Abstract
DNA methylation is a well-recognized epigenetic mechanism that has been the subject of a growing body of literature typically focused on the identification and study of profiles of DNA methylation and their association with human diseases and exposures. In recent years, a number of unsupervised clustering algorithms, both parametric and non-parametric, have been proposed for clustering large-scale DNA methylation data. However, most of these approaches do not incorporate known biological relationships of measured features, and in some cases, rely on unrealistic assumptions regarding the nature of DNA methylation. Here, we propose a modified version of a recursively partitioned mixture model (RPMM) that integrates information related to the proximity of CpG loci within the genome to inform correlation structures from which subsequent clustering analysis is based. Using simulations and four methylation data sets, we demonstrate that integrating biologically informative correlation structures within RPMM resulted in improved goodness-of-fit, clustering consistency, and the ability to detect biologically meaningful clusters compared to methods which ignore such correlation. Integrating biologically-informed correlation structures to enhance modeling techniques is motivated by the rapid increase in resolution of DNA methylation microarrays and the increasing understanding of the biology of this epigenetic mechanism.
Keywords: finite mixture models epigenetics, genomic data, model-based clustering
Introduction
DNA methylation has emerged as one of the most widely studied epigenetic states due to its role in regulating gene expression and gene expression potential. While DNA methylation is a normal and essential process for human development, aberrant methylation patterns have been linked to pathogenesis and progression of various human diseases, as well as a wide variety of exposures (Joubert et al., 2012; Langevin et al., 2012; Zhai et al., 2012). Altered methylation patterns in the context of human health have highlighted the possibility of using DNA methylation for the purposes of diagnostics, in which profiles of DNA methylation are used for risk assessment, early disease detection, and disease recurrence monitoring (Laird, 2003). Similar to analyses involving microarray-based gene expression data, unsupervised clustering of DNA methylation data is often used to identify altered methylation profiles. Although a number of clustering methods have been applied to DNA methylation data (Houseman et al., 2008; Grigoriu et al., 2011; Mousa et al., 2012), many of the methods used to date do not incorporate known biological relationships of measured features, and in some cases, make unrealistic assumptions regarding the underlying biology of DNA methylation data. The rapid emergence of epigenetics literature and the increasing interest in the use of profiles of DNA methylation for diagnostic purposes, underscores the importance of continued advances in analytical tools that incorporate known features of DNA methylation.
Genome-wide DNA methylation is often studied using large-scale microarrays. The Illumina GoldenGate and Infinium Human Methylation27 arrays (Illumina, San Diego, CA, USA) simultaneously measure cytosine methylation at 1505 and 27,578 CpG sites, respectively, providing a glimpse of DNA methylation in important regulatory regions. Illumina's most recent methylation assay, the 450K Infinium Methylation BeadChip, interrogates the methylation status of 485,533 CpG sites per sample at single-nucleotide resolution, covering 96% of CpG Islands, with additional coverage in island shores (<2 Kb from CpG Islands) and the regions flanking them. Methylation measurements from the Illumina technologies are typically quantified using the average β value, a continuous variable, calculated as the average of several replicates (i.e., several beads per sample), and lying between zero (unmethylated) and one (methylated).
Unsupervised clustering of DNA methylation data is often used for the identification of methylation subgroups, or groups of samples with a similar methylation profile across a collection CpGs. Although there is no universal consensus on the best clustering method for array-based DNA methylation data, Siegmund et al. (2003) argue that model-based methods for clustering via finite mixture models are preferred to their non-parametric counterparts. Along these lines, Houseman et al. (2008) proposed the recursively partitioned mixture model (RPMM), a computationally efficient model-based hierarchical method of clustering high-dimensional data. This methodology has been shown to perform effectively for DNA methylation data and has to date, been applied in a number of different settings (Christensen et al., 2011; Hinoue et al., 2012; Koestler et al., 2012). One principal advantage of this method is that it provides a convenient framework for robustly estimating the number of classes K or clusters in the data, a fundamental issue in problems involving clustering (Chen, 1995). Moreover, RPMM allows for the attainment of subject-specific posterior probabilities of class membership, which can be helpful in understanding a subjects relative propensity within each of the predicted classes, as demonstrated in Koestler et al. (2010). Despite these advantages, RPMM is limited by its reliance on the assumption of class conditional independence (i.e., the methylation status of CpG sites are assumed to be independent conditional on class membership), which when violated, may lead to an overestimation the true number of classes, resulting in an over-fit solution (Lindsay et al., 1991). We further note that metric-based hierarchical clustering algorithms using the Euclidean distance-metric remain unaffected by correlation between features, as the expected value of the Euclidean distance depends only on the trace of the variance-covariance matrix (hence only the diagonal terms). This is further described in the Appendix (Section 6).
The assumption of class conditional independence provides an opportunity to advance the existing RPMM framework for DNA methylation data, for which correlation of methylation between neighboring probes may be pronounced. Indeed several recently published studies have reported high correlation in the methylation status of neighboring CpG sites, which is most pronounced between pairs of closely located CpG sites and decreases as function of their distance in base pairs (Ehrich et al., 2008; Nautiyal et al., 2010). In a study of DNA methylation among 27 epithelial ovarian tumors and 15 ovarian cancer cell lines, Houshdaran et al. (2010) reported that DNA methylation measurements from multiple probes representing different CpG sites associated with the same gene (related probes) exhibited large correlation (mean Pearson correlation: 0.64 for related pairs of probes and 0.04 for unrelated pairs). Consistent with this finding, we observed distinct distributions of correlation between related pairs of probes and unrelated pairs using methylation data from 158 mesothelioma tumors (Christensen et al., 2009) (mean Pearson correlation: 0.40 for related pairs of probes and 0.07 for unrelated pairs). Although several recently published works have proposed statistical approaches that incorporate the dependency structure between neighboring CpGs (Laurila et al., 2011; Kuan and Chiang, 2012), very little attention has been given toward the application of such information within unsupervised clustering methods. Given the prominent role of unsupervised clustering in revealing underlying structure in large-scale genomic data and the promise of such techniques for identifying clinically/biologically important profiles of DNA methylation (Banister et al., 2011; Marsit et al., 2011; Koestler et al., 2012), we aimed to understand whether incorporating correlation between pairs related probes within the framework of RPMM improves clustering performance (as measured by accurate estimation of the true number of clusters, model goodness-of-fit, and clustering consistency) and the ability to detect biologically meaningful clusters.
Here, we propose Gaussian- and beta-distributed RPMMs that incorporate correlation between neighboring CpG sites. Motivated by the findings of Houshdaran et al. (2010), our proposed methodologies (1) permit the methylation status among CpG sites associated with the same gene to be correlated, (2) while assuming independence between CpG sites associated with different genes. We examine the clustering performance and computational complexity of the proposed methods compared to alternative model-based and non-parametric clustering techniques in simulation as well as in the analysis of four independent array-based methylation data sets. We also investigate the ability of our proposed methodology to identify biologically meaningful clusters using case/control data collected from two independent studies of cancer: a head and neck squamous cell carcinoma (HNSCC) (Langevin et al., 2012) and a bladder cancer data set (Marsit et al., 2011). The methods described were implemented in R version 2.13 (http://cran.r-project.org/) statistical language and are freely available on the first authors website. See Appendix for details.
