Abstract
Background:
Aberrant CXC chemokine receptor 4 (CXCR4) expressions in malignant tissues have been reported; however, its role in kidney cancer prognosis remains unknown. The aim of this study was to determine the prognostic value of CXCR4 expression in patients with clear-cell renal cell carcinoma (ccRCC).
Methods:
The study included 225 patients with ccRCC. The cohort was split into a training set (n=125) and a validation set (n=100). CXC chemokine receptor 4 expression was analysed by immunohistochemical staining and its correlations with clinicopathologic features and prognosis were evaluated.
Results:
CXCR4-staining intensity increased gradually accompanied with disease progression from TNM stages I to IV in 225 patients with ccRCC. Moreover, high CXCR4 expression indicated reduced overall survival (OS) in the training (P<0.001) and validation (P<0.001) sets, especially for patients with early-stage (TNM stage I+II) diseases. Furthermore, CXCR4 expression was identified as an independent prognostic factor for OS, and combining TNM stage with CXCR4 expression showed a better prognostic value for OS in both sets.
Conclusions:
High CXCR4 expression, an independent adverse prognostic factor, could be combined with TNM stage to generate a predictive nomogram for clinical outcome in patients with ccRCC.
Keywords: ccRCC, CXCR4, overall survival, nomogram, prognostic model
Renal cell carcinoma (RCC), which has an incidence of 63 920 new cases and ∼13 860 deaths annually in the United States, accounts for 2–3% of all malignant diseases in adults (Siegel et al, 2014). Its major histologic subtype is clear-cell RCC (ccRCC), with a proportion of 70–85% in RCC (Escudier et al, 2012). About 20–40% localised ccRCC relapse even after curative nephrectomy, usually leading to incurable disease (Stewart et al, 2011). Metastatic RCC, characterised by high resistance to radiotherapy and chemotherapy, has a poor prognosis, with a 5-year survival rate ranging from 0–20% (Figlin, 1999).
TNM stage, Fuhrman grade, and Eastern Cooperative Oncology Group performance status (ECOG-PS) remain the most commonly used predictors of clinical outcome for patients with RCC. Integrated systems, such as University of California Integrated Staging System (UISS), and Mayo Clinic stage, size, grade and necrosis (SSIGN) score are also used to predict clinical outcome. In ccRCC, patients with similar clinical and pathological features may have diverse outcomes. Improved predictors of survival of RCC are needed (Eichelberg et al, 2009; Sun et al, 2011).
In ccRCC, inactivation of the von Hippel-Lindau gene is a landmark. However, it failed to interpret the diverse biological behaviours of ccRCC. The CXC chemokine receptor 4 (CXCR4) and its chemokine ligand 12 (CXCL12) are two key factors in the cross-talking between cancer cells and their microenvironment, which makes them promising therapeutic targets for cancer. They were first defined as regulators of lymphocyte trafficking and localisation of B cells (Burger et al, 1999; Mohle et al, 1999). Currently, CXCR4 overexpression is found in more than 20 human tumour types, including ovarian cancer (Hall and Korach, 2003), prostate cancer (Taichman et al, 2002), oesophageal cancer (Kaifi et al, 2005), melanoma (Kim et al, 2008), neuroblastoma (Geminder et al, 2001), and RCC (Zagzag et al, 2005; D'Alterio et al, 2010a). A growing number of evidence have shown that the CXCR4/CXCL12 axis has a vital role in tumour proliferation, survival, and metastasis (Teicher and Fricker, 2010; Cho et al, 2013). CXCR4 has also been validated to be a prognostic predictor in various cancer types, such as breast cancer (Mendeloff and Burns, 2012), prostate cancer (Jung et al, 2011), and melanoma (Scala et al, 2005).
In this study, we sought to explore the clinical and prognostic value of CXCR4 in ccRCC. Our results demonstrated that high expression of CXCR4 was associated with a poor prognosis in patients with ccRCC. CXCR4 can further stratify early-stage ccRCC (TNM stage I+II) patients into two risk groups with significantly different overall survival (OS). In addition, the predictive accuracy of the TNM stage was improved when CXCR4 expression was incorporated.
Materials and Methods
Patients and clinical database
We recruited 225 ccRCC patients undergoing nephrectomy at the Zhongshan Hospital, Fudan University, Shanghai, China, between 1999 and 2006. This study was approved by the hospital's ethics committee and informed consent was obtained from each patient. Inclusion criteria included the following: no history of previous anticancer therapy, no history of other malignancies, patients after radical or partial nephrectomy, and histopathologically proven ccRCC. Exclusion criteria were as follows: mixed type of primary renal cancer as confirmed histopathologically, tumours with necrosis >80%, and patients who died within the first month after surgery due to surgical complications. Computer-generated random numbers were used to assign 125 of these patients to a training set, and 100 patients to a validation set. Patients' baseline clinical and pathological data and information about disease outcome, including date of death or last follow-up, were recorded. Tumour size was recorded as the longest diameter described in the pathology report. Tumour necrosis was defined as microscopic coagulative necrosis and was recorded as either present or absent. Presence of nodal and metastatic disease was defined according to intraoperative, pathologic, and radiographic findings. Patients were staged using radiographic reports and postoperative pathological data and were reassigned according to the 2010 AJCC TNM classification (Edge and Compton, 2010). Patients were followed up postoperatively with physical examinations, laboratory studies, chest imaging, and abdominal ultrasound or CT scans every 6 months for the first 2 years and annually thereafter for 5 years. We calculated OS from the date of nephrectomy to the date of death from all causes.
