Abstract
Background:
Helicobacter pylori antimicrobial resistance is an important factor responsible for treatment failure. The purpose of this study was evaluating the prevalence of point mutations in clarithromycin-resistant clinical isolates of H. pylori in Isfahan city of Iran.
Materials and Methods:
Thirty isolates of H. pylori from 130 biopsy specimens were isolated by culture and confirmed by biochemical and PCR tests. The MIC of clarithromycin antibiotic for 30 clinical isolates of H. pylori was determined by E-test method. The point mutations in the 288 bp of 23S rRNA gene of H. pylori were investigated in four clarithromycin-resistant clinical isolates by PCR followed by sequencing.
Results:
Among 30 isolates of H. pylori, 4 cases were resistant to clarithromycin. One point mutation was found at position T2243C in the 23S rRNA gene in all resistance isolates.
Conclusions:
In our study, H. pylori resistance to clarithromycin associated with point mutation at position 2243 (T2243C).
Keywords: Clarithromycin resistance, Helicobacter pylori, point mutations
INTRODUCTION
Helicobacter pylori is a Gram-negative, spiral-shaped bacterium involved in gastric diseases such as gastritis, peptic ulcer, and two forms of stomach cancer, adenocarcinoma and MALT lymphoma.[1,2,3,4]
H. pylori colonizes the stomachs of more than 50% of the world's population. The prevalence of H. pylori infection varies among different countries from 25 to 50% in developed countries to more than 80% in the developing world.[5,6]
Eradication of H. pylori infection by a triple or quadruple therapy regimen is recommended but antibiotic resistance is an important problem for the treatment of diseases.[7,8] Clarithromycin is recognized as the main component of combination therapy. However, clarithromycin resistance in H. pylori is an important cause of the failure treatment.[9] Clarithromycin is a bacteriostatic antibiotic that complete reversibly attached to domain V of the 23S rRNA gene in the 50S ribosomal sub-unit of microorganisms and inhibit protein synthesis.[10] H. pylori is resistant to clarithromycin associated with a point mutation in domain V of the 23S rRNA gene. Mainly mutations occurred at positions 2142 or 2143 in domain V of the 23S rRNA gene.[11] However, other point mutations outer of domain V were reported with H. pylori resistant to clarithromycin, such as G2224A, C2245T, T2289C, and T2182C.[3] The size of 23S and 5S rRNA gene in gene bank accession number u27270 is 3857 bp that from position 372 to 3339 is related to 23S rRNA gene. At the present study, part of 23S rRNA gene (2365-2653), which is include domain V of this rRNA, were selected to evaluate the common important point mutations.
The present study aimed to evaluate the common important point mutations in 23S rRNA gene which could be associated with clarithromycin resistance isolates in patients with gastrointestinal disorders in Isfahan city — Iran.
MATERIALS AND METHODS
Bacteria and culture conditions
Thirty H. pylori isolates were obtained from 130 patients who referred to endoscopy section of the three centers in Isfahan Hospitals from March 2011 to July 2012. Biopsy samples were cultured on Brucella agar supplemented with 5% human blood, 7% fetal calf serum (FCS) (Bahar Afshan – Iran), vancomycin (2 mg/L) (Merck, Germany), polymyxin B (0.05 mg/L) (Merck, Germany), L-cysteine 2% (Merck, Germany), trimethoprim (1 mg/L) (Merck, Germany), and amphotericin B (5 mg/L). Then, the plates in the microaerophilic atmosphere (6% O2, 10% CO2, and 84% N2) and at 37°C were incubated for 3 to 5 days in MART system (ANOXOMAT, Lichtenvoorde, the Netherlands). Growth bacteria were identified as H. pylori based on colony morphology, gram staining, and positive biochemical reactions such as urease, catalase, and oxidase. Positive clinical isolates were stored at −80°C in Brucella broth supplemented with 20% glycerol, 7% FCS until susceptibility tests.
Antibiotic susceptibility tests
Minimum inhibitory concentrations (MICs) were determined by the E-test strips. For this purpose, suspensions of pure bacteria were prepared in sterile saline equivalent to standard three McFarland (9 × 108 CFU/mL). The suspension was spread on Brucella agar supplemented with 5% human blood and 7% fetal FCS. Clarithromycin strips were put on the plates and incubated in the micro-aerophilic atmosphere (6% O2, 10% CO2, and 84% N2) at 37°C for 3 to 5 days in MART system. Strains were considered resistant to clarithromycin if the MIC was ≥1 μg/mL (AB Biodisk, Solna, Sweden).[12]
PCR amplification and sequencing of 23S rRNA gene
DNA was extracted from four H. pylori isolates by using Qiagen DNA Mini kit for DNA extraction according to the manufacturer's instruction (QIA amp DNA mini kit, USA). Primers shown in Table 1 were used in this study.[13]
Table 1.