Statistical methods
As previously mentioned, methylation levels in Illumina methylation assays are quantified by the β value and are approximately continuous distributed, lying between zero and one, with values of zero indicating an unmethylated locus and values of one representing a methylated locus. To this end, the beta distribution is a natural distribution for modeling the observed β values. However, the maximum likelihood estimatior of the beta distribution parameters does not have a closed form and thus relies on numerical methods such as the Newton-Raphson or Fisher scoring algorithm (Ji et al., 2005). An alternative approach involves modeling the transformed β values [i.e., arcsine square-root transformation as in Rocke (1993); Houseman et al. (2009) or logit transformation as in Kuan et al. (2012)] using a normal distribution. While there are moderate gains in computational efficiency inherent to the later approach, the log-likelihood of the transformed-normal distribution may be more sensitive near the boundaries compared to the log-likelihood of the beta distribution (Verkuilen and Smithson, 2012). We consider both of the above approaches – specifically, we propose a class of modified RPMMs that incorporate correlation in the methylation values between CpG sites associated with the same gene where a (1) modified Gaussian RPMM is assumed and fit to the resulting transformed β values and (2) a modified beta RPMM is assumed and fit to the untransformed β values.
Gaussian distributed RPMM with within-gene correlation
Let Yij represent the methylation β value for subject i ∈ {1,2,...,n} and CpG locus . Let represent the subset of CpG loci that are associated with gene g, where g ∈ {1,2,...,G} and G is the total number of genes. Assuming that there are Jg elements contained in , we define Zij to be the appropriately transformed methylation β value for subject i, loci j (i.e., ). Then Zi is a J×1 vector of transformed methylation values for subject i and Zig represents Jg×1 vector corresponding to the Jg transformed methylation β values for subject i among loci associated with gene g. We assume the following distribution,
where θkj is a vector of parameters that depends on both class k and gene g. Note that the present version of the Gaussian RPMM assumes that where is the variance of loci j associated with gene g for class k. Our modified version assumes a more flexible covariance structure for Σgk. Specifically, letting , where j=1,2,...,Jg, we assume (class notation suppressed),
–
– Cov(zigj,zigj′)=ρgjj′σjσj′, where j ≠ j′
– Cov(zigj,zig′j′)=0
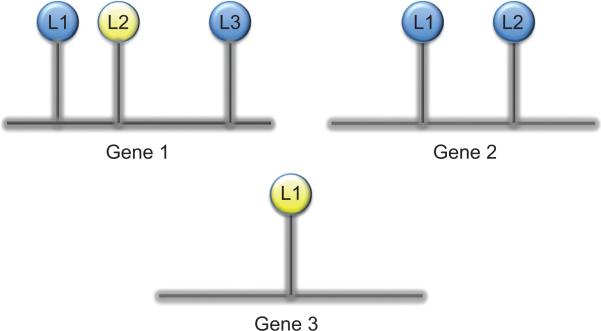
where g ≠ g′ and j ≠ j′ where |ρgjj′|≤1 represents the correlation between loci j and j′, both of which are associated with gene g. Figure 1 helps make these ideas more transparent. This figure depicts three genes, each of which has varying numbers of CpG sites for which methylation measurements are available. Our modeling strategy allows for correlation in the methylation among CpG sites associated with the same gene, while assuming the methylation of CpG sites associated with different genes to be independent. For instance, within Gene 1, we allow there to be correlation between the methylation levels at CpG sites L1, L2, and L3, but assume the methylation of these sites to be independent of CpG sites L1 and L2 among Gene 2.
Figure 1.
Illustration of the assumed covariance structure for DNA methylation data. The methylation status was assessed for three CpG loci associated with Gene 1 (L1, L2, and L3), two CpG sites associated with Gene 2 (L1 and L2), and a single CpG site associated with Gene 3 (L1). We allow correlation between the methylation of CpG sites associated with the same gene, however assume the methylation status of CpG sites associated with different genes to be independent. Blue represents a methylated CpG site and yellow represents an unmethylated CpG site.
Under the assumption that Ci=k with probability ηk and , and that the methylation status of unrelated probes is independent conditional on class membership, the likelihood contribution from subject i is given by
| (1) |
where ς=(η1,..., ηK–1, θ11,..., θ1G, θ21,..., θKG) is a vector of model parameters. With observed data , the conventional mixture model approach involves maximizing the full-data log-likelihood,
| (2) |
with respect to ς. This is easily achieved using an expectation-maximization (EM) algorithm (Dempster et al., 1977). Briefly, this involves initializing the procedure with an N×K matrix of weight W=(wik) whose rows sum to one. The rows reflect initial guesses at class membership probabilities across for each subject. Thus, for each k, we set and maximizing the quantity (2) with respect to ς. At each iteration we recompute weights wik and iterate until does not change. The final weight, wik represents the posterior probability that subject i belongs to class k.
Since the number of classes K is typically unknown, one might decide on the number of classes by fitting mixture models for a range of possible values of K, computing the resulting BIC statistics and selecting the value of K that corresponds to the minimum BIC. The entire operation has approximate complexity , where Kmax is the maximum number of classes attempted. Houseman et al. (2008) proposed a recursive alternative to conventional mixture model approach based on a weighted-likelihood version of (2) that typically has complexity no more than NJK log K. Briefly, RPMM recasts the conventional mixture model formulation into a hierarchical framework [a model-based version of the HOPACH algorithm van der Laan and Pollard (2003)], where the first step of RPMM, representing the top of the tree, involves fitting a 1-class model to the entire dataset. The BIC from the resulting model is then computed and compared to the BIC resulting from a 2-class mixture model fit to the entire data (first branch of the tree). If the BIC from the 2-class model is less than the BIC from the 1-class model, we continue recursion. Under the assumption that the resulting classes from the previous 2-class model can be further split, and that each subject belongs to the subsequent splits only with probability equal to the weight assigned from the previous split, the weighted-likelihood EM algorithm is applied recursively to obtain two new classes (next branch in the tree). As before we compare the BIC from the previous split to the new split and continue recursion if the BIC from the new split is less than the BIC from the previous split, suggesting a more parsimonious representation of the data. As previously described, recursion can be terminated early if the split leads to a less parsimonious representation of the data or if the classes under consideration comprise a small number of pseudo-subjects. The later is used as a safeguard, due to the fact that mixture models become unstable with small weights (representing a small number of pseudo-subjects).
The final clustering solution consists of K classes, with the final assembled from the individual vectors , as well as the posterior probabilities of class membership for each subject among each of the terminal classes (i.e., ).