Immunohistochemistry
Immunohistochemical staining was performed on tissue microarray (TMA). Tissue microarrays were established as previously described (Zhu et al, 2008). Primary antibody against human CXCR4 (1 : 400 dilution, R&D systems, Minneapolis, MN, USA) was applied in the procedure. The sections were heated at 70 °C for 1 h, dewaxed in xylene, and dehydrated through a gradient concentration of alcohol. After retrieving and blocking the endogenous peroxidase and non-specific staining with 3% (v/v) HR2ROR2R and normal goat serum, the sections were incubated with anti-CXCR4 antibody overnight at 4 °C. The slides were then incubated with HRP-conjugated goat anti-mouse IgG secondary antibody for 10 min at 37 °C. Finally, the sections were visualised by DAB solution (DAKO, Carpinteria, CA, USA) and counterstained with haematoxylin (DAKO). Staining intensities and percentages of positive tumour cells were scored independently by two pathologists who were blind to the patients' outcome. A five-staged score (0, 1, 2, 3, and 4) was deducted from these two parameters according to a previously described scheme (Went et al, 2005).
Statistical analysis
For statistical analyses, CXCR4 staining was dichotomized into two groups (high (scores 0, 1, and 2) and low (scores 3 and 4)). We compared two groups using χ2 or Fisher's exact test for categorical variables and the t-test for continuous variables. Survival curves were established using the Kaplan–Meier method, and log-rank test was applied to compare the difference between the curves. The Cox proportional hazards regression model was applied to perform univariate and multivariate analyses, and those parameters that demonstrated a statistically significant effect on OS in the univariate analysis were included in the multivariate analysis. The sensitivity and specificity for the prediction of OS were analysed by receiver operating characteristic (ROC) curves. The area under the curve (AUC) was used to measure prognostic or predictive accuracy. Data were analysed using SPSS Statistics 21.0 (SPSS Inc., Chicago, IL, USA). All statistical tests were two-sided and statistical significance was set at 0.05. We used the R software version 3.0.2 and the ‘rms' package (R Foundation for Statistical Computing, Vienna, Austria) to perform the nomogram analysis.
Results
Immunohistochemical CXCR4 intensity and its association with pathological characteristics
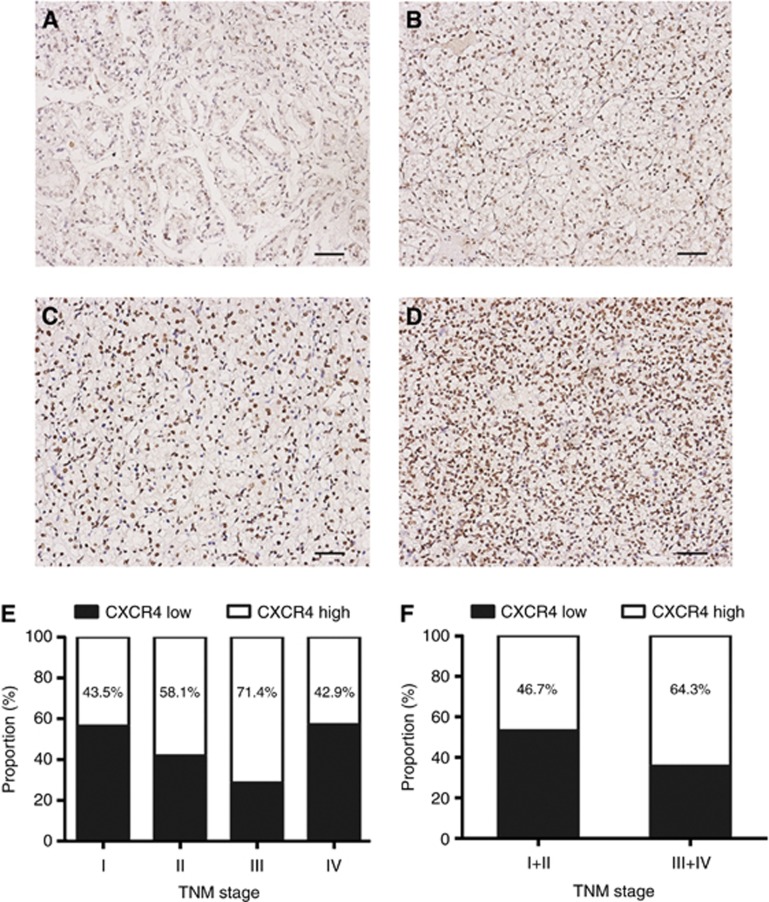
The characteristics of patients were listed in Table 1. The median follow-up was 62 months (range: 7–116 months) and 68 months (range: 8–117 months) for the training and validation sets, respectively. The two sets were well matched for pathological characteristics. CXCR4-positive staining was predominantly located in the nuclei of tumour cells. A total of 53.6% (67 of 125) and 43% (43 of 100) tumours were scored as high CXCR4 expression in the training and validation sets, respectively. CXCR4-staining density increased gradually accompanied with disease progression from TNM stage I to IV (Figure 1A–D), even though the correlation did not reach significant difference (Figure 1E and F). CXCR4 expression was significantly associated with tumour size (P=0.010 and P=0.037, respectively; Table 1). Additionally, in the validation set, CXCR4 expression was associated with tumour necrosis (P<0.001; Table 1).