Primers used for polymerase chain reaction amplification

Polymerase chain reaction was performed for the amplification of the 288 bp fragment of 23S rRNA gene. PCR amplification reaction mixture (25 μL) contained 17.5 mL sterile deionized water, 1.25 μL MgCl2 (50 mM), 2.5 μL 10x PCR buffer, 0.5 μL dNTPs (10 mM), 0.25 μL Ex-Taq polymerase (5 u/μL), 1 μL of each oligonucleotide primer (2 μL totally), and 1 μL template DNA.
PCR conditions were as follows: Initial denaturation, 94°C for 5 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 1 min, and final extension step at 72°C for 10 min. The PCR products were observed on 1.5% agarose gels and then were sent for sequencing (Bioneer, Korea).
RESULTS
In this study, the MIC of four isolates resistant to clarithromycin were MIC = 1.5, 2, 2, and 4 μg/mL. Sequences related to 23S rRNA resistance gene (288 bp) against clarithromycin in mentioned isolates were amplified in our study [Figure 1]. After sequencing process, DNA sequence analysis was performed by MEGA 4 software. In all of four clarithromycin-resistant H. pylori isolates, point mutation was observed at position T2243C [Figure 2]. The updated numbering of point mutation in the H. pylori 23S rRNA gene sequence was designated as shown in Taylor et al.[14] in which position 373A in GenBank number U27270 was defined as the 5 end of the H. pylori 23S rRNA. The numbering of nucleotide position was calculated (2615 − 373 + 1 = 2243) based on Taylor et al. method.[14] The control positive clarithromycin-resistant H. pylori strain was S251 in NCBI data bank. Our registered gene accession no. in GenBank of NCBI is KC417289.
Figure 1.
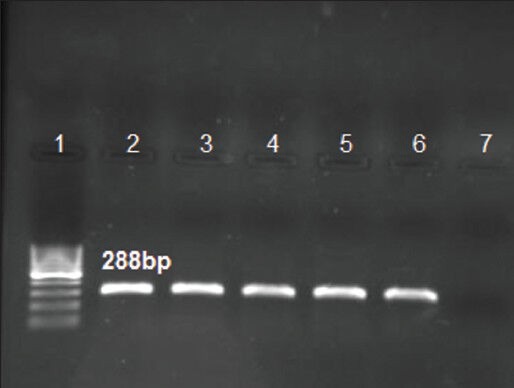
Line 7: Negative control; extracted DNA of P. aeruginosa ATCC27853, Line 1: Ladder100 bp DNA, Lines 2-5: 288 bp fragment, Line 6: Positive control; extracted DNA of H. pylori strain 26695
Figure 2.

The arrow shows the point mutation at position T2243C. The numbering of nucleotide position calculated (2615 – 373 + 1 = 2243) based on Taylor et al. method.[14] The control positive clarithromycin-resistant H. pylori strain was S251
DISCUSSION
H. pylori is a Gram-negative bacteria found in the stomach about half of the world's population and is associated with gastrointestinal diseases.[15,16] Various guidelines are recommended for eradication of H. pylori from the patients. However, in last years, the most recommended treatment is triple therapy regimen that contains a proton pump inhibitor (PPI) or ranitidine bismuth citrate combined with two antibiotics such as metronidazole and/or clarithromycin and/or amoxicillin. Several factors are involved in the treatment failure of H. pylori infection. One of the most factors are emergence of resistance to antibiotics by H. pylori.[9,17]
One of the important antimicrobial agents that are used in combination with other antibiotics in the eradication of H. pylori is clarithromycin. However, recent studies in different parts of the world showed that bacterial resistance to clarithromycin were increasing and thus are causing concern.[18,19,20]
Studies have shown clarithromycin resistance in H. pylori isolates to be associated with three common point mutations at positions A2142C, A2142G, and A2143G of the 23S rRNA gene.[7] In studies conducted in Italy in 2004, Chile in 2007, Italy in 2002, and China in 2004 in addition to point mutations above ones, they reported point mutations at positions 2144 (A→T), 1939 (G→A), 1942 (T→C), 2147 (C→G), 2717 (T→C), 2245 (C→T), 2224 (G→A), and 2289 (T→C), respectively.[1,10,11,21]