Beta distributed RPMM with within-gene correlation
We consider an approach based on a generalized linear mixed effects model (GLMM) formulation for integrating within-gene correlation among CpG sites within a beta distributed RPMM. Using the notation introduced in Section 2.1, and assuming that subject i belongs to class k (i.e., Ci=k, k ∈ {1,2,...,K}), we have that Yij ~ β(αkj,βkj), where αkj>0 and βkj>0. The parameters αkj and βkj can be formulated in terms of the mean, μkj, and the dispersion parameter, ψkj>0 in the following way:
Therefore, a beta distribution can also be uniquely determined by its mean and dispersion. Considering a set of Jg CpG loci, all of which are associated with gene g, we propose the following random intercept model:
| (3) |
where h(.)is an appropriate link function (e.g., logit, probit, complementary log-log, etc.), , and . In model (3), μikgj represents the mean methylation for loci among subject i belonging to class k, vkgj corresponds to the overall mean methylation for locus j within class k, and aikg is a random effect term, representing the subject-specific deviation from the CpG-specific population mean (among CpG sites associated with gene g) for class k. In model (3), the methylation values for subject i are independent only conditional on the random effect aikg. Following the typical development of GLMM models (Breslow and Clayton, 1993), we integrate out the random effects and maximize the marginal log-likelihood:
such that,
| (4) |
where ς is a vector of model parameters, p(aigk) is the pdf of a normal distribution with mean 0 and variance and is the likelihood of yigj given aigk. The integral in equation (4) can be approximated by Gaussian-Hermite quadrature (Kennedy and Gentle, 1980). The marginal log-likelihood expressed above can then be incorporated into the RPMM framework described above and estimation proceeds as discussed in Section 2.1. As before, the final clustering solution provides an estimate K, , and the posterior probabilities of class membership for each subject among each of the terminal classes.
Implementation
Simulation studies
We considered a simulation study to assess the clustering performance and computational efficiency of the proposed within-gene correlated RPMM relative to the standard RPMM, where “standard RPMM” refers to the RPMM methodology that assumes class-conditional independence. Our simulation study used the mesothelioma cancer data set described in Christensen et al. (2009) to simulate realistic methylation data. Briefly, this dataset consisted of 158 subjects diagnosed with mesothlelioma. Each of the 158 subjects were profiled for the methylation status of approximately 1500 CpG loci associated with over 800 cancer related genes using the Illumina GoldenGate methylation array. We began by selecting two genes (ZMYND10 and ZP3), each of which contained DNA methylation measurements for two CpG loci (thus, a total of four CpG sites were selected). For the most part, the selection of these specific genes was arbitrary, however we sought to select genes with CpG sites whose methylation exhibited moderate to high correlation. For both the selected genes, the correlation in methylation among the CpG sites associated with that gene was 0.43. Note that the correlation of DNA methylation at individual CpGs between these two genes was low, consistent with other studies (Houshdaran et al., 2010). We next fit a beta-distributed RPMM to the mesothelioma cancer data subset consisting of the CpG loci from the two selected genes. This resulted in four classes, which were then trimmed to two classes by considering only the top-level split. We then estimated the within-class mean and dispersion parameters for each of the considered CpG loci as well as the variance of the random effect terms using the random intercept model described in Section 2.2. Based on the para meter estimates, we then simulated methylation data for two classes (50 subjects per class) and proceeded by fitting standard Gaussian and beta RPMMs (StanGaussian and StanBeta, respectively), Gaussian and beta RPMMs fit to the within-gene average methylation value across CpGs (AvgGaussian and AvgBeta, respectively), the within-gene correlated Gaussian and beta RPMMs (CorrGaussian and CorrBeta, respectively), and Ward's hierarchical clustering (Ward, 1963) to the simulated data. For Ward's hierarchical clustering we used the Euclidean distance metric and assigned class labels to samples based on pruning the resulting tree dendogram to two classes. For the the StanGaussian and CorrGaussian methods, the simulated methylation β values were transformed to an approximate Gaussian distribution using an arcsine square-root transformation (Rocke, 1993).
We considered a total of 100 simulations; for each simulation we assessed the clustering performance and computational efficiency of the seven considered methods. We used the adjusted Rand index to access the similarity between the true class membership and predicted class membership (Rand, 1971) and for each method (excluding Ward's hierarchical clustering), compared the predicted number of classes, K̂, to the true number of classes (i.e., K=2). The computational efficiency was determined by recording the amount of time (in seconds) it took to cluster a single simulated dataset. All analyses were performed in R version 2.11 using a computing cluster containing quad-core Nehalem processors and 24 GB of DDR-3 memory (1333 GHz).
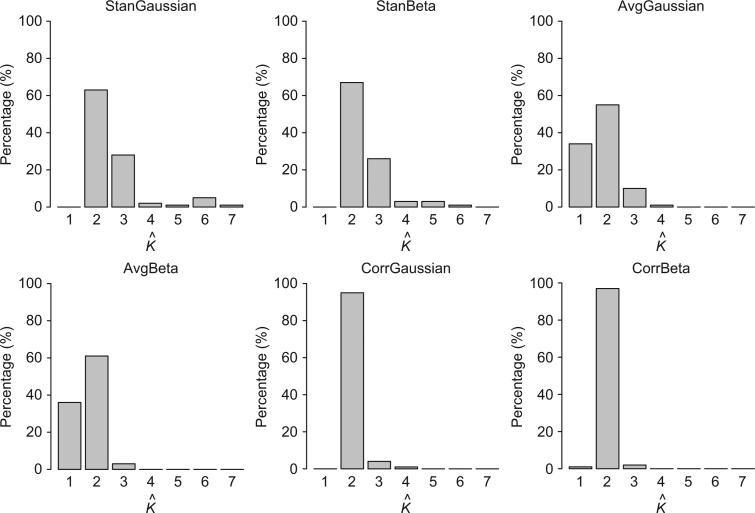
Table 1 contains the average adjusted Rand index and computational time for each of the seven methods considered. Most notably, we see that the StanGaussian, StanBeta, CorrGaussian, and CorrBeta have substantially improved adjusted Rand index compared to Ward's hierarchical clustering and the RPMM methods fit to the average methylation values for each gene (i.e., AvgGaussian and AvgBeta). The later result is not entirely surprising, as clustering subjects using the average within-gene methylation would be expected to result in loss of information, which is even more pronounced given the small number of genes used in our simulation. Also, although the data used in this simulation study were simulated from a beta random intercept model, consistent with the within-gene correlated beta RPMM described in Section 2.2, the CorrGaussian method demonstrated the best clustering performance across the four methods. In particular, the adjusted Rand index was significantly higher for the CorrGaussian method compared to all other methods (P<0.0001 for all) using a Wilcoxon signed-rank test. Although the adjusted Rand index was appreciably higher for the CorrBeta model than for the StanGaussian (P=0.0022) and StanBeta (P=0.0681), this was only at the expense of much greater computational cost, having a computational time that was 200–2000 times greater compared to the other RPMM methods. We further note that CorrGaussian and CorrBeta methods exhibited substantial improvement in estimating the true number of classes (>95% accuracy for both) (Figure 2). This contrasts with the StanGaussian and StanBeta methods which tended to overestimate the true number of classes and the AvgGaussian and AvgBeta methods which tended to underestimate the number of classes. The former is expected based on the results of Lindsay et al. (1991), which suggest overestimation of the true number of classes when the assumption of class-conditional independence is violated.
Table 1.