Table 1. Correlations between CXCR4 expression and clinical characteristics in ccRCC patients.
|
Training set (n=125) |
Validation set (n=100) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
Patients |
CXCR4 expression |
|
Patients |
CXCR4 expression |
|
||||
| Characteristic | No. | % | Low | High | P | No. | % | Low | High | P |
|
Mean age, years | ||||||||||
| Mean±s.d. |
57.6±12.7 |
55.9±12.4 |
59.0±12.9 |
0.166 |
60.5±11.5 |
59.4±11.3 |
62.0±11.7 |
0.266 |
||
| Gender |
|
|
|
|
0.705 |
|
|
|
|
0.687 |
| Female | 41 | 32.8 | 20 | 21 | 35 | 35.0 | 38 | 27 | ||
| Male |
84 |
67.2 |
38 |
46 |
|
65 |
65.0 |
19 |
16 |
|
|
Tumour size, cm | ||||||||||
| Mean±s.d. |
5.1±2.8 |
4.5±2.3 |
5.7±3.1 |
0.010 |
5.1±2.8 |
4.5±2.6 |
5.7±2.9 |
0.037 |
||
| T stage |
|
|
|
|
0.239 |
|
|
|
|
0.234 |
| T1 | 77 | 61.6 | 40 | 37 | 65 | 65.0 | 41 | 24 | ||
| T2 | 16 | 12.8 | 5 | 11 | 10 | 10.0 | 5 | 5 | ||
| T3 |
32 |
25.6 |
13 |
19 |
|
24 |
24.0 |
11 |
14 |
|
| N status |
|
|
|
|
1.000 |
|
|
|
|
0.576 |
| N0 | 122 | 97.6 | 57 | 65 | 97 | 97.0 | 56 | 41 | ||
| N1 |
3 |
2.4 |
1 |
2 |
|
3 |
3.0 |
1 |
2 |
|
| M status |
|
|
|
|
1.000 |
|
|
|
|
1.000 |
| M0 | 122 | 97.6 | 57 | 65 | 95 | 95.0 | 54 | 41 | ||
| M1 |
3 |
2.4 |
1 |
2 |
|
5 |
5.0 |
3 |
2 |
|
| TNM stage |
|
|
|
|
0.314 |
|
|
|
|
0.173 |
| I | 85 | 68.0 | 44 | 41 | 69 | 69.0 | 43 | 26 | ||
| II | 26 | 20.8 | 10 | 16 | 17 | 17.0 | 8 | 9 | ||
| III | 11 | 8.8 | 3 | 8 | 10 | 10.0 | 3 | 7 | ||
| IV |
3 |
2.4 |
1 |
2 |
|
4 |
4.0 |
3 |
1 |
|
| Fuhrman grade |
|
|
|
|
0.484 |
|
|
|
|
0.175 |
| 1 | 28 | 23.2 | 15 | 13 | 22 | 22.0 | 12 | 10 | ||
| 2 | 47 | 36.8 | 24 | 23 | 40 | 40.0 | 27 | 13 | ||
| 3 | 36 | 28.8 | 14 | 22 | 28 | 28.0 | 15 | 13 | ||
| 4 |
14 |
11.2 |
5 |
9 |
|
10 |
10.0 |
3 |
7 |
|
| Tumour necrosis |
|
|
|
|
0.115 |
|
|
|
|
<0.001 |
| Absent | 77 | 61.6 | 40 | 37 | 56 | 56.0 | 41 | 15 | ||
| Present |
48 |
38.4 |
18 |
30 |
|
44 |
44.0 |
16 |
28 |
|
| ECOG-PS |
|
|
|
|
0.097 |
|
|
|
|
0.128 |
| 0 | 81 | 64.8 | 42 | 39 | 62 | 62.0 | 39 | 23 | ||
| ⩾1 | 44 | 35.2 | 16 | 28 | 38 | 38.0 | 18 | 20 | ||
Abbreviations: CXCR4=CXC chemokine receptor 4; ECOG-PS=Eastern Cooperative Oncology Group performance status.
Figure 1.
CXCR4 expression in ccRCC tissues. CXCR4 expression in TNM stage I (A), TNM stage II (B), TNM stage III (C), TNM stage IV (D). Scale bar=50.0 μm. Frequencies of CXCR4 expression level in each TNM stage (E), TNM stage I+II and III+IV (F).
Prognostic value of CXCR4 intensity for clinical outcome of ccRCC patients
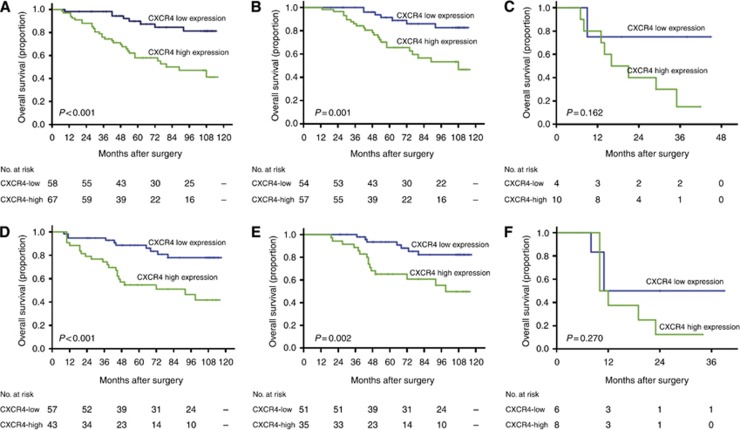
We applied Kaplan–Meier survival analysis to compare OS according to the CXCR4 expression. Patients with high CXCR4 expression had a significantly poorer OS than those with a low CXCR4 expression in the two sets (P<0.001 and P<0.001, respectively; Figure 2A and D). We further examined whether CXCR4 expression could stratify patients with early-stage and late-stage (stage III+IV) diseases. When the analysis was restricted to early-stage RCC, patients could be significantly stratified by CXCR4 expression (P=0.001 and P=0.002, respectively; Figure 2B and E). However, the CXCR4 expression was not predictive in late-stage RCC in our study (P=0.162 and P=0.270, respectively; Figure 2C and F).
Figure 2.
Kaplan–Meier analysis of OS according to the expression of CXCR4 in patients with ccRCC. Kaplan–Meier analysis of OS of all patients in the training set (A), patients with early-stage ccRCC in the training set (B), patients with late-stage ccRCC in the training set (C); all patients in the validation set (D), patients in early-stage ccRCC within the validation set (E), patients with late-stage ccRCC in the validation set (F). P-value was calculated by log-rank test.