Studies have been conducted in Iran to identify the mechanism of H. pylori resistance to clarithromycin. In these studies, point mutations at positions 2142 or 2143 of the 23S rRNA gene have been identified.[22,23] Point mutations identified in our study were different with those studies in Iran. In comparison with the sequence of strain u27270 H. pylori, we found one new point mutation at position T2243C (GenBank KC417289). We did not find any point mutations at positions 2142 and 2143. It seems that the existence of geographical differences and number of patients in their studies were effective to determine point mutations. Similar results in agreement with our study have been shown in study conducted by Fontana et al. In Fontana et al.'s studies, point mutations at positions 2142 and 2143 were not found but reported T2717C point mutation. They divided clarithromycin-resistant strains into two groups showing different phenotypes and genotypes. Low-level resistant strains with MIC = 0.5-1 μg/mL was one of the groups that do not show point mutations at position 2142 and 2143 of the 23S rRNA gene. T2243C point mutation in our isolates with MIC = 1.5, 2, 2, and 4 μg/mL, more similar to group 1 (low-level resistant strains) in Fontana torrent category.[1] However, the point mutations listed by various studies were not the only causes of H. pylori resistance to clarithromycin, other point mutations, and other factors also contribute to this resistance. Hirata et al. have suggested efflux pump as another mechanism for H. pylori resistance to clarithromycin.[24]
The various studies demonstrated that the genetic characteristics of H. pylori in various geographical areas are different. So, more research is needed to indicate environmental differences and genetic characteristics of H. pylori isolates.
It is interesting that all clarithromycin-resistant isolates had T2243C mutation in our region. Based on this finding, it is possible that T2243C plays a major role of clarithromycin resistance but other mutations may affect. Although the role of three main point mutations was verified in the various studies, further study is needed to clarify the presence of a single mutation such as T2243C in clinical isolates.
In conclusion, our study showed the novel point mutation, T2243C in H. pylori 23S rRNA gene that contributed to clarithromycin resistance in our isolates that appears a regional differences pattern.
It is necessary to study the whole gene of 23S rRNA for finding the other point mutations in future.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Fontana C, Favaro M, Minelli S, Criscuolo AA, Pietroiusti A, Galante A, et al. New site of modification of 23S rRNA associated with clarithromycin resistance of Helicobacter pylori clinical isolates. Antimicrob Agents Chemother. 2002;46:3765–9. doi: 10.1128/AAC.46.12.3765-3769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG. Helicobacter pylori and antimicrobial resistance: Molecular mechanisms and clinical implications. Lancet Infect Dis. 2006;6:699–709. doi: 10.1016/S1473-3099(06)70627-2. [DOI] [PubMed] [Google Scholar]
- 3.Jones KR, Cha JH, Merrell DS. Who's winning the war? Molecular mechanisms of antibiotic resistance in Helicobacter pylori. Curr Drug Ther. 2008;3:190–203. doi: 10.2174/157488508785747899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rimbara E, Noguchi N, Kijima H, Yamaguchi T, Kawai T, Sasatsu M. Mutations in the 23S rRNA gene of clarithromycin-resistant Helicobacter pylori from Japan. Int J Antimicrob Agents. 2007;30:250–4. doi: 10.1016/j.ijantimicag.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Hosseini E, Poursina F, de Wiele TV, Safaei HG, Adibi P. Helicobacter pylori in Iran: A systematic review on the association of genotypes and gastroduodenal diseases. J Res Med Sci. 2012;17:280–92. [PMC free article] [PubMed] [Google Scholar]
- 6.Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441–52. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- 7.Baglan PH, Bozdayi G, Ozkan M, Ahmed K, Bozdayi AM, Ozden A. Clarithromycin resistance prevalence and Icea gene status in Helicobacter pylori clinical isolates in Turkish patients with duodenal ulcer and functional dyspepsia. J Microbiol. 2006;44:409–16. [PubMed] [Google Scholar]