Average adjusted Rand index and computing time for seven considered methods.
| Method | Avg. adjusted Rand index (s.e.) | Avg. computation time (s) |
|---|---|---|
| StanGaussian | 0.71 (0.014) | 2 |
| StanBeta | 0.73 (0.013) | 8 |
| AvgGaussian | 0.13 (0.141) | 0.71 |
| AvgBeta | 0.11 (0.132) | 0.73 |
| CorrGaussian | 0.79 (0.010) | 22 |
| CorrBeta | 0.76 (0.010) | 4549 |
| Ward | 0.29 (0.017) | 0.004 |
Figure 2.
Distribution of the number of estimated classes (i.e., K̂) across 100 simulations for RPMM-based methods. The true number of classes is two (i.e., K=2).
This simulation study represented a relatively small scale analysis, using only a total four CpG loci for subsequent clustering. In general clustering of array-based methylation data would include many more CpG loci (Houseman et al., 2008; Christensen et al., 2009, 2011), rendering the CorrBeta method as a substantial computational burden given that the complexity of RPMM is on the order of J, where J represents the number of CpG loci used for clustering. For this reason and because the within-gene correlated Gaussian RPMM demonstrated greater promise with respect to predicting true class membership, we focus our attention on only the CorrGaussian method in our subsequent data application.
Data application
Description of data sets
We compared the clustering performance of the proposed within-gene correlated Gaussian RPMM (CorrGaussian) against the other RPMM-based methods using four array-based DNA methylation data sets. To gain an understanding of the robustness of the proposed methodology to array type and to provide insight regarding clustering performance and consistency across multiple methylation data sets, we considered array-based methylation data sets that were acquired using both the Illumina GoldenGate and the Infinium Human Methylation27 array technologies. As previously described, the GoldenGate methylation array simultaneously profiles the methylation status of over 1500 CpG loci, associated with approximately 800 cancer related genes, whereas the Infinium Human Methylation27 methylation array assesses the methylation status of over 27,000 CpG loci, associated with approximately 14,000 genes.
The first data set, which we refer to as the Glioma data set, is described in detail in Christensen et al. (2011). Briefly, the Glioma data set consisted of 131 subjects with glioma (glioblastomas, astrocytomas, oligodendrogliomas, oligoastrocytomas, ependymomas, and pilocytic astrocytomas). Each of the 131 samples in the Glioma data set were profiled for the methylation status within the tissue of origin using the Illumina GoldenGate methylation array. The second data set, which we refer to as the Mesothelioma data set, consisted of 158 mesothelioma tumor samples derived from two, independent series of mesothelioma cases (Christensen et al., 2009). Similar to the Glioma data set, methylation data on each of the 158 samples in the Mesothelioma data set were obtained using the Illumina GoldenGate methylation array. The third data set we considered, which we refer to as the Bladder data set, is described in Marsit et al. (2011) and consisted of 223 incident bladder cancer cases and 237 controls. For each of the subjects in the Bladder data set, DNA methylation was assessed in peripheral blood using the Illumina Infinium Human Methylation27 methylation array. The fourth data set was consisted of methylation data on 92 head and neck squamous cell carcinoma (HNSCC) cases and 92 controls (Langevin et al., 2012). We hereafter refer to this data set as the HNSCC data set. Similar to the Bladder data set, each subject was profiled for the status of DNA methylation in peripheral blood using the Illumina Human Methylation27 methylation array.
Mahalanobis distance was used to screen outliers and all CpG loci on X and Y chromosomes were excluded from the analysis, leaving a final 1413 autosomal CpG loci representing 773 unique genes and 26,486 autosomal CpG loci representing 13,890 unique genes, for the GoldenGate and Infinium Human Methylation27 arrays respectively (Houseman and Coull, 2004). As our analysis compares the proposed within-gene correlated Gaussian RPMM to the standard Gaussian RPMM, methylation β values were transformed using an arcsine square-root transformation to more closely approximate a Gaussian distribution (Rocke, 1993; Houseman et al., 2009). The untransformed methylation β values were used to fit the standard beta RPMM.
Description of data analysis
We investigated both the clustering performance and ability of the CorrGaussian method to detect biologically meaningful clusters. To evaluate the clustering performance, we computed and compared model goodness-of-fit and clustering consistency across the RPMM-based methods. Specifically, for each of the four methylation data sets considered (Glioma, Mesothelioma, Bladder, and HNSCC) we considered 100 separate analyses for a range of different numbers of randomly selected genes. More precisely, for each of the 100 separate analyses, we randomly selected M unique genes (M={10, 50, 100, 500}), from the total number of genes for that data set. Subsequent clustering analysis by CorrGaussian, StanGaussian, and StanBeta was then based on CpG loci associated with those genes. For the AvgGaussian and AvgBeta methods, Gaussian- and beta-distributed RPMMs were fit to the within-gene average methylation, computed as the mean methylation among the CpGs associated with a given gene. We then compared the model goodness-of-fit and clustering consistency for each of the four data sets across the 100 separate analyses for each selection of M.
Model goodness-of-fit statistics were using to provide insight toward model preference given the data. As RPMM-based methods are likelihood-based clustering algorithms, we compared the goodness-of-fit of the StanGaussian and CorrGaussian methods using the Bayesian information criterion [BIC, Schwartz (1978)], which has been widely used for model selection in mixture model problems (Dasgupta and Raftery, 1998; Fraley and Raftery, 2002). The StanBeta, AvgGaussian, and AvgBeta methods were not included in this comparison as the BICs obtained from these methods is not directly comparable to the BICs for the StanGaussian and CorrGaussian methods. It should be noted that for each of the considered methods, the BIC used to assess goodness-of-fit was computed based on the terminal nodes, or otherwise, the final clustering representation. In addition to comparing the model goodness-of-fit among CorrGaussian and StanGaussian methods, we also computed and compared the number of estimated methylation classes for these two methods as well as the other RPMM-based methods.
Though in general, the resulting clustering solutions will not be identical for a different group of randomly selected genes, there should be some degree of consistency between the clustering solutions (depending on the particular data used for clustering and the total number of features used to cluster), representing the true underlying structure of the data. Generally speaking, for a particular clustering algorithm, the higher the degree of clustering consistency across a different group of randomly selected genes, the greater the propensity of that algorithm for identifying and representing the true underlying structure of the data. As in our simulation study, we used the adjusted Rand index for assessing clustering consistency. In addition to comparing the clustering consistency between the various RPMM methods, we also compared the clustering consistency among several non-parametric alternatives. We included two versions of Ward's hierarchical clustering as well as the Hierarchical Ordered Partitioning and Collapsing Hybrid (HOPACH) clustering algorithm (Ward, 1963; van der Laan and Pollard, 2003). The two versions of Ward's hierarchical clustering that we considered, referred to as Ward1 and Ward2, were based on two different methods of pruning the resulting hierarchical clustering solution. For a particular analysis, the Ward1 and Ward2 methods consisted of assigning class labels to subjects by cutting the resulting tree dendogram at the appropriate height such that the number of resulting clusters were equal to the number of estimated clusters based on the StanGaussian and CorrGaussian methods, respectively. We also included HOPACH in this comparison, since it represents a non-parametric hierarchical clustering method that combines the strengths of both partitioning and agglomerative clustering methods. Similar to the methods based on RPMM, HOPACH automatically estimates the number of clusters K. For each of the non-parametric methods considered, Euclidean distance was used as the dissimilarity metric.