To investigate whether CXCR4 expression was an independent prognostic predictor of OS, univariate and multivariate analyses were performed. As shown in Table 2, univariate analysis demonstrated that CXCR4 was significantly associated with OS in the training set (HR 4.32, 95% CI 1.98–9.42, P<0.001, ) and validation set (HR 3.57, 95% CI 1.69–7.56, P=0.001). Furthermore, multivariate analysis showed that CXCR4 remained an independent prognostic indicator for OS in the training set (HR 3.38, 95% CI 1.49–7.68, P=0.004) and validation set (HR 2.88, 95% CI 1.26–6.59, P=0.012), as did TNM stage (P=0.002 and P<0.001, respectively), Fuhrman grade (P<0.001 and P=0.002, respectively), and ECOG-PS (P=0.047 and P=0.015, respectively).
Table 2. Univariate and multivariate Cox regression analyses of potential prognostic factors for overall survival.
| |
Overall survival |
|||
|---|---|---|---|---|
| |
Training set |
Validation set |
||
| Variable | HR (95% CI) | P | HR (95% CI) | P |
|
Univariate | ||||
| Tumour size |
1.22 (1.12–1.34) |
<0.001 |
1.33 (1.22–1.46) |
<0.001 |
| T stage |
|
0.001 |
|
<0.001 |
| 2 vs 1 |
3.61 (1.56–8.35) |
0.003 |
5.46 (2.01–14.84) |
0.001 |
| 3 vs 1 |
3.71 (1.81–7.62) |
<0.001 |
4.85 (2.22–10.57) |
<0.001 |
| N status (N1 vs N0) |
4.48 (1.06–18.93) |
0.042 |
31.90 (6.87–148.18) |
<0.001 |
| M status (M1 vs M0) |
21.21 (4.09–109.93) |
<0.001 |
9.59 (3.18–28.93) |
<0.001 |
| TNM stage |
|
<0.001 |
|
<0.001 |
| II vs I |
3.39 (1.65–6.96) |
0.001 |
2.84 (1.21–6.64) |
0.016 |
| III vs I |
17.69 (6.46–48.43) |
<0.001 |
30.95 (9.42–101.69) |
<0.001 |
| IV vs I |
59.07 (10.34–337.40) |
<0.001 |
59.85 (12.65–283.10) |
<0.001 |
| Fuhrman grade |
|
<0.001 |
|
<0.001 |
| 2 vs 1 |
0.65 (0.20–2.12) |
0.470 |
1.01 (0.19–5.22) |
0.994 |
| 3 vs 1 |
2.74 (1.00–7.50) |
0.049 |
7.18 (1.66–9.31.15) |
0.008 |
| 4 vs 1 |
8.43 (2.96–24.01) |
<0.001 |
16.09 (3.39–76.43) |
0.005 |
|
Tumour necrosis | ||||
| Present vs absent |
1.97 (1.04–3.71) |
0.037 |
6.22 (2.82–13.74) |
<0.001 |
| ECOG-PS (⩾1 vs 0) |
2.21 (1.16–4.22) |
0.016 |
4.40 (2.09–9.25) |
<0.001 |
| CXCR4 (high vs low) |
4.32 (1.98–9.42) |
<0.001 |
3.57 (1.69–7.56) |
0.001 |
|
Multivariate | ||||
| Tumour size |
1.03 (0.92–1.16) |
0.588 |
1.06 (0.92–1.21) |
0.409 |
|
TNM stage | ||||
| III+IV vs I+II |
4.96 (1.78–13.84) |
0.002 |
9.06 (2.89–28.37) |
<0.001 |
|
Fuhrman grade | ||||
| 3+4 vs 1+2 |
4.16 (1.95–8.87) |
<0.001 |
4.53 (1.71–12.00) |
0.002 |
|
Tumour necrosis | ||||
| Present vs absent |
1.27 (0.57–2.84) |
0.504 |
1.90 (0.71–5.08) |
0.325 |
| ECOG-PS (⩾1 vs 0) |
2.11 (1.01–4.40) |
0.047 |
2.76 (1.22–6.26) |
0.015 |
| CXCR4 (high vs low) | 3.38 (1.49–7.68) | 0.004 | 2.88 (1.26–6.59) | 0.012 |
Abbreviations: CI=confidence interval; CXCR4=CXC chemokine receptor 4; ECOG-PS=Eastern Cooperative Oncology Group performance status; HR=hazard ratio.
