–2290. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18 (10:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Chen X, Hu ZB, Wang WJ, Ba Y, Ma LJ, Zhang CN, Wang C, Ren ZJ, Zhao Y, Wu SJ, Zhuang R, Zhang YX, Hu H, Liu CZ, Xu L, Wang J, Shen HB, Zhang JF, Zen K, Zhang CY. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130 (7:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127 (12:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374 (9688:477–490. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helicobacter and Cancer Collaborative Group Gastric cancer and Helicobacter pylori: a combined analysis of 12 case control studies nested within prospective cohorts. Gut. 2001;49 (3:347–353. doi: 10.1136/gut.49.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28 (10:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao Y, Tang J, Chen X, Dai J, Wei Q, Zhang C, Shen H. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis. 2012;33 (4:828–834. doi: 10.1093/carcin/bgs030. [DOI] [PubMed] [Google Scholar]
- Ichikawa D, Komatsu S, Konishi H, Otsuji E. Circulating microRNA in digestive tract cancers. Gastroenterology. 2012;142 (5:1074–1078. doi: 10.1053/j.gastro.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, Kim VN. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37 (5:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi H, Ichikawa D, Komatsu S, Shiozaki A, Tsujiura M, Takeshita H, Morimura R, Nagata H, Arita T, Kawaguchi T, Hirashima S, Fujiwara H, Okamoto K, Otsuji E. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br J Cancer. 2012;106 (4:740–747. doi: 10.1038/bjc.2011.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods (San Diego, Calif.) 2010;50 (4:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, Wu KC, Wu DC, Sollano J, Kachintorn U, Gotoda T, Lin JT, You WC, Ng EK, Sung JJ. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol. 2008;9 (3:279–287. doi: 10.1016/S1470-2045(08)70072-X. [DOI] [PubMed] [Google Scholar]
- Lin SW, Freedman ND, Hollenbeck AR, Schatzkin A, Abnet CC. Prospective study of self-reported diabetes and risk of upper gastrointestinal cancers. Cancer Epidemiol Biomarkers Prev. 2011;20 (5:954–961. doi: 10.1158/1055-9965.EPI-10-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang W, Ma Y, Xiao H. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012;316 (2:196–203. doi: 10.1016/j.canlet.2011.10.034. [DOI] [PubMed] [Google Scholar]
- Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R, Deng T, Liu H, Yin J, Wang S, Zen K, Ba Y, Zhang CY. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer (Oxford, England: 1990) 2011;47 (5:784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105 (30:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HK, Tan AL, Das K, Ooi CH, Deng NT, Tan IB, Beillard E, Lee J, Ramnarayanan K, Rha SY, Palanisamy N, Voorhoeve PM, Tan P. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res. 2011;17 (9:2657–2667. doi: 10.1158/1078-0432.CCR-10-3152. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13 (3:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, Hastings ML, Duelli DM. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5 (10:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah MA, Ajani JA. Gastric cancer—an enigmatic and heterogeneous disease. JAMA. 2010;303 (17:1753–1754. doi: 10.1001/jama.2010.553. [DOI] [PubMed] [Google Scholar]
- Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143 (1:35–47. doi: 10.1053/j.gastro.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Song S, Ajani JA. The role of microRNAs in cancers of the upper gastrointestinal tract. Nat Rev Gastroenterol Hepatol. 2013;10 (2:109–118. doi: 10.1038/nrgastro.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steevens J, Schouten LJ, Goldbohm RA, van den Brandt PA. Vegetables and fruits consumption and risk of esophageal and gastric cancer subtypes in the Netherlands Cohort Study. Int J Cancer. 2011;129 (11:2681–2693. doi: 10.1002/ijc.25928. [DOI] [PubMed] [Google Scholar]
- Su Y, Ni Z, Wang G, Cui J, Wei C, Wang J, Yang Q, Xu Y, Li F. Aberrant expression of microRNAs in gastric cancer and biological significance of miR-574-3p. Int Immunopharmacol. 2012;13 (4:468–475. doi: 10.1016/j.intimp.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11 (2:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9 (6:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11 (9:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106 (11:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, Lu X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70 (18:7176–7186. doi: 10.1158/0008-5472.CAN-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.