We assessed the clustering consistency across the various methods by taking the average of the pairwise adjusted Rand index across each of the 100 separate analyses [i.e., 100×(100–1)/2 pairwise comparisons] for each specification of the number of randomly selected genes for clustering, M. Significant differences with respect to the adjusted Rand index across the clustering methods was assessed using a bootstrap procedure (Arcones and Gine, 1992). Specifically, letting Q represent the total number of separate analyses (Q=100), we sampled Q times with replacement from the q={1,2,...,Q} separate analyses, computed the corresponding Q(Q–1)/2 pairwise adjusted Rand indices and from those estimates, calculated the resulting average adjusted Rand index for each bootstrap sample. This effectively resulted in a distribution of average adjusted Rand index for each of the considered clustering methods.
Using the two case/control cancer data sets (i.e., Bladder and HNSCC), we evaluated the ability of the RPMM-based methods for detecting biologically meaningful clusters. Specifically, within the Bladder and HNSCC data sets, CpG loci were rank-ordered based on variance and the genes associated with the top (M={10, 50, 100, 500}) most variable CpG loci were selected for subsequent clustering analysis. The resulting clusters tested for their association with cancer case/control status using a χ2 test. P-values were recorded for each selection of M and compared across the RPMM-based methods.
Data application results
Table 2 and Figure 3 contain the results for goodness-of-fit and clustering consistency, respectively, for our data application. As noted in Table 2, for each of the considered datasets and across varying numbers of randomly selected genes (i.e., M={10, 50, 100, 500}), the CorrGaussian method consistently outperformed the StanGaussian method in terms of goodness-of-fit. For a fixed data set, the differences in goodness-of-fit between these two methods appeared to increase a function of the number of randomly selected genes used for subsequent clustering analysis. This is likely due the potential of the CorrGaussian method to capture the features of DNA methylation data, which becomes more pronounced when applied to a larger pool of selected genes. To put the results on goodness-of-fit between the CorrGaussian and StanGaussian methods into a possibly more meaningful context, we note that across the various analyses considered in our data application, the CorrGaussian had a lower BIC compared to StanGaussian on average, 94% of the time. We further note the tendency of CorrGaussian method to result in a fewer number of estimated classes compared to the StanGaussian and StanBeta methods. This is to be expected as violations of the assumption of class-conditional independence result in overestimation of the true number of classes.
Table 2.
Results comparing the goodness of fit of the and number of classes estimated for the RPMM-based methods. Δ BIC represents the mean difference in BIC between the CorrGaussian and StanGaussian methods and K̂ represent the mean over 100 separate analyses.
| Dataset | M | Δ BIC | Avg. K̂ |
||||
|---|---|---|---|---|---|---|---|
| StanGaussian | StanBeta | AvgGaussian | AvgBeta | CorrGaussian | |||
| Glioma | 10 | –427 | 9.4 | 7.8 | 5.8 | 5.1 | 7.6 |
| Glioma | 50 | –17,741 | 14.1 | 11.4 | 9.8 | 8.3 | 12.1 |
| Glioma | 100 | –5825 | 15.7 | 11.6 | 11.8 | 9.6 | 12.2 |
| Glioma | 500 | –20,100 | 13.1 | 12.4 | 11.6 | 10.5 | 3.1 |
| Meso | 10 | –491 | 7.2 | 6.2 | 4.6 | 4.2 | 5.1 |
| Meso | 50 | –2287 | 8.4 | 6.8 | 6.1 | 5.5 | 6.5 |
| Meso | 100 | –6111 | 9.1 | 6.9 | 6.5 | 5.8 | 6.8 |
| Meso | 500 | –46,521 | 11.5 | 7.9 | 8.0 | 5.5 | 4.0 |
| Bladder | 10 | –899 | 13.7 | 13.1 | 10.9 | 10.7 | 11.4 |
| Bladder | 50 | –6138 | 18.5 | 17.2 | 19.1 | 17.6 | 13.2 |
| Bladder | 100 | –13,415 | 19.2 | 16.8 | 20.2 | 18.5 | 12.4 |
| Bladder | 500 | –45,565 | 19.2 | 17.6 | 18.2 | 17.6 | 6.6 |
| HNSCC | 10 | –308 | 6.1 | 6.0 | 5.8 | 5.8 | 5.2 |
| HNSCC | 50 | –819 | 6.1 | 6.0 | 7.1 | 6.8 | 4.9 |
| HNSCC | 100 | –1647 | 6.2 | 5.5 | 7.1 | 6.6 | 4.4 |
| HNSCC | 500 | –7327 | 5.7 | 5.8 | 7.9 | 6.0 | 4.0 |
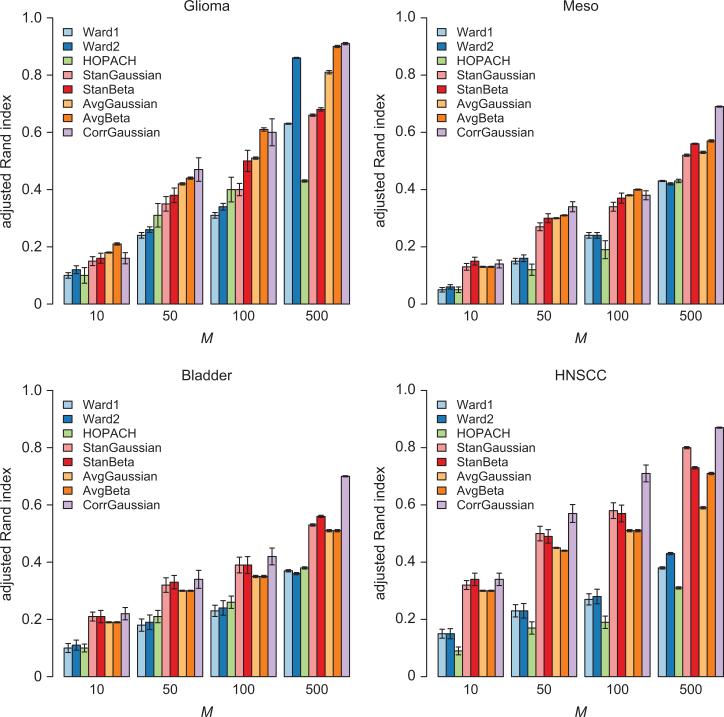
Figure 3.
Results comparing the clustering consistency of Ward1, Ward2, HOPACH, StanGaussian, StanBeta and CorrGaussian for M randomly selected genes. The adjusted rand-index represents the mean pairwise adjusted rand-index across 100 separate analyses. The 95% CI for the adjusted rand-index is displayed for each of the methods.