Extension of the TNM stage prognostic model with CXCR4 intensity for ccRCC patients
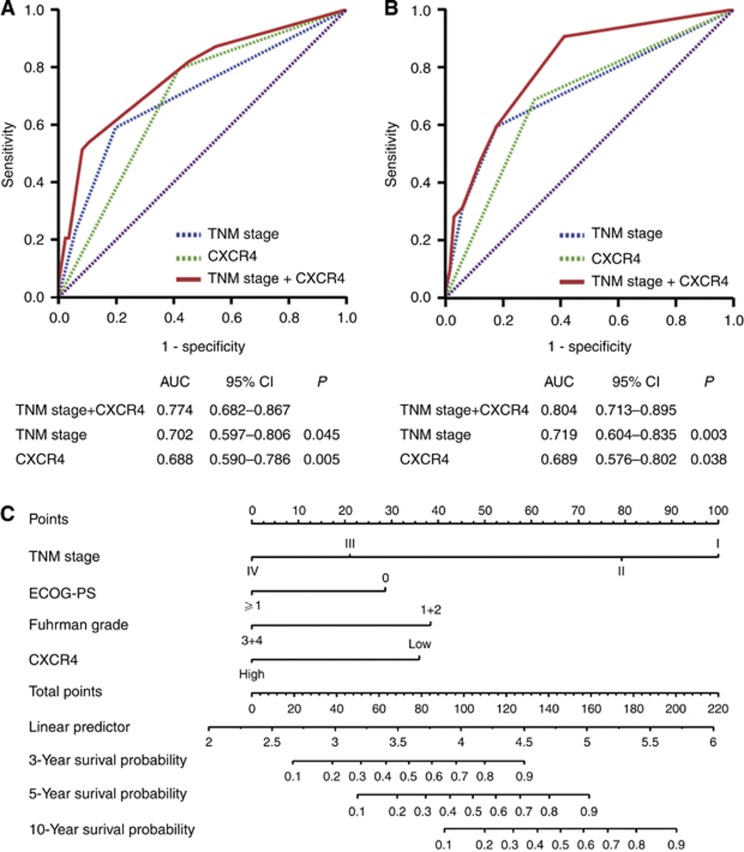
On the basis of above-mentioned findings, we investigated whether incorporation of the CXCR4 expression into the TNM stage would improve its predictive accuracy. The combination of CXCR4 expression and TNM stage (AUC 0.774 and AUC 0.804, respectively) showed a better prognostic value than TNM stage alone (AUC 0.702, P=0.045; AUC 0.719, P=0.003, respectively) or CXCR4 expression alone (AUC 0.688, P=0.005; AUC 0.689, P=0.038, respectively) in both sets (Figure 3A and B). Finally, we constructed a nomogram to predict OS at 3, 5, and 10 years, with a predictive accuracy of 88.9, 85.6, and 83.8%, respectively (Figure 3C). Our results rising from the training set yielded a multivariate model, which constituted the basis of the nomogram. The predictors included TNM stage, ECOG-PS, Fuhrman grade, and CXCR4 expression, all of which were independent prognostic indicators for OS in multivariate analysis in the training set.
Figure 3.
ROC analysis and nomogram generation for the prediction of OS in patients with ccRCC. ROC analysis of the sensitivity and specificity for the prediction of OS in the training set (A), in the validation set (B) by the TNM stage and CXCR4 combined model, the TNM stage model, and the CXCR4 expression model. P-values show statistical significance of the AUC of the TNM stage and CXCR4 combined model, the TNM stage model, and the CXCR4 expression model. (C) Nomogram to predict risk of OS in training set.
Discussion
Our study confirms the biologic and prognostic significance of CXCR4 for patients with ccRCC and provides support for their use to stratify patients with different prognoses after nephrectomy. Chemokine receptors form a superfamily that can mediate chemotaxis of cells towards a gradient of chemokine. CXCR4 is a G protein-coupled chemokine receptor (Caruz et al, 1998). It exerts its biological effect by binding to its ligand CXCL12, activating the downstream protein kinase B (AKT)/mitogen-activated protein kinases (MAPKs) signalling pathway (Teicher and Fricker, 2010). The previous studies demonstrated that CXCR4 was mediated by the hypoxia-inducible transcription factor (HIF) pathway and had a critical role in tumour initiation and metastasis in patients with RCC (Staller et al, 2003; Vanharanta et al, 2013). A recent paper reported that galectin-1 could promote tumour progression through upregulation of CXCR4 via NF-κB in RCC (Huang et al, 2014). In this study, CXCR4-positive staining was predominantly located in the nuclei of ccRCC. However, Zagzag et al (2005) demonstrated that all the samples of RCC showed a cytoplasmic staining, with additional staining in the nuclei and membranes. CXCR4 nuclear expression was reported in breast cancer (Woo et al, 2008), lung cancer (Na et al, 2008), and colorectal cancer (Wang et al, 2010), indicating an increased risk of lymph node metastasis. Wang et al (2009) demonstrated that CXCR4 nuclear localisation may be responsible for certain metastatic changes in ccRCC. In our study, the analysis of correlations between lymph node involvement and CXCR4 expression was not applicable due to limited cases (n=6).
A growing body of evidence indicated that CXCR4 expression was upregulated in a series of malignant tumours and CXCR4 had crucial roles in the metastasis of many solid tumours (Burger and Kipps, 2006). Thyroid (De Falco et al, 2007), prostate (Taichman et al, 2002), and colon (Zeelenberg et al, 2003) cancer models showed that directed metastasis of cancer cells was mediated by CXCR4 activation towards a concentration gradient of CXCL12. CXCL12 expression levels are high in the liver, lung, and bone marrow, making them common metastasis sites. In our study, CXCR4 expression was positively correlated with tumour size, implying that CXCR4 expression might promote tumour growth. It has been reported that CXCR4 could enhance tumour cell proliferation in many cancers (Bajetto et al, 2006; Shen et al, 2010; Luo et al, 2014). The growth-promoting role of CXCR4 could also be confirmed by showing that CXCR4 antagonists inhibit tumour growth in multiple experiment orthotopic (Smith et al, 2004; Cho et al, 2013) and subcutaneous human xenograft (De Falco et al, 2007; Porvasnik et al, 2009). Moreover, we also observed an association between high CXCR4 level and tumour necrosis, which might be due to rapid tumour growth mediated by CXCR4 upregulation.