- 8.Wang G, Wilson TJ, Jiang Q, Taylor DE. Spontaneous mutations that confer antibiotic resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2001;45:727–33. doi: 10.1128/AAC.45.3.727-733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agudo S, Pérez-Pérez G, Alarcón T, López-Brea M. High prevalence of clarithromycin-resistant Helicobacter pylori strains and risk factors associated with resistance in Madrid, Spain. J Clin Microbiol. 2010;48:3703–7. doi: 10.1128/JCM.00144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrido L, Toledo H. Novel genotypes in Helicobacter pylori involving domain V of the 23S rRNA gene. Helicobacter. 2007;12:505–9. doi: 10.1111/j.1523-5378.2007.00506.x. [DOI] [PubMed] [Google Scholar]
- 11.Toracchio S, Aceto GM, Mariani‐Costantini R, Battista P, Marzio L. Identification of a novel mutation affecting domain V of the 23S rRNA gene in Helicobacter pylori. Helicobacter. 2004;9:396–9. doi: 10.1111/j.1083-4389.2004.00267.x. [DOI] [PubMed] [Google Scholar]
- 12.Solna, Sweden: AB Biodisk; 2000. E-Test Technical Guide 8. Susceptibility testing of Helicobacter pylori. [Google Scholar]
- 13.Kim JM, Kim JS, Kim N, Kim YJ, Kim IY, Chee YJ, et al. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J Microbiol Biotechnol. 2008;18:1584–9. [PubMed] [Google Scholar]
- 14.Taylor DE, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–8. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay CY, Mitchell H, Dong Q, Goh KL, Dawes IW, Lan R. Population structure of Helicobacter pylori among ethnic groups in Malaysia: Recent acquisition of the bacterium by the malay population. BMC Microbiol. 2009;9:126. doi: 10.1186/1471-2180-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yula E, Nagiyev T, Kaya OA, Inci M, Celik MM, Köksal F. Detection of primary clarithromycin resistance of Helicobacter pylori and association between cagA (+) status and clinical outcome. Folia Microbiol (Praha) 2013;58:141–6. doi: 10.1007/s12223-012-0192-8. [DOI] [PubMed] [Google Scholar]
- 17.Kadayifci A, Uygun A, Kilciler G, Kantarcioglu M, Kara M, Ozcan A, et al. Low efficacy of clarithromycin including sequential regimens for Helicobacter pylori infection. Helicobacter. 2012;17:121–6. doi: 10.1111/j.1523-5378.2011.00924.x. [DOI] [PubMed] [Google Scholar]
- 18.Ho SL, Tan EL, Sam CK, Goh KL. Clarithromycin resistance and point mutations in the 23S rRNA gene in Helicobacter pylori isolates from Malaysia. J Dig Dis. 2010;11:101–5. doi: 10.1111/j.1751-2980.2010.00423.x. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Rimbara E, Thirumurthi S, Trespalacios A, Reddy R, Sabounchi S, et al. Detection of clarithromycin resistance in Helicobacter pylori following noncryogenic storage of rapid urease tests for 30 days. J Dig Dis. 2012;13:54–9. doi: 10.1111/j.1751-2980.2011.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirzaei N, Poursina F, Faghri J, Talebi M, Khataminezhad MR, Hasanzadeh A, et al. Prevalence of resistance to Helicobacter pylori strains to selected antibiotics in Isfahan, Iran. Jundishapur J Microbiol. 2013;6:e6342. [Google Scholar]
- 21.Hao Q, Li Y, Zhang ZJ, Liu Y, Gao H. New mutation points in 23S rRNA gene associated with Helicobacter pylori resistance to clarithromycin in northeast China. World J Gastroenterol. 2004;10:1075–7. doi: 10.3748/wjg.v10.i7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kargar M, Baghernejad M, Doosti A, Ghorbani-Dalini S. Clarithromycin resistance and 23S rRNA mutations in Helicobacter pylori isolates in Iran. African J Microbiol Res. 2011;5:853–6. [Google Scholar]
- 23.Savari M, Abdollahi H, Zahedi MJ, Darvish Moghadam S, Hayatbakhsh Abasi M. Detection of A2142C, A2142G, and A2143G mutations in 23s rRNA gene conferring resistance to clarithromycin among Helicobacter pylori isolates in Kerman, Iran. Iran J Med Sci. 2011;36:104–10. [PMC free article] [PubMed] [Google Scholar]
- 24.Hirata K, Suzuki H, Nishizawa T, Tsugawa H, Muraoka H, Saito Y, et al. Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J Gastroenterol Hepatol. 2010;25(Suppl 1):S75–9. doi: 10.1111/j.1440-1746.2009.06220.x. [DOI] [PubMed] [Google Scholar]