As illustrated in Figure 3, for each of the considered datasets and across varying numbers of randomly selected genes (i.e., M={10, 50, 100, 500}), the methods based on RPMM consistently resulted in more favorable clustering consistency compared to their non-parametric counterparts. Most notably though was the clustering consistency performance of the CorrGaussian method relative to the other methods. We note that the standard errors for the adjusted Rand index may be overly optimistic; as such, a bootstrap procedure was used to test for significant differences in adjusted Rand index between the various methods. Using the bootstrap method described above, considering 1000 bootstrap samples for each method, we tested the significance of the adjusted Rand index obtained from each of the methods. With some exceptions, the CorrGaussian method resulted in a significantly higher adjusted Rand index compared to the other methods across the various data sets and numbers of randomly selected genes M. For all data sets (M=10), there was no significant difference in the adjusted Rand index between the StanBeta, StanGaussian and CorrGaussian methods, however the AvgBeta method exhibited significantly higher clustering consistency in the Glioma data set. In addition, for the Mesothelioma and Baldder data sets (M=50, 100), there was no significant difference in the adjusted Rand index between the StanBeta, AvgGaussian, and CorrGaussian methods.
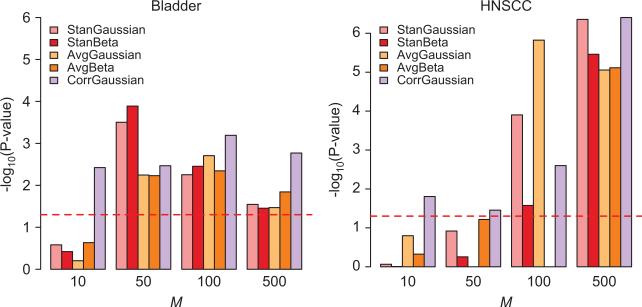
While our assessment of clustering performance demonstrated that CorrGaussian has improved goodness-of-fit over the StanGaussian, estimates on average, a fewer number of clusters compared to the methods that do not incorporate within-gene correlation, and in general, improved clustering consistency relative to competing approaches, the biological relevance of clusters identified by this method remains unclear. To examine this more thoroughly we focused our attention on the two cancer case/control data sets – Bladder and HNSCC – and investigated the extent to which the CorrGaussian method identifies clusters that are more strongly related to cancer case control status. Despite the selection of M and across both data sets, the CorrGaussian method tended to consistently identify clusters that were significantly associated cancer case/control status (100% with P<0.05) Figure 4. While the other RPMM-based identified clusters that were significantly associated with case/control status, this was typically only observed in instances where the number of genes used for clustering was relatively large (i.e., M={100, 500}) Figure 4. We further note that the clusters identified by CorrGaussian method had the smallest P-value 75% of the time, which suggests that incorporating within-gene correlation may lead to an improved ability to differentiate biologically meaningful clusters.
Figure 4.
Bar plots depicting the –log10(P-value) obtained from testing the association between the identified classes and cancer case/control status. Red-dotted line represents the –log10(0.05).
Discussion
DNA methylation is a widely studied epigenetic mechanism that has substantially contributed to our understanding of the role epigenetic alterations play in the development and progression of a vast array of different human diseases. In particular, profiles of DNA methylation have served to elucidate the mechanisms by which epigenetic changes might lead to aberrant gene expression patterns and disease (Laird, 2010). Recent advancements in high-throughput technology have enabled the simultaneous assessment of DNA methylation at single-site CpG resolution for thousands to hundreds of thousands of CpG loci. Such data is amenable to studying profiles of DNA methylation based on hundreds or thousands or more CpG loci. Compared to the identification of methylation makers at individual CpG dinucleotides, profiles of DNA methylation enable an understanding of the co-regulation of methylation across many CpG sites, often facilitating a more thorough understanding of the underlying biological processes and cellular pathways critical in states of human health and disease.
Unsupervised clustering of samples on the basis of their methylation information is a common approach for the identification of profiles of DNA methylation. Presently, there are a number of different methods for clustering high-dimensional array-based DNA methylation data, both non-parametric and parametric model-based methods; however, many of these existing methods do not incorporate known biological relationships of measured features and often rely on questionable assumptions regarding the biology of DNA methylation. We and others (Ehrich et al., 2008; Houshdaran et al., 2010; Nautiyal et al., 2010) have observed distinct patterns in the correlation of the methylation status between pairs of related probes (neighboring CpG sites) and pairs of unrelated probes (non-neighboring CpG sites). In light of these observations and since most array-based methylation platforms contain information regarding the genomic location of measured features, we proposed a modified version of RPMM that incorporates known biological features of DNA methylation data. As the existing RPMM framework assumes class-conditional independence of methylation for CpG sites, we proposed two modified versions of RPMM (a Beta and Gaussian version) which (1) allow correlation in methylation for neighboring CpG sites (i.e., CpG sites associated with the same gene), but (2) assume independence in methylation for non-neighboring CpG sites (i.e., CpG sites associated with different genes). While it would be ideal to assume a fully unstructured covariance structure, the nature of array-based DNA methylation prevents us from such ( setting).
We evaluated the clustering performance of our proposed modified within-gene correlated RPMMs relative to the standard RPMMs using both simulations and four array-based methylation data sets. In our comparisons, we also included a naive approach to handling the observed within-gene correlation between CpG sites, wherein a standard RPMM was fit to the within-gene average methylation, calculated as the mean methylation among the CpGs associated with that gene (AvgGaussian and AvgBeta methods). Briefly, our simulation study suggested improved clustering performance of the modified within-gene correlated RPMMs relative to the RPMMs that assume class-conditional independence of features. The within-gene correlated Gaussian RPMM had most favorable clustering performance among the considered clustering methods followed by the within-gene correlated Beta RPMM. However, the principal limitation of our proposed modified RPMMs was their computational efficiency relative to the standard RPMMs. This was quite pronounced for the within-gene correlated Beta RPMM, which had an average computational time of 200–2000 times that of the standard RPMMs. What was particularly concerning was the performance of the AvgGaussian and AvgBeta methods simulation, which exhibited the poorest performance in terms of correctly identifying true class membership.
Using four array-based methylation data sets, we assessed the clustering performance, via goodness-of-fit and clustering consistency, for the RPMM-based methods. The results indicated improved goodness-of-fit for CorrGaussian compared to StanGaussian across the different data sets and for different numbers of randomly selected genes for clustering. These findings are significant in that they demonstrate robustness in goodness-of-fit for of our proposed method across different data sets and across different platforms for array-based DNA methylation profiling (Illumina GoldenGate and Infinium Human Methylation27 arrays). We also compared the clustering consistency across the RPMM-based methods and included in this comparison: two methods based on Ward's hierarchical clustering, and HOPACH. With some exceptions, our results demonstrated significantly improved clustering consistency RPMM-based methods over their non-parametric counterparts, and above all, notable gains among CorrGaussian. While these results speak to the potential for improvements in model-fit and clustering consistency for the CorrGaussian method, what is most important to researchers is the extent to which incorporating within-gene correlation leads to an improved ability to detect biologically meaningful clusters. In both data sets considered, the CorrGaussian method consistently identified clusters that were significantly associated with cancer case/control status. Although the other RPMM-based methods did well in this respect, they tended to fall short when the number of genes used was low. A potential reason for this discrepancy is that differences in the correlation among CpGs that define biologically important subgroups would be missed by methods that do not incorporate correlation – a feature that the proposed CorrGaussian method is well suited to detect.