CXCR4 has been validated to be a prognostic factor in a variety of cancers, such as breast cancer (Mendeloff and Burns, 2012) and prostate cancer (Jung et al, 2011). D'Alterio et al (2010a, 2010b) demonstrated that CXCR4 expression is independently associated with disease-free survival in patients with RCC. In ccRCC, despite being diagnosed with early stage, ∼25–40% of RCC patients who underwent partial or radical nephrectomy eventually experienced recurrence (Stewart et al, 2011). This implied that defining molecular subgroups may identify patients who could benefit from targeted therapies and personalised treatment. In our study, we found that high CXCR4 expression was an independent poor predictor for OS of ccRCC. Early-stage ccRCC patients could be stratified according to CXCR4 expression levels. These data indicated that CXCR4 expression may be a potential prognostic marker of ccRCC with regard to counselling patients, selecting patients for adjuvant therapies after surgery. Furthermore, incorporation of CXCR4 expression into the TNM stage system could improve prognostic value for OS. These data suggested that the CXCR4 expression might increase prognostic information for patients with ccRCC and lead to a more accurate classification under the TNM stage system. However, the effects of integration of CXCR4 expression into the current TNM stage model and the potential clinical practice changing should be validated in a larger population. We proposed a nomogram that can be used to predict OS at 3, 5, and 10 years. Although the nomogram is useful for visualising our predictive models, it needs to be tested on independent patient populations.
As CXCR4 had such crucial roles in tumour growth, invasion, migration, and survival, optimal use of CXCR4 inhibition might be a part of potential targeted therapy. There is evidence that blocking CXCR4 can inhibit the proliferation, invasion, and metastasis of tumour cells (Rubin et al, 2003; Smith et al, 2004). CXCR4 antagonist AMD3100 is the most studied among the agents that inhibit CXCR4 signalling. AMD3100 is a small synthetic inhibitor of the CXCL12-binding chemokine receptors CXCR4 and CXCR7 (Pusic and DiPersio, 2010). It could interrupt the interaction of tumour cells with the microenvironment of distant metastasis sites and enhance the sensitivity to therapy (Azab et al, 2009). Previous study has demonstrated that CXCR4 antagonists may be useful for the treatment of peritoneal carcinomatosis of gastric cancer in a mouse model (Yasumoto et al, 2006). These data suggested that targeting CXCL12/CXCR4 signalling with AMD3100 may be a novel and efficient strategy for the treatment of ccRCC. The profound molecular roles of CXCR4 and its antagonist in ccRCC remain far from being fully elucidated and need further investigation.
Several limitations of this study should be acknowledged. First, we have no external validation. The clinical and prognostic implications of CXCR4 in ccRCC remain to be investigated in a larger cohort in the future. Second, the previous research demonstrated that CXCR4 expression was associated with metastasis. However, owing to a small number of patients with metastatic disease, the relationship between CXCR4 expression and tumour metastasis was not established in our study. In addition, CXCR7 is also a receptor for CXCL12 that binds this chemokine with greater affinity (Sun et al, 2010), the clinical and prognostic implications of the CXCL12/CXCR4/CXCR7 axis in patients with ccRCC remain to be evaluated.
In conclusion, our study clearly demonstrated that high CXCR4 expression was strongly associated with a poor outcome. CXCR4 can further stratify early-stage ccRCC patients with significantly different OS. This finding provides a novel independent predictor for prognosis and may improve current predictive systems in terms of counselling patients, selecting patients for adjuvant therapies, and customising follow-up after surgery.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (31100629, 31270863, 81372755), Program for New Century Excellent Talents in University (NCET-13-0146) and Shanghai Rising-Star Program (13QA1400300).
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Azab AK, Runnels JM, Pitsillides C, Moreau AS, Azab F, Leleu X, Jia X, Wright R, Ospina B, Carlson AL, Alt C, Burwick N, Roccaro AM, Ngo HT, Farag M, Melhem MR, Sacco A, Munshi NC, Hideshima T, Rollins BJ, Anderson KC, Kung AL, Lin CP, Ghobrial IM. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009;113 (18:4341–4351. doi: 10.1182/blood-2008-10-186668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajetto A, Barbieri F, Dorcaratto A, Barbero S, Daga A, Porcile C, Ravetti JL, Zona G, Spaziante R, Corte G, Schettini G, Florio T. Expression of CXC chemokine receptors 1-5 and their ligands in human glioma tissues: role of CXCR4 and SDF1 in glioma cell proliferation and migration. Neurochem Int. 2006;49 (5:423–432. doi: 10.1016/j.neuint.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94 (11:3658–3667. [PubMed] [Google Scholar]
- Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107 (5:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- Caruz A, Samsom M, Alonso JM, Alcami J, Baleux F, Virelizier JL, Parmentier M, Arenzana-Seisdedos F. Genomic organization and promoter characterization of human CXCR4 gene. FEBS Lett. 1998;426 (2:271–278. doi: 10.1016/s0014-5793(98)00359-7. [DOI] [PubMed] [Google Scholar]
- Cho KS, Yoon SJ, Lee JY, Cho NH, Choi YD, Song YS, Hong SJ. Inhibition of tumor growth and histopathological changes following treatment with a chemokine receptor CXCR4 antagonist in a prostate cancer xenograft model. Oncol Lett. 2013;6 (4:933–938. doi: 10.3892/ol.2013.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alterio C, Cindolo L, Portella L, Polimeno M, Consales C, Riccio A, Cioffi M, Franco R, Chiodini P, Carteni G, Mirone V, Longo N, Marra L, Perdona S, Claudio L, Mascolo M, Staibano S, Falsaperla M, Puglisi M, Martignoni G, Ficarra V, Castello G, Scala S. Differential role of CD133 and CXCR4 in renal cell carcinoma. Cell Cycle. 2010;9 (22:4492–4500. doi: 10.4161/cc.9.22.13680. [DOI] [PubMed] [Google Scholar]
- D'Alterio C, Consales C, Polimeno M, Franco R, Cindolo L, Portella L, Cioffi M, Calemma R, Marra L, Claudio L, Perdona S, Pignata S, Facchini G, Carteni G, Longo N, Pucci L, Ottaiano A, Costantini S, Castello G, Scala S. Concomitant CXCR4 and CXCR7 expression predicts poor prognosis in renal cancer. Curr Cancer Drug Targets. 2010;10 (7:772–781. doi: 10.2174/156800910793605839. [DOI] [PubMed] [Google Scholar]
- De Falco V, Guarino V, Avilla E, Castellone MD, Salerno P, Salvatore G, Faviana P, Basolo F, Santoro M, Melillo RM. Biological role and potential therapeutic targeting of the chemokine receptor CXCR4 in undifferentiated thyroid cancer. Cancer Res. 2007;67 (24:11821–11829. doi: 10.1158/0008-5472.CAN-07-0899. [DOI] [PubMed] [Google Scholar]
- Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17 (6:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- Eichelberg C, Junker K, Ljungberg B, Moch H. Diagnostic and prognostic molecular markers for renal cell carcinoma: a critical appraisal of the current state of research and clinical applicability. Eur Urol. 2009;55 (4:851–863. doi: 10.1016/j.eururo.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Escudier B, Eisen T, Porta C, Patard JJ, Khoo V, Algaba F, Mulders P, Kataja V, Group EGW Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23 (Suppl 7:vii65–vii71. doi: 10.1093/annonc/mds227. [DOI] [PubMed] [Google Scholar]
- Figlin RA.1999Renal cell carcinoma: management of advanced disease J Urol 161(2381–386.; discussion 386–387. [DOI] [PubMed] [Google Scholar]
- Geminder H, Sagi-Assif O, Goldberg L, Meshel T, Rechavi G, Witz IP, Ben-Baruch A. A possible role for CXCR4 and its ligand, the CXC chemokine stromal cell-derived factor-1, in the development of bone marrow metastases in neuroblastoma. J Immunol. 2001;167 (8:4747–4757. doi: 10.4049/jimmunol.167.8.4747. [DOI] [PubMed] [Google Scholar]
- Hall JM, Korach KS. Stromal cell-derived factor 1, a novel target of estrogen receptor action, mediates the mitogenic effects of estradiol in ovarian and breast cancer cells. Mol Endocrinol. 2003;17 (5:792–803. doi: 10.1210/me.2002-0438. [DOI] [PubMed] [Google Scholar]
- Huang CS, Tang SJ, Chung LY, Yu CP, Ho JY, Cha TL, Hsieh CC, Wang HH, Sun GH, Sun KH.2014Galectin-1 Upregulates CXCR4 to Promote Tumor Progression and Poor Outcome in Kidney Cancer J Am Soc Nephrole-pub ahead of print 7 February 2014. [DOI] [PMC free article] [PubMed]
- Jung SJ, Kim CI, Park CH, Chang HS, Kim BH, Choi MS, Jung HR. Correlation between chemokine receptor CXCR4 expression and prognostic factors in patients with prostate cancer. Korean J Urol. 2011;52 (9:607–611. doi: 10.4111/kju.2011.52.9.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifi JT, Yekebas EF, Schurr P, Obonyo D, Wachowiak R, Busch P, Heinecke A, Pantel K, Izbicki JR. Tumour-cell homing to lymph nodes and bone marrow and CXCR4 expression in esophageal cancer. J Natl Cancer Inst. 2005;97 (24:1840–1847. doi: 10.1093/jnci/dji431. [DOI] [PubMed] [Google Scholar]
- Kim SY, Lee CH, Midura BV, Yeung C, Mendoza A, Hong SH, Ren L, Wong D, Korz W, Merzouk A, Salari H, Zhang H, Hwang ST, Khanna C, Helman LJ. Inhibition of the CXCR4/CXCL12 chemokine pathway reduces the development of murine pulmonary metastases. Clin Exp Metastasis. 2008;25 (3:201–211. doi: 10.1007/s10585-007-9133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HN, Wang ZH, Sheng Y, Zhang Q, Yan J, Hou J, Zhu K, Cheng Y, Xu YL, Zhang XH, Xu M, Ren XY. MiR-139 targets CXCR4 and inhibits the proliferation and metastasis of laryngeal squamous carcinoma cells. Med Oncol. 2014;31 (1:789. doi: 10.1007/s12032-013-0789-z. [DOI] [PubMed] [Google Scholar]
- Mendeloff J, Burns R. States with low non-fatal injury rates have high fatality rates and vice-versa. Am J Ind Med. 2012;56 (5:509–519. doi: 10.1002/ajim.22047. [DOI] [PubMed] [Google Scholar]
- Mohle R, Failenschmid C, Bautz F, Kanz L. Overexpression of the chemokine receptor CXCR4 in B cell chronic lymphocytic leukemia is associated with increased functional response to stromal cell-derived factor-1 (SDF-1) Leukemia. 1999;13 (12:1954–1959. doi: 10.1038/sj.leu.2401602. [DOI] [PubMed] [Google Scholar]
- Na IK, Scheibenbogen C, Adam C, Stroux A, Ghadjar P, Thiel E, Keilholz U, Coupland SE. Nuclear expression of CXCR4 in tumor cells of non-small cell lung cancer is correlated with lymph node metastasis. Hum Pathol. 2008;39 (12:1751–1755. doi: 10.1016/j.humpath.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Porvasnik S, Sakamoto N, Kusmartsev S, Eruslanov E, Kim WJ, Cao W, Urbanek C, Wong D, Goodison S, Rosser CJ. Effects of CXCR4 antagonist CTCE-9908 on prostate tumor growth. Prostate. 2009;69 (13:1460–1469. doi: 10.1002/pros.21008. [DOI] [PubMed] [Google Scholar]