As previously described, the principal limitations of the within-gene correlated RPMMs is their computational efficiency relative to the standard RPMMs. While this was not a major issue with respect to the within-gene correlated Gaussian RPMM, it was a substantial limitation for the within-gene correlated Beta RPMM. The computational burden of the within-gene correlated Beta RPMM is largely due to fact that it relies upon numerical quadrature for evaluating the integral in equation (4) and is particularly prone to flat likelihoods arising from little information regarding the variance component, . Since each level of recursion within the RPMM framework may consist of numerous weighted EM iterations, thus multiple maximization of likelihoods involving quadrature, the computational efficiency of the methodology becomes severely compromised. While the methods we proposed allow for correlation in methylation between CpG sites associated with the same gene, the framework we have developed is also extensible for situations in which one seeks to model correlation in methylation of CpG sites as a function of their genomic distance. For instance, assuming CpG sites are ordered based on their genomic location, one could use Gaussian or exponential autoregressive covariance structure that captures desired relationships in methylation. Although this method was not used here, preliminary investigations demonstrate the feasibility of such methods. We also note that the methods we propose could be combined with a Semi-Supervised RPMM (Koestler et al., 2010) procedure for Illumina's most recent 450K Infinium Methylation BeadChip; particularly, to identify patterns of methylation among biologically important genomic regions (as opposed to individual CpGs). This represents a relevant application of the proposed modified RPMM, as the class-conditional independence assumption will obviously fail for denser arrays.
The modified versions of RPMM we proposed incorporate important biological relationships between profiled CpG sites, information which is often ignored by other methods. The proposed within-gene correlated Gaussian RPMM demonstrated appreciable gains in clustering performance, goodness-of-fit, and clustering consistency relative to the standard versions of RPMM and therefore represents an attractive method for clustering array-based DNA methylation data.
Appendix
The formula below makes explicit two facts about the Euclidean metric: (1) it remains unaffected by autocorrelated loci (since its expectation depends on the variance-covariance matrix only through the diagonal); and (2) it is influenced by all loci, including those that are non-informative and possibly noisy (with noisy loci contributing the most, even if they are not informative).
For independent random vectors Y1 and Y2,
With δj=1(θ1j=θ2j), the following equations make clear that in correctly-specified mixture models, non-informative loci have no influence on classification (via posterior class membership probability):
where . Consequently, terms that depend on factor out of the empirical Bayes formula for classification via posterior class membership probability:
Code for implementing the proposed methods was written in the R statistical language (http://cran.r-project.org/) and be found on the first author's website (http://bio-epi.hitchcock.org/faculty/koestler.html). Instructions for downloading and usage are provided there.
Contributor Information
Devin C. Koestler, Department of Community and Family Medicine, Geisel School of Medicine at Dartmouth, 1 Medical Center Dr., Lebanon, NH 03756, USA.
Brock C. Christensen, Department of Community and Family Medicine, Geisel School of Medicine at Dartmouth College, Lebanon, NH, USA; and Department of Pharmacology and Toxicology, Geisel School of Medicine at Dartmouth College, Hanover, NH, USA
Carmen J. Marsit, Department of Community and Family Medicine, Geisel School of Medicine at Dartmouth College, Lebanon, NH, USA; and Department of Pharmacology and Toxicology, Geisel School of Medicine at Dartmouth College, Hanover, NH, USA
Karl T. Kelsey, Department of Pathology and Laboratory Medicine, Brown University, Providence, RI, USA; and Department of Epidemiology, Brown University, Providence, RI, USA
E. Andres Houseman, Department of Public Health, Oregon State University, Corvallis, OR, USA.
References
- Arcones M, Gine E. “On the bootstrap of u and v statistics,”. Ann. Stat. 1992;20(2):655–674. [Google Scholar]
- Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA, Marsit CJ. “Infant growth restriction is associated with distinct patterns of dna methylation in human placentas,”. Epigenetics. 2011;6:920–927. doi: 10.4161/epi.6.7.16079. URL http://dx.doi.org/10.4161/epi.6.7.16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow NE, Clayton DG. “Approximate inference in generalized linear mixed models,”. J. Am. Stat. Assoc. 1993;88:9–25. [Google Scholar]
- Chen J. “Optimal rate of convergence for finite mixture models,”. Ann. Stat. 1995;23:221–233. [Google Scholar]
- Christensen BC, Houseman EA, Godleski JJ, Marsit CJ, Longacker JL, Roelofs CR, Karagas MR, Wrensch MR, Yeh R-F, Nelson HH, Wiemels JL, Zheng S, Wiencke JK, Bueno R, Sugarbaker DJ, Kelsey KT. “Epigenetic profiles distinguish pleural mesothelioma from normal pleura and predict lung asbestos burden and clinical outcome,”. Cancer Res. 2009;69:227–234. doi: 10.1158/0008-5472.CAN-08-2586. URL http://dx.doi.org/10.1158/0008-5472.CAN-08-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Smith AA, Zheng S, Koestler DC, Houseman EA, Marsit CJ, Wiemels JL, Nelson HH, Karagas MR, Wrensch MR, Kelsey KT, Wiencke JK. “Dna methylation, isocitrate dehydrogenase mutation, and survival in glioma,”. J. Natl. Cancer Inst. 2011;103:143–153. doi: 10.1093/jnci/djq497. URL http://dx.doi.org/10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A, Raftery A. “Detecting features in spatial point processes with clutter via model-based clustering,”. J. Am. Stat. Assoc. 1998;93:294–302. [Google Scholar]
- Dempster A, Laird N, Rubin D. “Maximum likelihhod from incomplete data via the em algorithm,”. J. R. Stat. Soc. B. 1977;39:1–38. [Google Scholar]
- Ehrich M, Turner J, Gibbs P, Lipton L, Giovanneti M, Cantor C, van den Boom D. “Cytosine methylation profiling of cancer cell lines,”. Proc. Natl. Acad. Sci. USA. 2008;105:4844–4849. doi: 10.1073/pnas.0712251105. URL http://dx.doi.org/10.1073/pnas.0712251105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley C, Raftery A. “Model-based clustering, discriminant analysis, and density estimation,”. J. Am. Stat. Assoc. 2002;97(458):611–631. [Google Scholar]
- Grigoriu A, Ferreira JC, Choufani S, Baczyk D, Kingdom J, Weksberg R. “Cell specific patterns of methylation in the human placenta,”. Epigenetics. 2011;6:368–379. doi: 10.4161/epi.6.3.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Lange CPE, Shen H, Byun H-M, Berg DVD, Malik S, Pan F, Noushmehr H, van Dijk CM, Tollenaar RAEM, Laird PW. “Genome-scale analysis of aberrant dna methylation in colorectal cancer,”. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. URL http://dx.doi.org/10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E, Coull B. “Cholesky residuals for assessing normal errors in a linear model with correlated outcomes,”. J. Am. Stat. Assoc. 2004;99(466):383–394. [Google Scholar]