- Pusic I, DiPersio JF. Update on clinical experience with AMD3100, an SDF-1/CXCL12-CXCR4 inhibitor, in mobilization of hematopoietic stem and progenitor cells. Curr Opin Hematol. 2010;17 (4:319–326. doi: 10.1097/MOH.0b013e328338b7d5. [DOI] [PubMed] [Google Scholar]
- Rubin JB, Kung AL, Klein RS, Chan JA, Sun Y, Schmidt K, Kieran MW, Luster AD, Segal RA. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc Natl Acad Sci USA. 2003;100 (23:13513–13518. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala S, Ottaiano A, Ascierto PA, Cavalli M, Simeone E, Giuliano P, Napolitano M, Franco R, Botti G, Castello G. Expression of CXCR4 predicts poor prognosis in patients with malignant melanoma. Clin Cancer Res. 2005;11 (5:1835–1841. doi: 10.1158/1078-0432.CCR-04-1887. [DOI] [PubMed] [Google Scholar]
- Shen X, Artinyan A, Jackson D, Thomas RM, Lowy AM, Kim J. Chemokine receptor CXCR4 enhances proliferation in pancreatic cancer cells through AKT and ERK dependent pathways. Pancreas. 2010;39 (1:81–87. doi: 10.1097/MPA.0b013e3181bb2ab7. [DOI] [PubMed] [Google Scholar]
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64 (1:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- Smith MC, Luker KE, Garbow JR, Prior JL, Jackson E, Piwnica-Worms D, Luker GD. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64 (23:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425 (6955:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- Stewart GD, O'Mahony FC, Powles T, Riddick AC, Harrison DJ, Faratian D. What can molecular pathology contribute to the management of renal cell carcinoma. Nat Rev Urol. 2011;8 (5:255–265. doi: 10.1038/nrurol.2011.43. [DOI] [PubMed] [Google Scholar]
- Sun M, Shariat SF, Cheng C, Ficarra V, Murai M, Oudard S, Pantuck AJ, Zigeuner R, Karakiewicz PI. Prognostic factors and predictive models in renal cell carcinoma: a contemporary review. Eur Urol. 2011;60 (4:644–661. doi: 10.1016/j.eururo.2011.06.041. [DOI] [PubMed] [Google Scholar]
- Sun X, Cheng G, Hao M, Zheng J, Zhou X, Zhang J, Taichman RS, Pienta KJ, Wang J. CXCL12 / CXCR4 / CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010;29 (4:709–722. doi: 10.1007/s10555-010-9256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taichman RS, Cooper C, Keller ET, Pienta KJ, Taichman NS, McCauley LK. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. 2002;62 (6:1832–1837. [PubMed] [Google Scholar]
- Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16 (11:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- Vanharanta S, Shu W, Brenet F, Hakimi AA, Heguy A, Viale A, Reuter VE, Hsieh JJ, Scandura JM, Massague J. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat Med. 2013;19 (1:50–56. doi: 10.1038/nm.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Z, Yang B, Yang Q, Sun Y. CXCR4 nuclear localization follows binding of its ligand SDF-1 and occurs in metastatic but not primary renal cell carcinoma. Oncol Rep. 2009;22 (6:1333–1339. doi: 10.3892/or_00000572. [DOI] [PubMed] [Google Scholar]
- Wang SC, Lin JK, Wang HS, Yang SH, Li AF, Chang SC. Nuclear expression of CXCR4 is associated with advanced colorectal cancer. Int J Colorectal Dis. 2010;25 (10:1185–1191. doi: 10.1007/s00384-010-0999-1. [DOI] [PubMed] [Google Scholar]
- Went P, Dirnhofer S, Salvisberg T, Amin MB, Lim SD, Diener PA, Moch H. Expression of epithelial cell adhesion molecule (EpCam) in renal epithelial tumors. Am J Surg Pathol. 2005;29 (1:83–88. doi: 10.1097/01.pas.0000.146028.70868.7a. [DOI] [PubMed] [Google Scholar]
- Woo SU, Bae JW, Kim CH, Lee JB, Koo BW. A significant correlation between nuclear CXCR4 expression and axillary lymph node metastasis in hormonal receptor negative breast cancer. Ann Surg Oncol. 2008;15 (1:281–285. doi: 10.1245/s10434-007-9595-1. [DOI] [PubMed] [Google Scholar]
- Yasumoto K, Koizumi K, Kawashima A, Saitoh Y, Arita Y, Shinohara K, Minami T, Nakayama T, Sakurai H, Takahashi Y, Yoshie O, Saiki I. Role of the CXCL12/CXCR4 axis in peritoneal carcinomatosis of gastric cancer. Cancer Res. 2006;66 (4:2181–2187. doi: 10.1158/0008-5472.CAN-05-3393. [DOI] [PubMed] [Google Scholar]
- Zagzag D, Krishnamachary B, Yee H, Okuyama H, Chiriboga L, Ali MA, Melamed J, Semenza GL. Stromal cell-derived factor-1alpha and CXCR4 expression in hemangioblastoma and clear cell-renal cell carcinoma: von Hippel-Lindau loss-of-function induces expression of a ligand and its receptor. Cancer Res. 2005;65 (14:6178–6188. doi: 10.1158/0008-5472.CAN-04-4406. [DOI] [PubMed] [Google Scholar]
- Zeelenberg IS, Ruuls-Van Stalle L, Roos E. The chemokine receptor CXCR4 is required for outgrowth of colon carcinoma micrometastases. Cancer Res. 2003;63 (13:3833–3839. [PubMed] [Google Scholar]
- Zhu XD, Zhang JB, Zhuang PY, Zhu HG, Zhang W, Xiong YQ, Wu WZ, Wang L, Tang ZY, Sun HC. High expression of macrophage colony-stimulating factor in peritumoral liver tissue is associated with poor survival after curative resection of hepatocellular carcinoma. J Clin Oncol. 2008;26 (16:2707–2716. doi: 10.1200/JCO.2007.15.6521. [DOI] [PubMed] [Google Scholar]