- Houseman EA, Christensen BC, Yeh R-F, Marsit CJ, Karagas MR, Wrensch M, Nelson HH, Wiemels J, Zheng S, Wiencke JK, Kelsey KT. “Model-based clustering of dna methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions,”. BMC Bioinformatics. 2008;9:365. doi: 10.1186/1471-2105-9-365. URL http://dx.doi.org/10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Christensen BC, Karagas MR, Wrensch MR, Nelson HH, Wiemels JL, Zheng S, Wiencke JK, Kelsey KT, Marsit CJ. “Copy number variation has little impact on bead-arraybased measures of dna methylation,”. Bioinformatics. 2009;25:1999–2005. doi: 10.1093/bioinformatics/btp364. URL http://dx.doi.org/10.1093/bioinformatics/btp364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houshdaran S, Hawley S, Palmer C, Campan M, Olsen MN, Ventura AP, Knudsen BS, Drescher CW, Urban ND, Brown PO, Laird PW. “Dna methylation profiles of ovarian epithelial carcinoma tumors and cell lines,”. PLoS One. 2010;5:e9359. doi: 10.1371/journal.pone.0009359. URL http://dx.doi.org/10.1371/journal.pone.0009359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Wu C, Liu P, Wang J, Coombes KR. “Applications of beta-mixture models in bioinformatics,”. Bioinformatics. 2005;21:2118–2122. doi: 10.1093/bioinformatics/bti318. URL http://dx.doi.org/10.1093/bioinformatics/bti318. [DOI] [PubMed] [Google Scholar]
- Joubert BR, Hberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun O, Cupul-Uicab LA, Ueland PM, Wu MC, Nystad W, Bell DA, Peddada SD, London SJ. “450k epigenome-wide scan identifies differential dna methylation in newborns related to maternal smoking during pregnancy,”. Environ. Health Perspect. 2012;120(10):1425–1431. doi: 10.1289/ehp.1205412. URL http://dx.doi.org/10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy W, Gentle J. Statistical computing. Marcel Dekker; New York: 1980. [Google Scholar]
- Koestler DC, Marsit CJ, Christensen BC, Karagas MR, Bueno R, Sugarbaker DJ, Kelsey KT, Houseman EA. “Semi-supervised recursively partitioned mixture models for identifying cancer subtypes,”. Bioinformatics. 2010;26:2578–2585. doi: 10.1093/bioinformatics/btq470. URL http://dx.doi.org/10.1093/bioinformatics/btq470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler DC, Marsit CJ, Christensen BC, Accomando W, Langevin SM, Houseman EA, Nelson HH, Karagas MR, Wiencke JK, Kelsey KT. “Peripheral blood immune cell methylation profiles are associated with nonhematopoietic cancers,”. Cancer Epidemiol. Biomarkers Prev. 2012;21:1293–1302. doi: 10.1158/1055-9965.EPI-12-0361. URL http://dx.doi.org/10.1158/1055-9965.EPI-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan PF, Chiang DY. “Integrating prior knowledge in multiple testing under dependence with applications to detecting differential dna methylation,”. Biometrics. 2012;68(3):774–783. doi: 10.1111/j.1541-0420.2011.01730.x. URL http://dx.doi.org/10.1111/j.1541-0420.2011.01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan PF, Wang S, Zhou X, Chu H. “A statistical framework for illumina dna methylation arrays,”. Bioinformatics. 2010;26:2849–2855. doi: 10.1093/bioinformatics/btq553. URL http://dx.doi.org/10.1093/bioinformatics/btq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. “The power and the promise of dna methylation markers,”. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. URL http://dx.doi.org/10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Laird PW. “Principles and challenges of genomewide DNA methylation analysis,”. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. URL http://dx.doi.org/10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Langevin SM, Koestler DC, Christensen BC, Butler RA, Wiencke JK, Nelson HH, Houseman EA, Marsit CJ, Kelsey KT. “Peripheral blood dna methylation profiles are indicative of head and neck squamous cell carcinoma: an epigenome-wide association study,”. Epigenetics. 2012;7:291–299. doi: 10.4161/epi.7.3.19134. URL http://dx.doi.org/10.4161/epi.7.3.19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurila K, Oster B, Andersen CL, Lamy P, Orntoft T, Yli-Harja O, Wiuf C. “A beta-mixture model for dimensionality reduction, sample classification and analysis,”. BMC Bioinformatics. 2011;12:215. doi: 10.1186/1471-2105-12-215. URL http://dx.doi.org/10.1186/1471-2105-12-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay B, Clogg CC, Grego J. “Semiparametric estimation in the rasch model and related exponential response models, including a simple latent class model for item analysis,”. J. Am. Stat. Assoc. 1991;86:96–107. [Google Scholar]
- Marsit CJ, Koestler DC, Christensen BC, Karagas MR, Houseman EA, Kelsey KT. “Dna methylation array analysis identifies profiles of blood-derived dna methylation associated with bladder cancer,”. J. Clin. Oncol. 2011;29:1133–1139. doi: 10.1200/JCO.2010.31.3577. URL http://dx.doi.org/10.1200/JCO.2010.31.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousa AA, Archer KJ, Cappello R, Estrada-Gutierrez G, Isaacs CR, Strauss JF, Walsh SW. “Dna methylation is altered in maternal blood vessels of women with preeclampsia,”. Reprod. Sci. 2012;19(12):1332–1342. doi: 10.1177/1933719112450336. URL http://dx.doi.org/10.1177/1933719112450336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal S, Carlton VEH, Lu Y, Ireland JS, Flaucher D, Moorhead M, Gray JW, Spellman P, Mindrinos M, Berg P, Faham M. “High-throughput method for analyzing methylation of cpgs in targeted genomic regions,”. Proc. Natl. Acad. Sci. USA. 2010;107:12587–12592. doi: 10.1073/pnas.1005173107. URL http://dx.doi.org/10.1073/pnas.1005173107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand W. “Objective criteria for the evaluation of clustering methods,”. J. Am. Stat. Assoc. 1971;66(336):846–850. [Google Scholar]
- Rocke DM. “On the beta transformation family,”. Technometrics. 1993;35:72–81. [Google Scholar]
- Schwartz G. “Estimating the dimension of a model,”. Ann. Stat. 1978;6(2):461–464. [Google Scholar]
- Siegmund K, Laird P, Laird-Offringa IA. “A comparison of cluster analysis methods using dna methylation data,”. Bioinformatics. 2003;20:1896–1904. doi: 10.1093/bioinformatics/bth176. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Pollard K. “A new algorithm for hybrid heirarchical clustering with visualization and the bootstrap,”. J. Stat. Plan. Infer. 2003;117:275–303. [Google Scholar]
- Verkuilen J, Smithson M. “Mixed and mixture regression models for continuous bounded responses using the beta distribution,”. J. Educ. Behav. Stat. 2012;37(1):82–113. [Google Scholar]
- Ward J. “Hierarchical grouping to optimize an objective function,”. J. Am. Stat. Assoc. 1963;58(301):236–244. [Google Scholar]
- Zhai R, Zhao Y, Su L, Cassidy L, Liu G, Christiani DC. “Genomewide dna methylation profiling of cell-free serum dna in esophageal adenocarcinoma and barrett esophagus,”. Neoplasia. 2012;14:29–33. doi: 10.1593/neo.111626. [DOI] [PMC free article] [PubMed] [Google Scholar]